Atomic Scale Structural Studies of Macromolecular Assemblies by Solid-state Nuclear Magnetic Resonance Spectroscopy
Summary
Structures of supramolecular protein assemblies at atomic resolution are of high relevance because of their crucial roles in a variety of biological phenomena. Herein, we present a protocol to perform high-resolution structural studies on insoluble and non-crystalline macromolecular protein assemblies by magic-angle spinning solid-state nuclear magnetic resonance spectroscopy (MAS SSNMR).
Abstract
Supramolecular protein assemblies play fundamental roles in biological processes ranging from host-pathogen interaction, viral infection to the propagation of neurodegenerative disorders. Such assemblies consist in multiple protein subunits organized in a non-covalent way to form large macromolecular objects that can execute a variety of cellular functions or cause detrimental consequences. Atomic insights into the assembly mechanisms and the functioning of those macromolecular assemblies remain often scarce since their inherent insolubility and non-crystallinity often drastically reduces the quality of the data obtained from most techniques used in structural biology, such as X-ray crystallography and solution Nuclear Magnetic Resonance (NMR). We here present magic-angle spinning solid-state NMR spectroscopy (SSNMR) as a powerful method to investigate structures of macromolecular assemblies at atomic resolution. SSNMR can reveal atomic details on the assembled complex without size and solubility limitations. The protocol presented here describes the essential steps from the production of 13C/15N isotope-labeled macromolecular protein assemblies to the acquisition of standard SSNMR spectra and their analysis and interpretation. As an example, we show the pipeline of a SSNMR structural analysis of a filamentous protein assembly.
Introduction
Advances in magic-angle spinning solid-state nuclear magnetic resonance spectroscopy (SSNMR) offer an efficient tool for the structural characterization of macromolecular protein assemblies at an atomic resolution. These protein assemblies are ubiquitous systems that play essential roles in many biological processes. Their molecular structures, interactions and dynamics are accessible by SSNMR studies, as has been shown for viral (capsids1) and bacterial infection mechanisms (secretion systems2,3, pili4), membrane protein complexes5,6,7,8 and functional amyloids 9,10,11. This type of molecular assembly can also provoke pathologies such as in neurodegenerative diseases where proteins assemble in misfolded, amyloid states and cause aberrant cell behavior or cell death 12,13. Protein assemblies are often built by the symmetric oligomerization of multiples copies of protein subunits into large supramolecular objects of various shapes including fibrils, filaments, pores, tubes, or nanoparticles. The quaternary architecture is defined by weak interactions between protein subunits to organize the spatial and temporal assembly and to allow for sophisticated biological functions. Structural investigations at an atomic scale on these assemblies are a challenge for high-resolution techniques since their intrinsic insolubility and very often their non-crystallinity restricts the use of conventional X-ray crystallography or solution NMR approaches. Magic-angle spinning (MAS) SSNMR is an emerging technique to obtain atomic resolution data on insoluble macromolecular assemblies and has proven its efficiency to resolve 3D atomic models for an increasing number of complex biomolecular systems including bacterial filaments, amyloid assemblies and viral particles 14,15,16,17,18,19,20,21,22. Technical advances on high magnetic fields, methodological developments and sample preparation has established MAS SSNMR into a robust method to investigate insoluble proteins in various environments, notably in their biologically-relevant macromolecular assembled state or in cellular membranes, making the technique highly complementary to cryo-electron microscopy. In many cases, a very high degree of symmetry characterizes such protein assemblies. MAS SSNMR exploits this feature, as all protein subunits in a homomolecular assembly would have the same local structure and therefore virtually the same SSNMR signature, drastically reducing the complexity of the analysis.
An efficient protocol for structural studies of macromolecular protein assemblies by moderate MAS (<25 kHz) SSNMR is presented in this video and can be subdivided into different steps (Figure 1). We will demonstrate the critical stages of the workflow of a SSNMR structural study exemplified on a filamentous protein assembly (see highlighted steps in Figure 1), with the exception of protein subunit purification, differing for each protein assembly but of critical importance for structural studies, and without going into the technical/methodological details of SSNMR spectroscopy and structure calculation for what specialized tutorials are available online. While the present protocol will primarily focus on solid-state NMR experiments performed under MAS conditions, the use of aligned biological environments 23,24,25,26,27, such as aligned bicelles, allow for the investigation of protein conformation and dynamic protein-protein interaction in membrane-like media without MAS technology. We will show the protein expression and assembly steps as well as the recording of the crucial SSNMR spectra and their analysis and interpretation. Our aim is to provide insights into the structural analysis pipeline enabling the reader to perform an atomic-resolution structural study of a macromolecular assembly by SSNMR techniques.
The protocol encompasses 3 sections:
1. Solid-state NMR sample production
As a prerequisite to a solid-state NMR analysis, the protein components of the macromolecular assembly need to be expressed, isotope-labeled, purified and assembled in vitro into the native-like complex state (for an example see Figure 2). To ensure high NMR sensitivity, isotope enrichment in 13C and 15N labeling is required through the use of minimal bacterial expression media supplemented with 13C and 15N sources, such as uniformly 13C-labeled glucose/glycerol and 15NH4Cl respectively. In the later stage of the protocol, selectively 13C-labeled samples produced with selectively 13C-labeled sources such as (1,3-13C)- and (2-13C)-glycerol (or (1-13C)- and (2-13C)-glucose) are used to facilitate the NMR analysis. Mixed labeled sample corresponding to an equimolar mixture of either 50% 15N- and 50% 13C-labeled or 50% (1,3-13C)- and 50% (2-13C)-glucose are introduced to describe the detection of intermolecular interactions. A high degree of protein purity as well as rigorous conditions during the assembly step are key factors to insure a homogeneous structural order of the final sample.
2. Preliminary structural characterization based on one-dimensional (1D) solid-state NMR
We present the essential experiments for a structural analysis by SSNMR. One-dimensional (1D) cross-polarization (CP) and INEPT / RINEPT28 experiments, detected on 13C nuclei are used to detect rigid and flexible protein segments in the assembly, respectively, and to estimate the degree of structural homogeneity and local polymorphism (for an example see Figure 3).
3. Conformational analysis and 3D structure determination
Subsections 1 and 2 concern the conformational analysis, which is based on the SSNMR resonance assignment of all rigid residues of the protein assembly, as the chemical shifts are very sensitive probes to the local environment and can be used to predict the phi/psi dihedral angles and thereby determine the secondary structure. Figure 4 illustrates an example of a sequential resonance assignment in the rigid core of a protein assembly. The 3D structure determination is based on the collection of structural data such as distance restraints encoding close proximities (<7 – 9 Å), containing both intra- and intermolecular information. Subsections 3 and 4 describe long-range distant restraint collection and interpretation. Long-range contacts are defined as intramolecular 13C-13C proximities arising from residue i to j, with |i-j| ≥4, defining thereby the tertiary protein fold of the monomeric subunit, or as intermolecular 13C-13C proximities, defining the intermolecular interfaces between protein subunits in the assembly. Intra- and intermolecular interfaces are illustrated in Figure 5. SSNMR restraints detected through 13C-13C and 15N-13C recoupling experiments usually encode for internuclear distances < 1 nm. Subsection 4 explains the detection of intermolecular distance restraints. In symmetrical protein assemblies, the use of homogeneously labeled samples (i.e. 100% uniformly or selectively labeled) for identifying intermolecular subunit-subunit interactions is limited, as both intra- and inter-molecular contacts lead to detectable signals. The unambiguous detection of intermolecular proximities is achieved by using mixed labeled samples, containing an equimolar mixture of two differently labeled samples, combined prior to aggregation. Subsection 5 briefly introduces structure modeling.
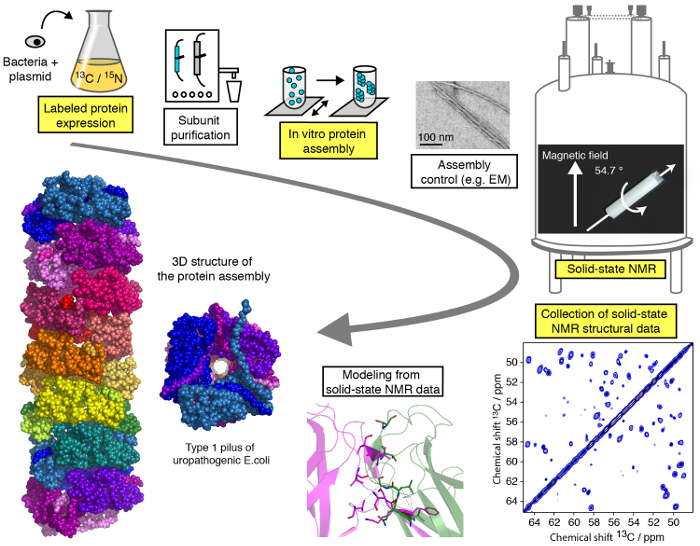
Figure 1: Workflow of an atomic-resolution structural study by solid-state NMR.13C, 15N isotope labeled protein production, subunit purification, subunit assembly, control of assembly formation, SSNMR experiments, SSNMR experiment analysis and extraction of distance restraints, and structure modeling are shown. Please click here to view a larger version of this figure.
Protocol
Representative Results
Discussion
Solid-state NMR (SSNMR) is a method of choice for characterizing macromolecular protein assemblies at an atomic level. One of the central issues in SSNMR-based structure determination is the spectral quality of the investigated system, that allows establishing 3D structural models of various precision, typically ranging from low-resolution models (containing the secondary structure elements and little 3D information) to pseudo-atomic 3D structures. The quantity and quality of structural information extracted from multi-dimensional SSNMR experiments is the key to compute a high-resolution NMR structure of the assembly.
The described protocol relies on the detection of 13C-13C and 15N-13C structural restraints requiring the recording of several 2D (and sometimes 3D) spectra with high signal-to-noise. At moderate MAS frequencies (<25 kHz), the sample is introduced into rotors with sizes of 3.2-4 mm diameter allowing for protein quantities of up to ~50 mg, dependent on the sample hydration. The amount of sample inside the rotor is directly proportional to the signal-to-noise ratio in SSNMR spectra, a decisive factor for the detection of long-range distance restraints and their unambiguous assignment.
The spectral resolution is a crucial parameter during the sequential resonance assignment and the restraints collection. To obtain optimal results, the sample preparation parameters need to be optimized, particularly in the purification of the subunit and the assembly conditions (pH, buffer, shaking, temperature, etc.). For sample optimization, it is recommended to prepare unlabeled samples for several distinct conditions for which assembly has been observed, and to record a 1D 1H-13C CP spectrum (described in step 2.1) on each prepared sample. The spectra serve to compare spectral resolution and dispersion between the different preparations, based upon which the optimal conditions can be determined.
The quality of the SSNMR data depends strongly on the choice of the NMR acquisition parameters, especially for the polarization transfer steps. The use of high magnetic field strengths (≥600 MHz 1H frequency) is essential for high sensitivity and spectral resolution, required when facing complex targets such as macromolecular protein assemblies.
A limiting factor in many cases is the spectrometer availability. Therefore, a judicious choice of the samples to be prepared should precede the spectrometer session. In any case, a uniformly 13C, 15N-labeled sample is a prerequisite to perform the sequential and intra-residual resonance assignment. For proteins assigned by solid-state NMR techniques see71. Structure determination of macromolecular assemblies at moderate MAS frequencies requires selectively 13C-labeled samples; for the detection of long-range 13C-13C and 13C-15N contacts samples based on 1,3-13C- and 2-13C-gylcerol and/or 1-13C – and 2-13C-glucose labeling are commonly used, as described above. The choice between the two labeling schemes is based on the spectral signal-to-noise ratio and resolution. To distinguish between intra- and intermolecular long-range contacts, mixed labeled and diluted samples have revealed efficient.
In short, the critical steps for an atomic SSNMR structural study are: (i) the preparation of the subunits and the assembly need to be optimized to obtain excellent sample quantity and quality, (ii) spectrometer field strength and acquisition parameters have to be chosen carefully; (iii) selective labeling strategies are required for a 3D structure determination and the amount of required data depends on data quality and the availability of complementary data.
Despite its applicability to a wide range of supramolecular systems ranging from membrane proteins to homomultimeric nano-objects, SSNMR is often limited by the need for mg-quantities of isotopically labeled material. The recent technological developments in ultra-fast MAS (≥100 kHz) SSNMR open up the avenue to 1H-detected NMR, and push the limit of minimal sample quantity to sub-mg 72,73,74. Nevertheless, for detailed structural studies 13C-labeled samples are indispensable, which limits the application of SSNMR to samples assembled in vitro or to systems expressed in organisms that survive on minimal medium where in-cell SSNMR is an emerging method (for reviews see 75,76,77,78).
An important factor in SSNMR application to obtain high-resolution 3D structures is the spectral resolution: intrinsic conformational heterogeneity in an assembly can limit spectral resolution and spectra analysis. Residue specific 13C labeling may in some cases provide an alternative to obtain specific distance information on strategic residues in order to obtain structural models (for a recent examples see 79,80).
SSNMR for 3D structure determination still requires the collection of several datasets with often long data collection times on sophisticated instruments, depending on the approach and the system several days to weeks on a 600-1000 MHz (1H frequency) spectrometer. Therefore, the access to spectrometer time can be a limiting factor in an in-depth SSNMR study.
In the case of homomultimeric protein assemblies, leading to SSNMR data of sufficient quality to identify a high number of structural restraints such as in 3,57,64,70, SSNMR still gives no access to the microscopic dimensions. Therefore, in a de novo SSNMR structure determination of a homomultimeric assembly, EM or mass-per-length (MPL) data ideally complement SSNMR data to derive the symmetry parameters. SSNMR data alone provide the atomic intra- and intermolecular interfaces
SSNMR is highly complementary with structural techniques such as EM or MPL measurements but the data can also perfectly be combined with atomic structures obtained by X-ray crystallography or solution NMR on mutated or truncated subunits. An increasing number of studies can be found in literature where the conjunction of different structural data has allowed for determining atomic 3D models of macromolecular assemblies (see Figure 6 for representative examples).
In the field of Structural Biology, SSNMR emerges as promising technique to study insoluble and non-crystalline assemblies at the atomic level, i.e. providing structural data at the atomic scale. In this respect, SSNMR is the pendant to solution NMR and X-ray crystallography for molecular assemblies, including membrane proteins in their native environment and protein assemblies such as viral envelopes, bacterial filaments or amyloids, as well as RNA and RNA-protein complexes (see for example81). Its highly versatile applications in vitro and in the cellular context, such as tracking secondary, tertiary and quaternary structural changes, identifying interaction surfaces with partner molecules on the atomic scale (for example 82) and mapping molecular dynamics in the context of assembled complexes, indicate the important potential of SSNMR in future structural studies on complex biomolecular assemblies.
| Component | M9 medium |
| NaCl | 0.5 g/L |
| KH2PO4 | 3 g/L |
| Na2HPO4 | 6.7 g/L |
| MgSO4 | 1 mM |
| ZnCl2 | 10 μM |
| FeCl3 | 1 μM |
| CaCl2 | 100 μM |
| MEM vitamin mix 100X | 10 mL/L |
| 13C-glucose | 2 g/L |
| 15NH4Cl | 1 g/L |
Table 1: Composition of minimal expression medium for recombinant protein production in E. coli BL21 cells.
Disclosures
The authors have nothing to disclose.
Acknowledgements
This work is funded by the ANR (13-PDOC-0017-01 to B.H. and ANR-14-CE09-0020-01 to A.L.), "Investments for the future" Programme IdEx Bordeaux/CNRS (PEPS 2016 to B.H.) reference ANR-10-IDEX-03-02 to B.H., the Fondation pour la Recherche Médicale (FRM-AJE20140630090 to A.L.), the FP7 program (FP7-PEOPLE-2013-CIG to A.L.) and the European Research Council (ERC) under the European Union's Horizon 2020 research and innovation program (ERC Starting Grant to A.L., agreement No 639020) and project "WEAKINTERACT."
Materials
| Instruments | |||
| NMR Spectrometer (> 11.7 Tesla) | Bruker | – | |
| triple resonance MAS SSNMR probehead | Bruker | – | |
| SSNMR rotors 4mm | Bruker | K1910 | |
| Centrifuge 5804 R | Eppendorf | 5805000629 | |
| GeneQuant 1300 spectrometer | Dutscher | 28-9182-13 | |
| IGS60 INCUBATEUR HERATHERM 75 L | Dutscher | 228001 | |
| MaxQ 4450 bench top orbital shaker | Dutscher | 78376 | |
| Tube Revolver Agitator | Dutscher | 79547 | |
| sonopuls HD 3100 | Bandelin | 3680 | |
| MicroPulser electroporator | Biorad | 165-2100 | |
| mini-PROTEAN tetra cell system | Biorad | 165-8000 | |
| AKTA pure system | GE Healthcare | 29-0182-24 | |
| capillary microman M25 pipet | Gilson | F148502 | |
| Name | Company | Catalog Number | Comments |
| Materials | |||
| amiconR ultra-15 | sigma | Z740199-8EA | |
| capillaries and pistons | Gilson | F148112 | |
| spatula | Fisher | 13263799 | |
| Name | Company | Catalog Number | Comments |
| Reagents | |||
| D-glucose 13C6 | Sigma | 389374 | |
| Ammonium-15N-chloride | Sigma | 299251 | |
| 1,3 13C2 glycerol | Sigma | 492639 | |
| 2 13C glycerol | Sigma | 489484 | |
| Kanamycin | Sigma | K1876 | |
| Carbenicillin | Sigma | C3416 | |
| Sodium phosphate dibasic | Sigma | S7907 | |
| Potassium phosphate monobasic | Sigma | P5655 | |
| Sodium chloride | Sigma | 71380 | |
| calcium chloride | Sigma | C1016 | |
| Magnesium sulfate | Sigma | 208094 | |
| Iron Chloride | Sigma | 157740 | |
| Zinc chloride | Sigma | 793523 | |
| MEM Vitamin Solution (100×) | Sigma | M68954 | |
| IPTG | Fisher | BP1755 | |
| Trizma base | Sigma | T1503 | |
| Tricine | Sigma | T0377 | |
| SDS | Sigma | 436143 | |
| sodium azide | sigma | 71289 | |
| 4,4-dimethyl-4-silapentane-1-sulfonic acid | Sigma | 178837 | |
| Name | Company | Catalog Number | Comments |
| Softwares | |||
| Unicorn 6.3 | GE Healthcare | Akta systems | |
| ccpNMR | CCPN | spectrometer systems |
References
- Morag, O., Sgourakis, N. G., Baker, D., Goldbourt, A. The NMR-Rosetta capsid model of M13 bacteriophage reveals a quadrupled hydrophobic packing epitope. Proc Natl Acad Sci U S A. 112 (4), 971-976 (2015).
- Loquet, A., et al. Atomic model of the type III secretion system needle. Nature. 486 (7402), 276-279 (2012).
- Demers, J. P., et al. High-resolution structure of the Shigella type-III secretion needle by solid-state NMR and cryo-electron microscopy. Nat Commun. 5 (4976), (2014).
- Habenstein, B., et al. Hybrid Structure of the Type 1 Pilus of Uropathogenic Escherichia coli. Angew Chem Int Ed Engl. 54 (40), 11691-11695 (2015).
- Cady, S. D., et al. Structure of the amantadine binding site of influenza M2 proton channels in lipid bilayers. Nature. 463 (7281), 689-692 (2010).
- Park, S. H., et al. Structure of the chemokine receptor CXCR1 in phospholipid bilayers. Nature. 491 (7426), 779-783 (2012).
- Kaplan, M., et al. Probing a cell-embedded megadalton protein complex by DNP-supported solid-state NMR. Nat Methods. 12 (7), 649-652 (2015).
- Wang, S., et al. Solid-state NMR spectroscopy structure determination of a lipid-embedded heptahelical membrane protein. Nat Methods. 10 (10), 1007-1012 (2013).
- Daskalov, A., et al. Signal transduction by a fungal NOD-like receptor based on propagation of a prion amyloid fold. PLoS Biol. 13 (2), e1002059 (2015).
- Daskalov, A., et al. Identification of a novel cell death-inducing domain reveals that fungal amyloid-controlled cell death is related to necroptosis. Proc Natl Acad Sci U S A. , (2016).
- Li, J., et al. The RIP1/RIP3 necrosome forms a functional amyloid signaling complex required for programmed necrosis. Cell. 150 (2), 339-350 (2012).
- Knowles, T. P., Vendruscolo, M., Dobson, C. M. The amyloid state and its association with protein misfolding diseases. Nat Rev Mol Cell Biol. 15 (6), 384-396 (2014).
- Aguzzi, A., Lakkaraju, A. K. Cell Biology of Prions and Prionoids: A Status Report. Trends Cell Biol. 26 (1), 40-51 (2016).
- Habenstein, B., Loquet, A. Solid-state NMR: An emerging technique in structural biology of self-assemblies. Biophys Chem. , (2015).
- Meier, B. H., Bockmann, A. The structure of fibrils from ‘misfolded’ proteins. Curr Opin Struct Biol. 30, 43-49 (2015).
- Miao, Y., Cross, T. A. Solid state NMR and protein-protein interactions in membranes. Curr Opin Struct Biol. 23 (6), 919-928 (2013).
- Tang, M., Comellas, G., Rienstra, C. M. Advanced solid-state NMR approaches for structure determination of membrane proteins and amyloid fibrils. Acc Chem Res. 46 (9), 2080-2088 (2013).
- Weingarth, M., Baldus, M. Solid-state NMR-based approaches for supramolecular structure elucidation. Acc Chem Res. 46 (9), 2037-2046 (2013).
- Loquet, A., Habenstein, B., Lange, A. Structural investigations of molecular machines by solid-state NMR. Acc Chem Res. 46 (9), 2070-2079 (2013).
- Yan, S., Suiter, C. L., Hou, G., Zhang, H., Polenova, T. Probing structure and dynamics of protein assemblies by magic angle spinning NMR spectroscopy. Acc Chem Res. 46 (9), 2047-2058 (2013).
- Tycko, R., Wickner, R. B. Molecular structures of amyloid and prion fibrils: consensus versus controversy. Acc Chem Res. 46 (7), 1487-1496 (2013).
- Hong, M., Zhang, Y., Hu, F. Membrane protein structure and dynamics from NMR spectroscopy. Annu Rev Phys Chem. 63, 1-24 (2012).
- Jelinek, R., Ramamoorthy, A., Opella, S. J. High-Resolution Three-Dimensional Solid-state NMR Spectroscopy of a Uniformly 15N-Labeled Protein. J Am Chem Soc. 117, 12348-12349 (1995).
- Xu, J., et al. Bicelle-enabled structural studies on a membrane-associated cytochrome B5 by solid-state MAS NMR spectroscopy. Angew Chem Int Ed Engl. 47 (41), 7864-7867 (2008).
- Durr, U. H., Gildenberg, M., Ramamoorthy, A. The magic of bicelles lights up membrane protein structure. Chem Rev. 112 (11), 6054-6074 (2012).
- Yamamoto, K., et al. Probing the transmembrane structure and topology of microsomal cytochrome-p450 by solid-state NMR on temperature-resistant bicelles. Sci Rep. 3, 2556 (2013).
- Huang, R., et al. Probing the transmembrane structure and dynamics of microsomal NADPH-cytochrome P450 oxidoreductase by solid-state NMR. Biophys J. 106 (10), 2126-2133 (2014).
- Durr, U. H., Yamamoto, K., Im, S. C., Waskell, L., Ramamoorthy, A. Solid-state NMR reveals structural and dynamical properties of a membrane-anchored electron-carrier protein, cytochrome b5. J Am Chem Soc. 129 (21), 6670-6671 (2007).
- Hong, M. Determination of multiple phi-torsion angles in proteins by selective and extensive (13)C labeling and two-dimensional solid-state NMR. J Magn Reson. 139 (2), 389-401 (1999).
- Lundstrom, P., et al. Fractional 13C enrichment of isolated carbons using [1-13C]- or [2- 13C]-glucose facilitates the accurate measurement of dynamics at backbone Calpha and side-chain methyl positions in proteins. J Biomol NMR. 38 (3), 199-212 (2007).
- Loquet, A., Lv, G., Giller, K., Becker, S., Lange, A. 13C spin dilution for simplified and complete solid-state NMR resonance assignment of insoluble biological assemblies. J Am Chem Soc. 133 (13), 4722-4725 (2011).
- Castellani, F., et al. Structure of a protein determined by solid-state magic-angle-spinning NMR spectroscopy. Nature. 420 (6911), 98-102 (2002).
- Higman, V. A., et al. Assigning large proteins in the solid state: a MAS NMR resonance assignment strategy using selectively and extensively 13C-labelled proteins. J Biomol NMR. 44 (4), 245-260 (2009).
- Bockmann, A., et al. Characterization of different water pools in solid-state NMR protein samples. J Biomol NMR. 45 (3), 319-327 (2009).
- Cavanagh, J., Fairbrother, W. J., Palmer, A. G., Skelton, N. J. . Protein NMR spectroscopy, principles and practice. , (1996).
- Hartman, S. R., Hahn, E. L. Nuclear Double Resonance in the Rotating Frame. Phys Rev. 128 (5), 2042-2053 (1962).
- Harris, R. K., et al. Further conventions for NMR shielding and chemical shifts IUPAC recommendations 2008. Solid State Nucl Magn Reson. 33 (3), 41-56 (2008).
- Wang, Y., Jardetzky, O. Probability-based protein secondary structure identification using combined NMR chemical-shift data. Protein Sci. 11 (4), 852-861 (2002).
- Shaka, A. J., Baker, P. B., Freeman, R. Computer-Optimized Scheme for Wideband Applications and Low-Level Operation. J Magn Reson. 64, 547-552 (1985).
- Szeverenyi, N. M., Sullivan, M. J., Maciel, G. E. Observation of Spin Exchange by Two-Dimensional Fourier-Transform C-13 Cross Polarization-Magic-Angle Spinning. J Magn Reson. 47, 462-475 (1982).
- Baldus, M., Petkova, A. T., Herzfeld, J., Griffin, R. G. Cross polarization in the tilted frame: assignment and spectral simplification in heteronuclear spin systems. Mol Phys. 95 (5), 1197-1207 (1998).
- Verel, R., Ernst, M., Meier, B. H. Adiabatic dipolar recoupling in solid-state NMR: the DREAM scheme. J Magn Reson. 150 (1), 81-99 (2001).
- . Sparky – NMR Assignment and Integration Software Available from: https://www.cgl.ucsf.edu/home/sparky/ (2017)
- Luca, S., et al. Secondary chemical shifts in immobilized peptides and proteins: a qualitative basis for structure refinement under magic angle spinning. J Biomol NMR. 20 (4), 325-331 (2001).
- Shen, Y., Bax, A. SPARTA+: a modest improvement in empirical NMR chemical shift prediction by means of an artificial neural network. J Biomol NMR. 48 (1), 13-22 (2010).
- Berjanskii, M. V., Neal, S., Wishart, D. S. PREDITOR: a web server for predicting protein torsion angle restraints. Nucleic Acids Res. 34 (Web Server issue), W63-W69 (2006).
- Bardiaux, B., Malliavin, T., Nilges, M. ARIA for solution and solid-state NMR. Methods Mol Biol. 831, 453-483 (2012).
- Guerry, P., Herrmann, T. Comprehensive automation for NMR structure determination of proteins. Methods Mol Biol. 831, 429-451 (2012).
- Vasa, S., et al. beta-Helical architecture of cytoskeletal bactofilin filaments revealed by solid-state NMR. Proc Natl Acad Sci U S A. 112 (2), E127-E136 (2015).
- He, L., et al. Structure determination of helical filaments by solid-state NMR spectroscopy. Proc Natl Acad Sci U S A. 113 (3), E272-E281 (2016).
- Tang, M., et al. High-resolution membrane protein structure by joint calculations with solid-state NMR and X-ray experimental data. J Biomol NMR. 51 (3), 227-233 (2011).
- Paravastu, A. K., Leapman, R. D., Yau, W. M., Tycko, R. Molecular structural basis for polymorphism in Alzheimer’s beta-amyloid fibrils. Proc Natl Acad Sci U S A. 105 (47), 18349-18354 (2008).
- Schutz, A. K., et al. Atomic-resolution three-dimensional structure of amyloid beta fibrils bearing the Osaka mutation. Angew Chem Int Ed Engl. 54 (1), 331-335 (2015).
- Sgourakis, N. G., Yau, W. M., Qiang, W. Modeling an in-register, parallel "iowa" abeta fibril structure using solid-state NMR data from labeled samples with rosetta. Structure. 23 (1), 216-227 (2015).
- Lewandowski, J. R., De Paepe, G., Griffin, R. G. Proton assisted insensitive nuclei cross polarization. J Am Chem Soc. 129 (4), 728-729 (2007).
- Carlon, A., et al. How to tackle protein structural data from solution and solid state: An integrated approach. Prog Nucl Magn Reson Spectrosc. 92-93, 54-70 (2016).
- Judge, P. J., Taylor, G. F., Dannatt, H. R., Watts, A. Solid-state nuclear magnetic resonance spectroscopy for membrane protein structure determination. Methods Mol Biol. 1261, 331-347 (2015).
- Wang, S., Ladizhansky, V. Recent advances in magic angle spinning solid state NMR of membrane proteins. Prog Nucl Magn Reson Spectrosc. 82, 1-26 (2014).
- Sborgi, L., et al. Structure and assembly of the mouse ASC inflammasome by combined NMR spectroscopy and cryo-electron microscopy. Proc Natl Acad Sci U S A. 112 (43), 13237-13242 (2015).
- Loquet, A., et al. Atomic structure and handedness of the building block of a biological assembly. J Am Chem Soc. 135 (51), 19135-19138 (2013).
- Walti, M. A., et al. Atomic-resolution structure of a disease-relevant Abeta(1-42) amyloid fibril. Proc Natl Acad Sci U S A. 113 (34), E4976-E4984 (2016).
- Colvin, M. T., et al. Atomic Resolution Structure of Monomorphic Abeta42 Amyloid Fibrils. J Am Chem Soc. 138 (30), 9663-9674 (2016).
- Xiao, Y., et al. Abeta(1-42) fibril structure illuminates self-recognition and replication of amyloid in Alzheimer’s disease. Nat Struct Mol Biol. 22 (6), 499-505 (2015).
- Tuttle, M. D., et al. Solid-state NMR structure of a pathogenic fibril of full-length human alpha-synuclein. Nat Struct Mol Biol. 23 (5), 409-415 (2016).
- Wasmer, C., et al. Amyloid fibrils of the HET-s(218-289) prion form a beta solenoid with a triangular hydrophobic core. Science. 319 (5869), 1523-1526 (2008).
- Van Melckebeke, H., et al. Atomic-resolution three-dimensional structure of HET-s(218-289) amyloid fibrils by solid-state NMR spectroscopy. J Am Chem Soc. 132 (39), 13765-13775 (2010).
- Lamley, J. M., et al. Solid-state NMR of a protein in a precipitated complex with a full-length antibody. J Am Chem Soc. 136 (48), 16800-16806 (2014).
- Agarwal, V., et al. De novo 3D structure determination from sub-milligram protein samples by solid-state 100 kHz MAS NMR spectroscopy. Angew Chem Int Ed Engl. 53 (45), 12253-12256 (2014).
- Stanek, J., et al. NMR Spectroscopic Assignment of Backbone and Side-Chain Protons in Fully Protonated Proteins: Microcrystals, Sedimented Assemblies, and Amyloid Fibrils. Angew Chem Int Ed Engl. 55 (50), 15504-15509 (2016).
- Baker, L. A., Baldus, M. Characterization of membrane protein function by solid-state NMR spectroscopy. Curr Opin Struct Biol. 27, 48-55 (2014).
- Luchinat, E., Banci, L. In-cell NMR: a topical review. IUCrJ. 4 (Pt 2), 108-118 (2017).
- Freedberg, D. I., Selenko, P. Live cell NMR. Annu Rev Biophys. 43, 171-192 (2014).
- Selenko, P., Wagner, G. Looking into live cells with in-cell NMR spectroscopy. J Struct Biol. 158 (2), 244-253 (2007).
- Qiang, W., Yau, W. M., Luo, Y., Mattson, M. P., Tycko, R. Antiparallel beta-sheet architecture in Iowa-mutant beta-amyloid fibrils. Proc Natl Acad Sci U S A. 109 (12), 4443-4448 (2012).
- Bateman, D. A., Tycko, R., Wickner, R. B. Experimentally derived structural constraints for amyloid fibrils of wild-type transthyretin. Biophys J. 101 (10), 2485-2492 (2011).
- Marchanka, A., Simon, B., Althoff-Ospelt, G., Carlomagno, T. RNA structure determination by solid-state NMR spectroscopy. Nat Commun. 6, 7024 (2015).
- Schutz, A. K., et al. The amyloid-Congo red interface at atomic resolution. Angew Chem Int Ed Engl. 50 (26), 5956-5960 (2011).

