Traction Force Microscopy to Study B Lymphocyte Activation
Summary
Here we present a protocol used to perform traction force microscopy experiments on B cells. We describe the preparation of soft polyacrylamide gels and their functionalization, as well as data acquisition at the microscope and a summary of data analysis.
Abstract
Traction force microscopy (TFM) enables the measurement of forces produced by a cell on a substrate. This technique infers traction force measurements from an experimentally observed displacement field produced by a cell pulling on an elastic substrate. Here, we adapted TFM to investigate the spatial and temporal structure of the force field exerted by B cells when activated by antigen engagement of the B cell receptor. Gel rigidity, bead density, and protein functionalization must be optimized for the study of relatively small cells (~ 6 µm) that interact with, and respond specifically to ligands for cell surface receptors.
Introduction
B cells are the antibody-producing cells of the immune system. To activate the adaptive immune response, they first acquire the antigen in a native form (i.e., non-processed) through a specific receptor called B cell receptor (BCR)1. This process occurs in the lymph node B cell zone. Even if some antigens can reach the B cell through lymphatic fluids, most antigens, especially with high molecular weight (>70 kDa, which is the limit size for lymphatic conduits) are indeed presented in their native form on the surface of an antigen presenting cell (APC), typically a subcapsular sinus macrophage or follicular dendritic cell, through lectin or Fc receptors (non-specific). The contact with this cell leads to the formation of an immune synapse where the BCR exerts force on the APC-associated antigens. The binding of an antigen to the BCR initiates BCR signaling, which may activate force-generating mechanisms. These forces could be important for amplifying BCR signaling, but are also essential for B cells to extract and then internalize the antigen.
Recent studies have shown that the BCR is indeed mechanosensitive2. For example, stiffer substrates elicit enhanced BCR signaling3. Moreover, force generated at the immune synapse pulls on single BCRs to probe its affinity to antigen and thereby ensure affinity discrimination4. It is therefore interesting to investigate the mechanical response of B cells to antigen presentation and to dissect this response in terms of type of receptors implicated (IgG/IgM)5, adhesion molecules (integrin ligands) or in pharmacologically and genetically modified cells (i.e., silencing of a protein downstream of BCR signaling or cytoskeleton dynamics)6.
A simple method to observe the response of a cell to a substrate of physiological rigidity and, at the same time, study forces exerted on the substrate is Traction Force Microscopy (TFM). TFM consists of observing the displacement field produced by the cell pulling on an elastic substrate. Originally the deformation of the gel was observed through wrinkles of the elastomer itself by phase-contrast microscopy7, but the insertion of fluorescence microbeads as fiducial markers allowed for better resolution and has since become the standard8. This method has been used to investigate the traction force exerted by adherent cells, tissues, and even organoids embedded in gels. Several variations of TFM have been developed9 including, combination with superresolution microscopy (i.e., STED10 or SRRF11), modification of the refractive index of the gel to allow for TIRF microscopy12, replacing beads by nano-printed patterns13, and using nanopillars instead of flat surface14. For a complete review of these variations, see Colin-York et al.15.
The protocol presented here describes a procedure to measure forces exerted by B cells on an antigen-coated substrate. These forces are applied on the ligands (antigen) in order to cluster them and subsequently extract them from the antigen-presenting substrate. We have adapted the standard TFM protocol to mimic the rigidity of physiological antigen-presenting substrates, the size and the relevant coating for the B cells. This protocol allows for the study of several cells simultaneously and can be used in conjunction with fluorescence microscopy techniques and chemical treatments. However, it does not aim to probe single molecule force measurements, for which optical tweezers16, molecular tension probes17,18, biomembrane force probes19, and atomic force microscopy20 are more suitable techniques. Compared to other single cell force measurement methods (e.g., micropipettes21 or microplates22) TFM allows for the reconstruction of a complete map of the forces exerted at the synapse with a resolution of ~300 nm. This is useful to identify spatio-temporal patterns in the forces exerted on the surface and, as the gel is compatible with confocal imaging, to correlate them with the recruitment of specific proteins (for example, cytoskeleton and signaling proteins).
Although 3D TFM is possible, it is not compatible with the rigidity and the setup we used. Deformations in 3D are achievable by other more complex setups such as protrusion force microscopy (AFM scanning a deformable membrane where the cells are plated)23,24 and elastic resonator interference stress microscopy (ERISM, a gel acting as resonating cavity for light and highlighting deformations of the substrate with accuracy of a few nanometers)25. Although these techniques are very promising, they have not yet been employed in B cells. Other types of TFM, such as on nanopillars14, could be used to have more reproducible substrates. However, this geometry is not adapted to soft cells as the cell interpenetrates the pillars, which complicates the analysis. This approach has indeed been used in T cells to observe the capability of the cell to build structures around the pillars26.
Despite its simplicity, TFM using polyacrylamide gels allows for the simultaneous observation of many cells and can be easily and inexpensively implemented in any lab equipped with a bench and an epifluorescence microscope (although we recommend confocal/spinning disk).
To mimic the physiological rigidity of an APC, we used polyacrylamide gels with a rigidity of ~500 Pa27 and functionalized the gel with activating antigens. In this protocol, we functionalized the surface of the polyacrylamide gel with hen egg lysozyme (HEL). This allows for the measurement of forces generated by stimulation of the BCR through engagement of the antigen binding site. The use of this antigen and the HEL-specific B cells from MD4 mice ensures relatively uniform force generation in response to antigen ligation28. However, other molecules (such as anti-IgM for B6 mice) can be grafted onto the gel, but the forces generated in these cases could be more heterogeneous and less intense. Because B cells are small cells (diameter ~6 µm), the number of beads has been optimized to be maximal but still trackable. For large cells that exert ~kPa forces on their substrates, one can achieve satisfactory results using relatively sparse beads or performing simple particle image velocimetry (PIV) to reconstruct the deformation field. However, for small cells such as B lymphocytes that exert stress as small as ~50 Pa, the use of single particle tracking is required (particle tracking velocimetry, PTV) to achieve the desired accuracy when reconstructing the deformation field. In order to reliably track beads individually, the magnification of the objective lens needs to be at least 60x and its numerical aperture around 1.3. Thus, the gels must be relatively thin (<50 µm), otherwise the beads are not visible as they are above the working distance of the objective.
The main protocol consists of three sections: gel preparation, gel functionalization and imaging; two more sections are optional and are dedicated to the antigen extraction quantification and imaging of fluorescent cells.
Protocol
1. Gel preparation
- Silanization of the gel support
- Activate the coverslip or glass-bottom Petri dish (which will be used as gel support) with a UV lamp for 2 min (wait 30 s before exposure to the UV lamp to avoid exposure to residual ozone).
- Silanize the coverslip/glass-bottom dish using 200 µL aminopropyltrimethoxysilane (APTMS) for 5 min. This will prepare the support for the covalent binding of the gel.
- Thoroughly wash the coverslip/glass-bottom dish with ultra-pure water.
- Dry the coverslip/glass-bottom dish using vacuum aspiration.
- Preparation of the 18mm coverslip used to flatten the gel
- To prepare the coverslips, first put them into a ceramic coverslip holder. Then put the coverslip holder into a small beaker (50 mL) and pour siliconizing reagent (stored at 4 °C, reusable) over the coverslips, being sure to completely cover them.
- Cover the beaker with aluminum foil and incubate for 3 min at room temperature. While waiting, fill a large beaker (500 mL) with ultra-pure water. After 3 min of incubation in siliconizing reagent, transfer the coverslip holder with coverslips to the beaker of water.
- Thoroughly rinse the coverslips with ultra-pure water, dry them well and keep on paper wipes. For best results, proceed immediately to the next section.
- Gel polymerization
- For gels of 0.5 kPa, mix 75 µL of 40% acrylamide with 30 µL of 2% bisacrylamide (crosslinker) and 895 µL of phosphate-buffered saline (PBS). This premix can be stored for up to one month at 4 °C.
- To 167 µL of 0.5 kPa gel premix, add 1% (1.67 µL) of beads, vortex and sonicate for 5 min in a bath sonicator (standard bench ultrasonic cleaner with power of 50–100 W and frequency 40 kHz). Keep the mix protected from light using aluminum foil.
NOTE: The premix does not polymerize until the initiator (TEMED) is added. - To catalyze polymerization, add 1% (1.67 µL) of 10% w/v ammonium persulfate (APS).
- To initiate polymerization, add 0.1% (0.2 µL) N,N,N′,N′-Tetramethylethylenediamine (TEMED). Mix with a pipette. Once APS and TEMED have been added, the gel rapidly polymerizes so proceed quickly to gel casting.
- Gel casting
- Pipet 9 µL of gel mix onto each coverslip/glass-bottom dish (drop in the center, Figure 1A)
- Place the silanized/hydrophobic coverslip and flatten the gel (Figure 1B). Using forceps, press the coverslip to ensure the gel spreads across the entire area of the coverslip (Figure 1C) until it starts leaking out.
- Invert the coverslip/glass-bottom dish into a large Petri dish and tap it on the bench to force beads going towards the gel surface (Figure 1D).
- Cover with aluminum foil and leave for 1 h to polymerize at room temperature in a humid chamber (i.e., put a wet tissue above the dish to prevent evaporation).
- After 1 h, add PBS to the sample to facilitate coverslip release. Carefully, remove the coverslip using a needle (the coating with different silanes should allow easy peeling off of the coverslip from the gel, Figure 1E).
- Leave the gel in PBS.
NOTE: Gels can now be stored in PBS at 4 °C for 5–7 days, but it is recommended to use them within 48 h.
2. Gel functionalization
- Prepare sulfosuccinimidyl 6-(4'-azido-2'-nitrophenylamino)hexanoate (Sulfo SANPAH) solution at 0.5 mg/mL in 10 mM HEPES buffer. This can be stored at 4 °C covered with aluminum foil for up to one week.
- Aspirate the PBS from gels.
- Add 150 µL of Sulfo SANPAH to the gel at room temperature (Figure 1F).
- Expose the gel to UV treatment for 2 min to photoactivate the sites of Sulfo SANPAH and make it stick to the gel surface.
- Wash with PBS three times (Figure 1G).
- Repeat steps 2.2–2.5.
- Add 250 µL of HEL (100 µg/mL) to each gel and incubate overnight in a humid chamber at 4 °C overnight while keeping covered with aluminum foil (Figure 1H).
- Remove HEL antigen and wash with PBS three times.
NOTE: HEL acts both as an antigen and as an adhesion molecule. It can be replaced by other molecules that bind to the receptor (e.g., an anti-mouse IgM, Bovine Serum Albumin, Ovalbumin) or mixed with integrin ligands (e.g., ICAM1 binding to LFA1). If needed, antigen extraction can be observed with a fluorescent version of the HEL (obtained by staining the molecule with a protein labeling kit, see step 4). Note that a given concentration in bulk might not yield the same surface concentration on the gel as on the glass: this needs to be quantified with secondary staining if direct comparison is required.
3. Cell loading and imaging
- Before imaging, remove PBS from the gels and add 500 µL of B cell media (RPMI 1640, 10% decomplemented fetal calf serum, 1% penicillin-streptomycin, 2% Sodium Pyruvate, 50uM Mercaptoethanol and 1X Non Essential Amino Acids) and let them to equilibrate to RT.
- Cell preparation
- Purify primary B cells from spleen according to a negative selection protocol (see Table of Materials). Typical final B cell yield is around 1 x 107 cells. Concentrate this to 3 x 106 cells/mL in B cell medium (RPMI-1640 supplemented with 10% fetal calf serum, 1% penicillin–streptomycin, 0.1% mercaptoethanol and 2% sodium pyruvate).
- Store cells as needed for up to 6 h at 4 °C.
- Keep the cells at 37 °C for 30 min before image acquisition.
- Imaging
- Use a confocal microscope with thermal and (possibly) CO2 control.
NOTE: Regardless of whether a confocal or spinning-disk microscope is used, it is important to use an objective/pinhole that allows a pixel size <200 nm to comfortably track the beads in the analysis phase (e.g., 60x, NA 1.3). Epifluorescence microscopy can also be used, however it provides lower signal to noise ratio and may make individual bead tracking harder. - Two main layers of beads will appear on the bottom and the top of the gel. Focus on the gel plane.
NOTE: A nice gel will appear as a starry sky, with beads approximately uniformly distributed on the same plane. - Program the acquisition for 30 min with a frame rate of 5 s (this is adaptable to the needs of the experiment, e.g., acquire other colors, acquire z stack, etc.)
- Aspirate the media from the gel, leaving about 200 µL of media on the gel. Position the gel on the microscope and find the surface layer of beads and a nice even area on the gel.
- Add 80 µL of cells (avoid touching the gel to maintain focus).
- Ensure that the focus is still correct and that cells can be seen descending in the area (under transmitted light). Launch the acquisition before the cells reach the gel.
- In case of accidental contact with gel, vibrations, or focus drift, adjust the focus.
NOTE: It is crucial to collect an image of the relaxed gel and this can be any image taken before the arrival of the cells on the gel.
- Use a confocal microscope with thermal and (possibly) CO2 control.
4. Fluorescent HEL extraction experiment
- Prepare fluorescent HEL by binding a fluorescent dye (of a color different from the beads one such as Alexa 555), see the Table of Materials.
- In step 2.7, replace conventional HEL with the fluorescent HEL.
- Acquire images with low illumination settings or low frame rate (e.g., 2 frames per minute) to avoid photo-bleaching.
- To quantify HEL extraction, compute the intensity integrated over the cell area for each frame I(t) corrected and normalized by the intensity I(0) of frame 0 according to the formula:
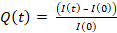
NOTE: The antigen conjugated with a fluorophore is not visible (probably due to quenching of the fluorophore at the gel surface), but its presence on the gel can be verified with an anti-HEL and a fluorescent secondary antibody. It can be verified that the fluorophore is indeed fluorescent when detached by stripping it from the gel with a coverslip coated with anti-HEL and revealing it with a secondary fluorescent antibody (on the coverslip)6. The signal of the extracted antigen is very dim and is sometimes masked by leaking of the beads. If one is interested only in antigen extraction, it is recommended to prepare the gel without beads (skip steps 1.3.2 and 1.4.3).
5. Fluorescence imaging
- Obtain fluorescent B cells by purifying B cell from the spleens of genetically modified mice as done for the wild type (e.g., from Lifeact-GFP or Myosin II GFP mice).
- For imaging fluorescent cells, use (if possible) a spinning disk microscope with a water immersion long-distance 40x–100x objective.
- Keep exposure duration and frame rate low to avoid bleaching.
NOTE: The point spread function in Z is highly degraded by the presence of the gel, hence we suggest using a water immersion objective. Live upright microscopy with water-dipping objectives suffers from strong spherical aberrations induced by the presence of the (spherical) cell (and cell nucleus) in the emission path.
6. Analysis
NOTE: Data analysis is in general performed by first correcting the whole stack for drift, finding the beads in each frame, tracking their movements with respect to a reference frame (taken in absence of cells), interpolating the displacement field and inverting the problem to obtain the stress using Fourier transform29. To this end, we suggest using a combination of ImageJ Macro and MATLAB programs downloadable from an online repository30.
- Open the movie in ImageJ as stack of images
- Run the macro “Crop_and_save.ijm”
- Select the regions of interest (ROI) with the “Rectangle” tool and add them to the ROI list using the ‘t’ key.
- When cropping the cell, be sure to include a region of at least 5–10 pixels of immobile beads. Exclude cells that are too close to the boundaries or to other cells from the analysis. When finished click on ‘OK’.
- The macro proposes a mask of the cell: if this is satisfactory click on “OK”. If not satisfactory, click on “Not ok” and then manually select a closed region with any selection tool (e.g. “Freehand” or “Oval”) and click on “Continue”.
- Open MATLAB and run “TFM_v1.m”.
- Input the required parameters: in particular check the image properties (pixel size, time interval of acquisition) and the gel properties (Young modulus E, Poisson ratio).
- The reference image is set to be the first by default. Set it to another frame if needed or set it to “0” to load an external file.
- Locate the outputs of the software in the same directory as the original file (for a description see the User_notice.pdf file). This includes a preliminary track of the beads (“FILENAME.fig”), a plot of the contractile energy over time (“FILENAME_energy.fig”), a table of several quantities integrated over the cell (energy, area, moments, etc) “FILENAME_finaltable.mat”, a structure containing the displacement and force field, movies of the bead, displacement field, stress and energy (that can be opened with any avi reader).
NOTE: In the input parameters, the “Window size” is the window over which the displacement is interpolated, hence the final resolution of the stress and displacement field. This is set to a few (by default four) pixels. It is not advisable to reduce this as it would artificially increase the resolution by interpolating regions where there are no beads.
Representative Results
Given the size of the cells, algorithms that extract the displacement map of the beads via correlative techniques (such as particle image velocimetry) are in general not very precise. However depending on the degree of resolution required, one can easily obtain qualitative results using a free Fiji/ImageJ plugin31,32. While this approach is sufficient to compare stimulating versus non-stimulating conditions, for a thorough analysis we recommend using our software downloadable from an online repository30, that tracks the beads individually and provides the displacement field map at a given time point as the interpolation of the individual bead displacements33. Several quantifications are possible at this point. For example (by assuming the displacement is caused only by stress tangential to the gel surface) the software also provides the stress at each point causing that specific displacement map. This is a type of “inversion problem”: the displacement at a certain point depends on the sum of all the forces applied all over the other points. The “inversion algorithm” takes into account the physical parameters of the substrate: its rigidity (Young modulus) and Poisson ratio. Direct algorithms are typically very accurate but computationally expensive. Algorithms based on Fourier transform, like ours, perform essentially a deconvolution in Fourier space and are more efficient but prone to some errors (mainly due to the interpolation step). These algorithms generally require the tuning of a parameter that prevents small local (and potentially artifactual) displacements to become too relevant in the computation of the stress field (Tikhonov regularization parameter8,29; “Regularization” variable in the dialog window; here we typically set equal to 5 x 10-19). For more advanced interpretation and analysis, such as spatio-temporal correlations, local movements, correlations with fluorescent channels, we recommend collaborating with experts in the field. For a review on computational methods see Schwarz et al.9.
As mentioned above, correct bead images look like a “starry sky”, a uniform and random distribution of bright spots (Figure 2A). Data and analysis are not reliable when the number of beads is too low (Figure 2B) or the image is out of focus (Figure 2C). Once B cells have settled onto the surface of the gel, the beads underneath the cells start to move due to the traction force exerted by the cell on the gel. Frames for which the beads are not trackable should be discarded.
As a check, it is possible to observe by eye the movement of beads comparing the “reference frame”, typically the one preceding the first contact of the cell with the substrate. Approximate results can be obtained from the single particle tracking (e.g., Trackmate, Fiji 34) as done in Figure 3A. The analysis provides a segmentation of the beads in the reference image (“FILENAME.fig”) as a control.
With the software we propose, one can obtain the displacement (Figure 3B) and stress field (the vector of the local stress at each pixel and each time point obtained by inversion from the displacement field, Figure 3C). Scalar product of the displacement and force fields integrated on the area of the cell provides total work exerted by the cell on the substrate (Figure 4A). This computation requires the mask of the cell introduced in step 6.2 of the protocol.
To compare two biological conditions (as activating HEL versus non-activating substrate BSA, or wild type versus knock-out) it is useful to compute the average curve (Figure 4B) or, even more synthetically, an average value over the last time points (20 min) where the energy reaches a plateau (Figure 4C). When the spatial information of the forces is relevant it is possible to compare single time points of each condition (Figure 4D). Refer to Kumari et al.6 for deeper analysis.
An example of fluorescence antigen extraction time lapse is shown in Figure 5A: the progressive appearance of fluorescence signals at the synapse indicated antigen detachment from the gel. The average extraction curve with its confidence interval (standard error of the mean) over 15 cells is shown in Figure 5B.
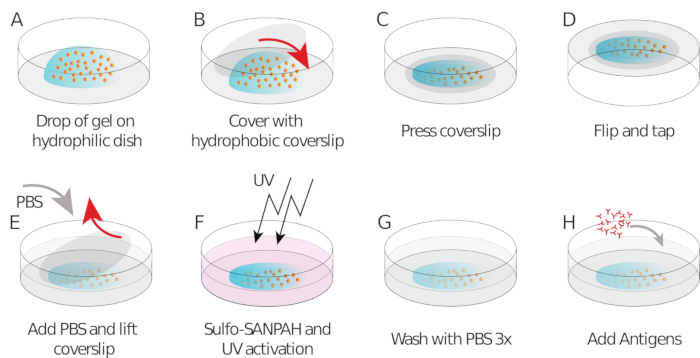
Figure 1: Schematic showing of the preparation of the gel and its functionalization. Steps are described in the protocol. Please click here to view a larger version of this figure.
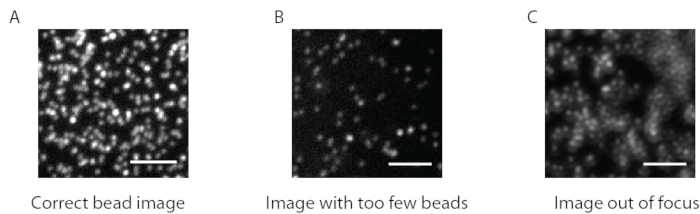
Figure 2: Three examples of bead images of different qualities. (A) Example of bead image with the correct signal to noise ratio and the correct density. (B) Examples of images with a too insufficient number of beads and (C) out of focus plane. Please click here to view a larger version of this figure.
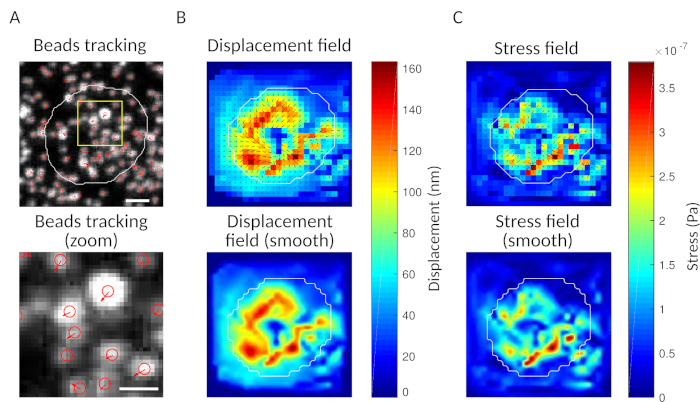
Figure 3: Processing of the images to extract the force field. (A) Example of an image of the beads (outline of the cell in white, extracted from the transmission image), bead tracking at time t = 5 min (red overlay) and displacement (arrows) relative to time t = 0 min (scale bar 5 µm). (B) Interpolated displacement field (represented as vector quiver and magnitude map, arrows are proportional to the displacement [nm]; see the color bar on the right); bottom: a smoother image of the magnitude (obtained by interpolation with a bicubic function). (C) Stress field from displacement field in panel B (represented as vector quiver and magnitude map; arrows are proportional to the shear stress [Pa]; see the color bar on the right); bottom: a smoother image of the magnitude (obtained by interpolation with a bicubic function). Please click here to view a larger version of this figure.
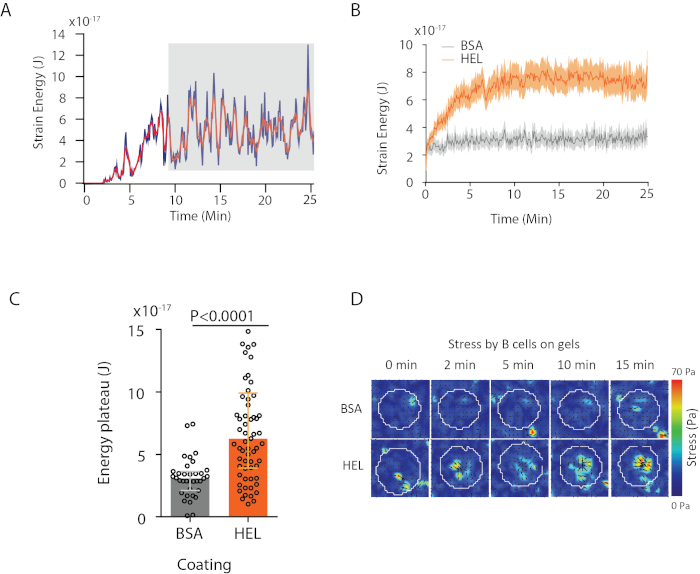
Figure 4: Example of information that can be extracted from force and displacement fields. (A) Example of evolution of energy in time for a single cell: a plateau phase (highlighted in gray) shows up after about 10 min. (B) Comparison of the average energy curves and (C) of the relative plateau levels for 65 cells plated on HEL (activating) coated gel and 35 cells on BSA (non-activating) coated gel (median ± interquartile ranges are shown, Mann-Whitney test was used for statistical significance). (D) Time-lapse color maps of stress for HEL and control BSA condition; both magnitude and quiver plots are shown. These images have been adapted from Kumari et al.6. Please click here to view a larger version of this figure.
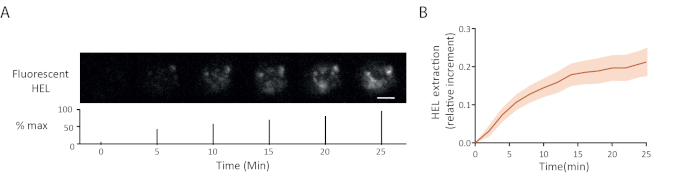
Figure 5: Example of experiments with fluorescent antigen. (A) Time lapse of the extraction of fluorescent HEL (below: percentage of the maximum, scale bar = 3µm). (B) Antigen gathering over time (Mean ± SEM, n = 15). These images have been adapted from Kumari et al.6. Please click here to view a larger version of this figure.
Discussion
The TFM method described here allows for the systematic study of the active mechanical capabilities of B cells. In the context of B cells, this is related to the ability to extract and internalize the antigen. Compared to other TFM methods, the protocol presented here is simple and rather reproducible: the rigidity, measured by indentation of a glass microsphere and using Hertz model, is between 400 and 600 Pa. Similar protocols have been successfully used not only for B cells35 but also for T cells36. In comparison to nanopillars (also used for T lymphocytes37) it provides a flat homogeneous surface, hence the results are easier to interpret as the interaction of the gel are mainly constrained to be tangential to the surface.
The protocol we described gives access to the spatiotemporal dynamics of the forces exerted by B cells on antigen-presenting substrates. On the spatial level this provides information of the localization of forces, and in combination with fluorescence microscopy, enables the experimenter to correlate local forces with the presence of specific molecules (i.e., components of the cytoskeleton or BCR signaling cascade). At the temporal level, it is possible to integrate quantities (such as total energy or total stress) to provide one value per time point and reduce the noise. This allows for observation of the evolution of the traction force in time (growth and plateau) and the presence of pulsatile patterns.
Critical experimental aspects for the analysis are described as following. (i) Cell density: to perform a correct analysis, cells should be sufficiently separated. We consider a cell to be analyzable if it has an empty region of its own size around it. (ii) Transmission image: it is advisable to collect at least a transmission image of the cells during the experiment to be used as a mask in the analysis. (iii) Number of beads in the image: we suggest analyzing only images where the number of beads in the synapse is between 30 and 200 (i.e., 1–8 beads/µm²). Lower densities do not allow for adequate map displacement reconstruction. High bead densities make single particle tracking unreliable. (iv) Number of beads should be constant during the experiment; however, fluctuations can occur due to small variability in the imaging conditions (especially in beads that are too close to each other). Focus drift, if occurring, must be corrected and problematic frames should be discarded. (v) Gel quality: gels with too many cracks, variability in beads distribution or gels that are too thick should be discarded. (vi) Depending on the cell type, after repeated exposures, cells at late time points (>300 frames) may suffer phototoxic effects. It is advisable to run the program on a mask devoid of cells as a “baseline” to be compared with the data. This provides a magnitude of the noise level solely due to the experimental conditions.
Gels used to measure traction force in classical adhesion allow for the investigation of processes that occur at the focal adhesion (actin flows and recruitment of signaling molecules)—the points where forces are applied38,39. However, forces at the synapse are not applied through focal adhesions. The spatiotemporal pattern of force generation at the B cell immune synapse has not been quantitatively investigated using this method until recently. Using TFM, we observed for the first time, force patterning at the B cell immune synapse, as presented in our recent study6, opening encouraging perspectives in the study of lymphocytes.
Notably, this method employs an image taken before the arrival of the cells on the gel as a reference image for the force computation. Usual TFM protocols suggest taking the reference image at the end of the experiment, after detaching the cells with trypsin; this allows the experimenter to look for a region rich in cells. Although this is possible here too, trypsin is rather inefficient at detaching B cells from antigen-coated gel, one needs to wait long for detachment and the risk of gel modification and movements (that make the whole data set unexploitable) is higher.
The method presented here is flexible and can be applied to study the effect of other signals at the immune synapse as it allows for grafting other proteins onto the gel surface (e.g., integrin ligands and immunoglobulins have been tested) and even fluorescent antigen (see section 4). Moreover, cells remain accessible to the experimenter for drug treatment and local perturbations. Finally, the method is also compatible with imaging fixed cells. For these observations, it is recommended to make the gel on a coverslip, stain the cells, glue the coverslip on a slide and only then add mounting media and another coverslip. Observation will then be done with the gel on top to avoid the degradation of the image through the gel.
Possible pitfalls are the variability in gel in polymerization and coating. Polymerization problems are mainly due to the quality of initiator/catalyst. Also, the gel can inflate, especially if not used right after assembly. This problem does not seem to dramatically affect the mechanical properties of the gel, but it can make the bead layer unreachable for the objective, effectively making the gel useless. We recommend preparing extra gels for each condition when this problem appears. There might be also a certain variability in the coating, and it is crucial to have freshly diluted Sulfo SANPAH.
In conclusion, we have described a simple, cheap and reproducible method to measure the forces exerted by B cells at the immunological synapse when activated by BCR ligand. It can be adapted to study the reaction to other ligands and other kinds of lymphocytes (memory B cells, T cells, etc.) with the use of the proper receptor ligand.
Disclosures
The authors have nothing to disclose.
Acknowledgements
The authors thank M. Bolger-Munro for critical reading and acknowledge the Nikon Imaging Center@CNRS-InstitutCurie and PICT-IBiSA, Institut Curie, Paris, member of the France-BioImaging national research infrastructure, for support in image acquisition and the Curie Animal Facility. PP was supported by CNRS. AK and JP were supported by Paris Descartes PhD fellowship and Ecole Doctorale FIRE—Programme Bettencourt. This project was funded by grants to PP (ANR-10-JCJC-1504-Immuphy) and AMLD (ANR-PoLyBex-12-BSV3-0014-001, ERC-Strapacemi-GA 243103).
Materials
| 3-aminopropyltrimethoxysilane (APTMS) | Sigma-Aldrich | 281778 | Store aliquoted, protected from humidity |
| 40% Acrylamide Solution | Biorad | 1610140 | |
| Alexa555 microscale protein labeling kit | Molecular Probes | A30007 | |
| Ammonium Persulfate (APS) | Sigma-Aldrich | A3678 | |
| B cell Isolation Kit, Mouse | Miltenyi Biotec | 130-090-862 | |
| B-mercaptoethanol | Gibco | 31350-010 | |
| 2% Bis Solution | Biorad | 161-0142 | |
| Bovine Serum Albumin (BSA) | Euromedex | 04-100-812-C | |
| Coverslip 18mm | VWR | 631-1580 | |
| Fetal calf serum | PAA | A15-151 | Decomplemented (40min @56°C) |
| Fluorodishes FD35 | World Precision Instruments, Inc | FD35100 | |
| Fluosphere: carboxylate-modified, 0.2um, dark red | Molecular Probes | F8807 | |
| Hen Egg Lysozyme | Sigma-Aldrich | L6876 | Stocked in aliquote 100mg/ml |
| MEM Non-Essential Amino Acids Solution (100X) | Thermofisher/Gibco | 11140035 | |
| N,N,N',N'-tetrametiletilendiammine (TEMED) | Euromedex | 50406-B | |
| PBS (Phosfate Buffer Saline) | Gibco | 10010-015 | |
| Penicillin–streptomycin | Gibco | 15140-010 | |
| RMPI 1640 – Glutamax I | Thermofisher | 61870-010 | |
| Sigmacote | Sigma-Aldrich | SL2 | |
| Sodium pyruvate | Gibco | 11360-039 | |
| sulfosuccinimidyl 6-(4'-azido-2'-nitrophenylamino)hexanoate (Sulfo-SANPAH) | Thermo Scientific | 22589 |
References
- Yuseff, M. -. I., Pierobon, P., Reversat, A., Lennon-Duménil, A. -. M. How B cells capture, process and present antigens: a crucial role for cell polarity. Nature Reviews. Immunology. 13 (7), 475-486 (2013).
- Spillane, K. M., Tolar, P. B cell antigen extraction is regulated by physical properties of antigen-presenting cells. The Journal of Cell Biology. 216 (1), 217-230 (2017).
- Shaheen, S., Wan, Z., et al. Substrate stiffness governs the initiation of B cell activation by the concerted signaling of PKCβ and focal adhesion kinase. eLife. 6, (2017).
- Natkanski, E., et al. B cells use mechanical energy to discriminate antigen affinities. Science. 340 (6140), 1587-1590 (2013).
- Wan, Z., Chen, X., et al. The activation of IgM- or isotype-switched IgG- and IgE-BCR exhibits distinct mechanical force sensitivity and threshold. eLife. 4, (2015).
- Kumari, A., Pineau, J., et al. Actomyosin-driven force patterning controls endocytosis at the immune synapse. Nature Communications. 10 (1), 2870 (2019).
- Dembo, M., Wang, Y. L. Stresses at the cell-to-substrate interface during locomotion of fibroblasts. Biophysical Journal. 76 (4), 2307-2316 (1999).
- Sabass, B., Gardel, M. L., Waterman, C. M., Schwarz, U. S. High resolution traction force microscopy based on experimental and computational advances. Biophysical Journal. 94 (1), 207-220 (2008).
- Schwarz, U. S., Soiné, J. R. D. Traction force microscopy on soft elastic substrates: A guide to recent computational advances. Biochimica et Biophysica Acta. 1853 (11), 3095-3104 (2015).
- Colin-York, H., Shrestha, D., et al. Super-Resolved Traction Force Microscopy (STFM). Nano Letters. 16 (4), 2633-2638 (2016).
- Stubb, A., Laine, R. F., Guzmán, C., Henriques, R., Jacquemet, G., Ivaska, J. Fluctuation-Based Super-Resolution Traction Force Microscopy. BioRxiv. , (2019).
- Gutierrez, E., Tkachenko, E., et al. High refractive index silicone gels for simultaneous total internal reflection fluorescence and traction force microscopy of adherent cells. Plos One. 6 (9), 23807 (2011).
- Bergert, M., Lendenmann, T., et al. Confocal reference free traction force microscopy. Nature Communications. 7, 12814 (2016).
- Schoen, I., Hu, W., Klotzsch, E., Vogel, V. Probing cellular traction forces by micropillar arrays: contribution of substrate warping to pillar deflection. Nano Letters. 10 (5), 1823-1830 (2010).
- Colin-York, H., Fritzsche, M. The future of traction force microscopy. Current Opinion in Biomedical Engineering. 5, 1-5 (2018).
- Feng, Y., et al. Mechanosensing drives acuity of αβ T-cell recognition. Proceedings of the National Academy of Sciences of the United States of America. 114 (39), 8204-8213 (2017).
- Spillane, K. M., Tolar, P. DNA-Based Probes for Measuring Mechanical Forces in Cell-Cell Contacts: Application to B Cell Antigen Extraction from Immune Synapses. Methods in Molecular Biology. 1707, 69-80 (2018).
- Stabley, D. R., Jurchenko, C., Marshall, S. S., Salaita, K. S. Visualizing mechanical tension across membrane receptors with a fluorescent sensor. Nature Methods. 9 (1), 64-67 (2011).
- Merkel, R., Nassoy, P., Leung, A., Ritchie, K., Evans, E. Energy landscapes of receptor-ligand bonds explored with dynamic force spectroscopy. Nature. 397 (6714), 50-53 (1999).
- Hinterdorfer, P., Dufrêne, Y. F. Detection and localization of single molecular recognition events using atomic force microscopy. Nature Methods. 3 (5), 347-355 (2006).
- Sawicka, A., Babataheri, A., et al. Micropipette force probe to quantify single-cell force generation: application to T-cell activation. Molecular Biology of the Cell. 28 (23), 3229-3239 (2017).
- Desprat, N., Guiroy, A., Asnacios, A. Microplates-based rheometer for a single living cell. Review of Scientific Instruments. 77 (5), 055111 (2006).
- Labernadie, A., Bouissou, A., et al. Protrusion force microscopy reveals oscillatory force generation and mechanosensing activity of human macrophage podosomes. Nature Communications. 5, 5343 (2014).
- Bouissou, A., Proag, A., et al. Protrusion force microscopy: A method to quantify forces developed by cell protrusions. Journal of Visualized Experiments. (136), 57636 (2018).
- Kronenberg, N. M., Liehm, P., et al. Long-term imaging of cellular forces with high precision by elastic resonator interference stress microscopy. Nature Cell Biology. 19 (7), 864-872 (2017).
- Basu, R., Whitlock, B. M., et al. Cytotoxic T cells use mechanical force to potentiate target cell killing. Cell. 165 (1), 100-110 (2016).
- Bufi, N., Saitakis, M., et al. Human Primary Immune Cells Exhibit Distinct Mechanical Properties that Are Modified by Inflammation. Biophysical Journal. 108 (9), 2181-2190 (2015).
- Goodnow, C. C., Crosbie, J., et al. Altered immunoglobulin expression and functional silencing of self-reactive B lymphocytes in transgenic mice. Nature. 334 (6184), 676-682 (1988).
- Butler, J. P., Tolić-Nørrelykke, I. M., Fabry, B., Fredberg, J. J. Traction fields, moments, and strain energy that cells exert on their surroundings. American Journal of Physiology. Cell Physiology. 282 (3), 595-605 (2002).
- . MBPPlab/TFM_v1: Software for Time dependent Traction Force Microscopy Available from: https://github.com/MBPPlab/TFM_v1 (2019)
- Tseng, Q., Duchemin-Pelletier, E., et al. Spatial organization of the extracellular matrix regulates cell-cell junction positioning. Proceedings of the National Academy of Sciences of the United States of America. 109 (5), 1506-1511 (2012).
- . ImageJ plugins by Qingzong TSENG Available from: https://sites.google.com/site/qingzongtseng/ (2019)
- Plotnikov, S. V., Sabass, B., Schwarz, U. S., Waterman, C. M. High-resolution traction force microscopy. Methods in Cell Biology. 123, 367-394 (2014).
- Schindelin, J., Arganda-Carreras, I., et al. Fiji: an open-source platform for biological-image analysis. Nature Methods. 9 (7), 676-682 (2012).
- Wang, J., Lin, F., et al. Profiling the origin, dynamics, and function of traction force in B cell activation. Science Signaling. 11 (542), (2018).
- Hui, K. L., Balagopalan, L., Samelson, L. E., Upadhyaya, A. Cytoskeletal forces during signaling activation in Jurkat T-cells. Molecular Biology of the Cell. 26 (4), 685-695 (2015).
- Bashour, K. T., Gondarenko, A., et al. CD28 and CD3 have complementary roles in T-cell traction forces. Proceedings of the National Academy of Sciences of the United States of America. 111 (6), 2241-2246 (2014).
- Gardel, M. L., Sabass, B., Ji, L., Danuser, G., Schwarz, U. S., Waterman, C. M. Traction stress in focal adhesions correlates biphasically with actin retrograde flow speed. The Journal of Cell Biology. 183 (6), 999-1005 (2008).
- Stricker, J., Sabass, B., Schwarz, U. S., Gardel, M. L. Optimization of traction force microscopy for micron-sized focal adhesions. Journal of Physics. Condensed Matter. 22 (19), 194104 (2010).

