The Citrobacter rodentium Mouse Model: Studying Pathogen and Host Contributions to Infectious Colitis
Summary
Citrobacter rodentium infection provides a valuable model to study enteric bacterial infections as well as host immune responses and colitis in mice. This protocol outlines the measurement of barrier integrity, pathogen load and histological damage allowing for the thorough characterization of pathogen and host contributions to murine infectious colitis.
Abstract
This protocol outlines the steps required to produce a robust model of infectious disease and colitis, as well as the methods used to characterize Citrobacter rodentium infection in mice. C. rodentium is a gram negative, murine specific bacterial pathogen that is closely related to the clinically important human pathogens enteropathogenic E. coli and enterohemorrhagic E. coli. Upon infection with C. rodentium, immunocompetent mice suffer from modest and transient weight loss and diarrhea. Histologically, intestinal crypt elongation, immune cell infiltration, and goblet cell depletion are observed. Clearance of infection is achieved after 3 to 4 weeks. Measurement of intestinal epithelial barrier integrity, bacterial load, and histological damage at different time points after infection, allow the characterization of mouse strains susceptible to infection.
The virulence mechanisms by which bacterial pathogens colonize the intestinal tract of their hosts, as well as specific host responses that defend against such infections are poorly understood. Therefore the C. rodentium model of enteric bacterial infection serves as a valuable tool to aid in our understanding of these processes. Enteric bacteria have also been linked to Inflammatory Bowel Diseases (IBDs). It has been hypothesized that the maladaptive chronic inflammatory responses seen in IBD patients develop in genetically susceptible individuals following abnormal exposure of the intestinal mucosal immune system to enteric bacteria. Therefore, the study of models of infectious colitis offers significant potential for defining potentially pathogenic host responses to enteric bacteria. C. rodentium induced colitis is one such rare model that allows for the analysis of host responses to enteric bacteria, furthering our understanding of potential mechanisms of IBD pathogenesis; essential in the development of novel preventative and therapeutic treatments.
Introduction
Infection by enteric bacterial pathogens triggers gastrointestinal (GI) inflammation, as well as intestinal pathology and pathophysiology, including diarrhea and intestinal epithelial barrier dysfunction. The virulence mechanisms by which bacterial pathogens colonize the GI tract of their hosts, as well as specific host responses that defend against such infections are poorly understood, however recent advances in the modeling of enteric bacterial infections have begun to aid our understanding of these processes. Enteric bacteria have also been linked to Inflammatory Bowel Diseases (IBDs). The IBDs Crohn’s Disease (CD) and UC are complex diseases of unknown etiology, characterized by chronic intestinal inflammation and tissue damage. Many mouse models of intestinal inflammation exist, from spontaneous inflammation in genetically modified strains, such as IL10 -/- mice, to chemical challenges with compounds, such as dextran sodium sulfate (DSS) and dinitrobenzene sulfonic acid (DNBS)1. It has been hypothesized that the maladaptive chronic inflammatory responses present in IBD patients develop in genetically susceptible individuals upon abnormal exposure of the intestinal mucosal immune system to enteric bacteria2, therefore the study of models of infectious colitis also offers significant potential for defining potentially pathogenic host responses to enteric bacteria. Citrobacter rodentium induced colitis is one of the rare models of infectious colitis that has been well characterized1,3, allowing for the analysis of host responses to enteric bacteria and further understanding of potential mechanisms of IBD pathogenesis; an essential step in developing novel preventative and therapeutic treatments.
C. rodentium is a gram negative attaching and effacing (A/E), murine specific bacterial pathogen that is closely related to the important human pathogens enteropathogenic E. coli (EPEC) and enterohaemorrhagic E. coli (EHEC)3-8. The family of A/E pathogens intimately attach to the apical host cell membrane of the cecal and colonic epithelium, forming a non-invasive pedestal-like structure on the host cell. Oral challenge with C. rodentium of 108-109 organisms produces a robust model of infectious colitis characterized by colonic hyperplasia or elongation of the crypts, mononuclear immune cell infiltration and goblet cell depletion3,4. The initial site of colonization, a few hours after challenge, is at the cecal patch, followed by progression to the distal colon 2 to 3 days after infection3. In immunocompetent mouse strains, clearance of the pathogen is achieved 3 to 4 weeks after infection1,3,4. However, many genetically modified strains, i.e. gene deficient or knockout (-/-) mice, have been found to display increased susceptibility to infection resulting in exaggerated damage and/or chronic infection and inflammation9-14. Use of this infectious colitis model in these knockout strains, many lacking innate signaling proteins, has been indispensible in revealing several host proteins integral to resolution of intestinal infection and inflammation.
Protocol
1. Preparation of Citrobacter rodentium Inoculum and Oral Gavage of Mice
- Prepare and autoclave Luria Bertani broth (LB).
- Obtain viable C. rodentium from a frozen glycerol stock and streak onto LB agar plate using a sterile inoculating loop or pipette tip. Incubate at 37 °C overnight. Inoculate 3 ml of sterile LB broth in a falcon culture tube with colonies from the LB plate using an inoculating loop or pipette tip. Preparation of the inoculum should be done using aseptic techniques.
- Incubate the C. rodentium culture aerobically at 37 °C overnight in a benchtop incubation shaker at 200 rpm. The inoculated LB broth should appear cloudy after overnight culture.
- Use a bulb tipped gastric gavage needle attached to a 1 ml syringe to gavage each mouse with 100 μl of the overnight C. rodentium culture. Gently scruff the mouse, by firmly grasping the loose skin over its neck and back with thumb and fingers. Pull back the animal’s head with your index finger to immobilize the head and straighten the esophagus for insertion of the gavage needle.
- Maintain the mouse in an upright position and direct the bulb tip of the gavage needle along the side of its mouth and over its tongue. Gently pass the needle along the roof of the mouth and advance it down the esophagus. If there is any resistance felt during this procedure, remove the gavage needle and re-insert it.
- Slowly inject 100 μl of the bacterial inoculum and gently remove the gavage needle. Monitor the breathing rate and behavior of the mouse after returning it to its cage.
Notes:
- In steps 2 and 3, also prepare a falcon culture tube using aseptic techniques with 3 ml of sterile LB broth to be cultured overnight to ensure that there is no bacterial contamination of the broth itself. The LB broth should appear clear after overnight culture.
- Gently agitate the culture within the falcon tube prior to loading syringes for gavage so that an equal dose of bacteria is delivered to each mouse, in step 4.
- Overnight culture should be discarded if it has been inoculated for over 24 hr.
- 모두 C. rodentium infections and housing of infected animals should be carried out in a Biosafety Level 2 facility.
2. Measuring Colonic Epithelial Barrier Permeability in C. rodentium-infected Mice
- Measure barrier integrity at a desired time point post-infection.
- On the day of the assay, prepare enough 4 kDa fluorescein isothiocyanate (FITC)-dextran dissolved in phosphate buffered saline (PBS) to a concentration of 80 mg ml-1 to gavage each mouse with 150 μl and to prepare the standard curve.
- Scruff the mouse and gavage with 150 μl of FITC-dextran. Withdraw food from the cage at this time.
- Anesthetize mice 4 hr after gavaging and collect as much blood as possible (~450 μl) through cardiac puncture. Add the blood to a final concentration of 3% acid-citrate dextrose in a microcentrifuge tube to deter coagulation. Euthanize the mice and collect colonic tissues in PBS for bacterial counts (steps 3.1 – 3.5) and in 10% formalin for tissue fixation and ultimately immunofluorescence staining (steps 4.1 – 4.8).
- Spin the blood samples at 1,000 x g for 12 min in a centrifuge at 4 °C. Collect the serum and dilute to 1/10 and 1/100 in PBS. Add 100 μl of each sample at these two dilutions to a 96 well plate in replicates.
- To prepare the standard curve, dilute the original 80 mg ml-1 FITC-dextran used to gavage the mice in PBS to the following concentrations: 800, 400, 200, 100, 50, 25, 12.5, 6.25, and 0 μg/ml. Add 100 μl of each concentration to a 96 well plate in triplicate.
- Quantify the fluorescence of each sample using a fluorometer at an excitation wavelength of 485 nm and 535 nm emission wavelength.
- Analyze the raw data by plotting the standard curve. Input the raw data for each sample into the equation of the line generated by the standard curve to determine the concentration of FITC-dextran in each sample.
Notes:
- From step 4 onward, keep blood samples on ice and minimize exposure to light.
- When measuring epithelial barrier integrity a time point at, or before, 7 days post-infection is optimal, as severe tissue damage occurs after this point. Measuring barrier integrity prior to severe damage occurring allows you to determine maximal differences in permeability during C. rodentium infection between mouse strains.
- Ensure that uninfected control mice are also tested for epithelial barrier integrity.
3. Measurement of Bacterial Load in Tissues of C. rodentium-infected Mice
- Add 1 ml sterile PBS and an autoclaved metal bead to a round bottomed 2 ml centrifuge tube using aseptic technique. Individual tissues collected from each animal will be placed in separate PBS tubes. Weigh the tubes prior to addition of the tissue.
- Remove the cecum and colon from the mouse. Separate the cecum from the colon and push out the luminal contents using forceps. Add the contents to a PBS tube. After separating 0.5 cm sections from the distal colon and cecum for 10% formalin fixation, cut open the remaining colon and cecum longitudinally and wash with sterile PBS before placing tissues in tubes.
- Weigh the PBS tubes with the tissue and then homogenize the samples using a bead beater for 6 min at 30 Hz.
- Using aseptic techniques, add 180 μl sterile PBS per well to a 96 well plate. Add 20 μl of each homogenized sample to the first well in each row. Mix well and serially dilute, adding 20 μl from each previous well to obtain dilutions from 10-1 (first well) to 10-6. Plate 10 μl of each dilution from each sample in triplicate on a square bottom LB plate with a grid. Incubate overnight at 37 °C.
- Count colony forming units (CFU), then average the 3 counts, and record the dilution at which each sample was counted. Multiply average CFU by 50, as 20 μl of the original 1 ml sample was used, and by the dilution factor at which the sample was counted resulting in CFU/ml. Finally, divide by the tissue weight to determine CFU/g.
4. Histological Assessment and Immunofluorescence Staining of Infected Colon Tissues
- Collect tissues as in step 3.2. Store tissues in formalin overnight, then wash with 70% ethanol. Embed tissues in paraffin and cut 5 μm sections for staining. Stain with hematoxylin & eosin for histological assessment and proceed to the following steps for immunofluorescence staining.
- To deparaffinize tissues prior to staining, place slides in a coplin staining jar and put in 65 °C water bath for 10 min. Next, place slides in xylene for four washes of 2 min each. Slides should then be rehydrated with two 5-minute washes in 100% ethanol, followed by one 5 min wash in each of the following: 95% ethanol, 75% ethanol and dH20. These washes should be done in a fume hood to avoid exposure to harmful vapors.
- Place slides in the coplin jar with pre-warmed sodium citrate buffer and then place into a steamer for 30 min, then let sit for 30 min. This process breaks the protein cross-links formed by formalin fixation retrieving potential antigenic sites.
- Wash with PBS azide for 5 min, three times. Dry slides and mark area around tissue with a PAP pen to create a hydrophobic barrier, keeping the staining reagents localized on the tissue.
- Block tissue for 1 hr at room temperature with blocking buffer (e.g. α-goat serum) in an immunostaining moisture chamber.
- Dilute primary antibody to desired concentration using antibody dilution buffer. Pour off blocking buffer and add 50-100 μl of primary antibody to tissues. Incubate at 4 °C overnight or at room temperature for 2 hr.
- Wash with PBS azide for 5 min, three times. Add 50-100 μl of secondary antibody diluted in antibody dilution buffer to the tissues and incubate at room temperature for 1 hr in the dark. Wash with dH2O for 5 min, three times.
- Dehydrate slides, add DAPI Prolong Gold mounting medium and apply a coverslip. View slides under a fluorescence microscope.
Notes:
- PBS azide can be substituted with freshly prepared PBS as an alternative to avoid bacterial growth in the wash medium from step 4 onward.
Representative Results
During a standard infection experiment, mice are infected with approximately 2.5 x 108 CFU through gavage of 100 μl overnight C. rodentium culture. Infection of C57BL/6 mice with C. rodentium results in modest and transient weight loss and diarrhea. Although a rare occurrence with C57BL/6 mice, animals may become ill and require euthanization. Therefore, mice should be monitored for degree of weight loss and symptoms of distress such as piloerect fur and hunched posture, to determine the extent to which different strains are affected by the infection.
Results presented in Figures 1 through 4 are representative of an infection done using an overnight culture prepared from a frozen glycerol stock. At 7 days post-infection, mice were anesthetized and blood was collected through cardiac puncture. Figure 1 shows FITC-dextran measured in the serum of C57BL/6 mice, as well as from Toll like receptor 2 (TLR2) knockout mice, which have previously been found to exhibit impaired epithelial barrier integrity during infection9. In the presence of an intact intestinal epithelial barrier, 4 kDa FITC-dextran is poorly permeable through this layer. Therefore, increased levels of FITC-dextran in serum suggest an impairment of intestinal epithelial barrier integrity during infection allowing this molecule to leak across. As a control, serum levels in uninfected mice gavaged with FITC-dextran are also presented.
By one week post-infection, bacterial loads in the colon have been found to peak around 109 CFU/g3,4. Bacterial loads can be measured at desired time points post-infection to confirm infection, as well as to assess if different knockout mice suffer from increased or decreased bacterial burdens. As C. rodentium is a luminal pathogen that can intimately adhere to intestinal epithelial cells, bacterial loads in the luminal contents, cecal, and colonic tissues can be measured. Figure 2 shows typical bacterial loads measured at day 7 post-infection cecal and colonic tissues as well as within the intestinal lumen of C57BL/6 mice.
Most of the pathology observed during C. rodentium infection in C57BL/6 mice occurs in the distal 2 cm of the colon11. Macroscopically by day 7 post-infection, a thickening of the colonic mucosa as well as a shortening in length of the colon is observed, with little overt damage seen in C57BL/6 mice. Normally during infection, C57BL/6 mice suffer only moderate inflammation and pathology characterized histologically by immune cell infiltration, elongation of colonic crypts and goblet cell depletion (Figure 3).
As seen in the H&E sections in Figure 3, several host responses are mounted during infection with C. rodentium. To further characterize these changes, immunofluorescence staining can be utilized to examine changes in proteins of interest, such as markers of proliferation, cell death, or innate and adaptive immune responses. Immunofluorescence staining is also valuable in examining aspects of the bacterial response, such as its localization within the infected tissue. An example of this staining technique, examining a host protein ki67 (red), a marker for cell proliferation and the C. rodentium protein tir, green, to examine bacterial localization at day 12 post-infection is presented in Figure 4.
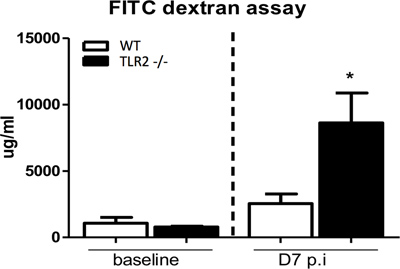
Figure 1. Measurement of serum FITC-dextran to assess epithelial barrier integrity. Mice were gavaged with 4 kDa FITC-dextran under uninfected conditions and at 7 days post-infection, following which serum FITC-dextran levels were determined. C57BL/6 (WT) mice exhibit a negligible increase in FITC-dextran serum levels at 7 days post-infection compared to uninfected conditions. In contrast, TLR2 -/- mice have significantly increased levels of FTIC-dextran in serum compared to baseline suggesting severely impaired barrier integrity in this strain during C. rodentium infection, as has previously been shown.
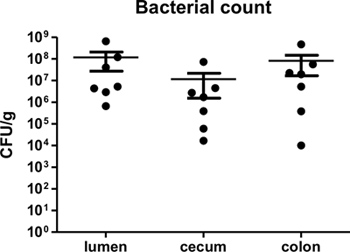
Figure 2. C. rodentium load in lumen, cecum, and colon of infected C57BL/6 mice at 7 days post-infection. Lumen, cecal, and colonic tissues were collected, homogenized, and plated in serial-dilution on LB agar. Each circle represents a single sample collected from individual animals. Solid lines indicate the geometric mean while vertical error bars indicate SEM.
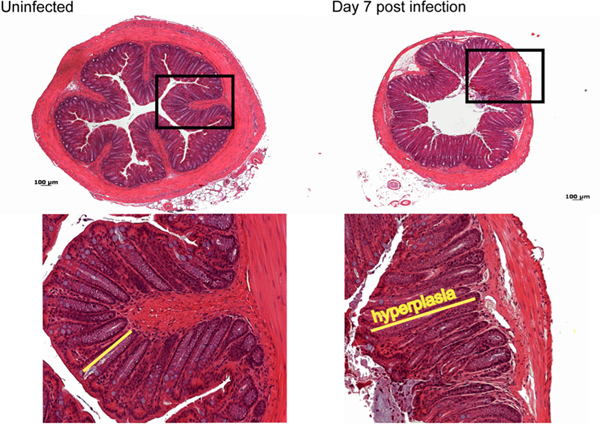
Figure 3. Histological analysis of damage caused by C. rodentium infection. By day 7 post-infection moderate immune cell infiltration, as well as elongation of crypts, goblet cell depletion and mild edema is observed compared to control uninfected tissue. Click here to view larger figure.
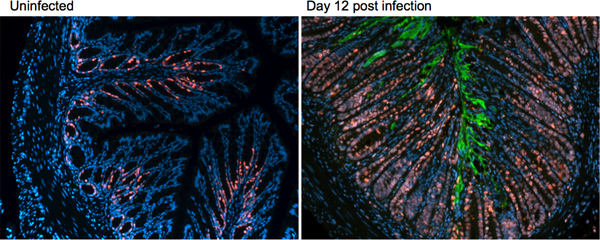
Figure 4. Immunofluorescence staining on uninfected and day 12 post-infection tissues of C57BL/6 mice. Distal colonic tissue is stained for the host protein ki67 (red) and the C. rodentium protein tir (green).
Discussion
Citrobacter rodentium infection provides a valuable model for the study of both infectious disease and colitis in mice. This unique model allows for the characterization of both host responses, as well as the pathogenic properties of bacteria. The steps outlined in this protocol detail the successful use of this model.
There are several critical steps in this protocol to keep in mind when inducing colitis and analyzing responses. First, the preparation of a fresh overnight C. rodentium inoculum in LB broth is required for successful infection of mice. As a dose of 108-109 organisms is required to infect most strains of mice, if the inoculum is left in the shaker or on the bench top at room temperature for extended periods, there will not be enough viable organisms remaining in the culture to induce colitis upon infection. It is also important to agitate the bacterial inoculum before loading the syringe for infection, to ensure that each mouse receives an equal dose of bacteria. When collecting tissues upon terminus of the infection, complete submersion of the colonic tissues in an ample volume of 10% formalin is required for proper tissue fixation to occur. It is suggested that tissues be added to formalin in a 5 ml vial, rather than into microcentrifuge tubes as a 10:1 ratio of fixative to tissue is recommended. Proper fixation is essential for later histological analysis, as well as immunofluorescence staining of these tissues. It is also important to keep in mind that different knockout strains will have varying responses upon induction of C. rodentium colitis. When infecting a new strain for the first time, mice should be closely monitored to determine a humane end-point for the experiment, if needed. Time points for analysis of epithelial barrier integrity, bacterial loads, and histology can be defined based on the response of the mouse strain of interest to the infection.
As plating of tissue homogenates on LB plates to determine bacterial loads is not an entirely selective method, performing PCR for a C. rodentium specific gene on bacterial colonies can be done to differentiate C. rodentium from other bacterialcolonies on the plate. Another option is to plate tissue homogenates on MacConkey agar, as this medium is more selective for C. rodentium than LB agar. An alternative approach is to use a streptomycin resistant wild-type C. rodentium strain instead.Substitution with a streptomycin resistant strain will result in the induction of the same colitis model described in this protocol. The benefit of using the streptomycin resistant strain is for easier analysis of bacterial loads, as tissue homogenates from these infections can be plated on LB agar supplemented with streptomycin rather than LB agar alone. This avoids potential growth of commensal microbes on the plate, making the CFU counting process easier and more accurate. Another alternative while measuring bacterial loads, is to incubate plates after plating dilutions of tissue homogenates on LB agar at either 37 °C overnight or at room temperature for 2-3 days, resulting in slower bacterial growth, before counting.
This protocol outlines the steps required to produce a robust model of infectious colitis, as well as the methods used to characterize C. rodentium infection in mice. Aside from using this model to examine pathogen-host interactions and immune responses, it can also be used to study how a bacterial infection can increase the risk of colon cancer. This can be done by exposing C. rodentium infected mice to the carcinogen azoxymethane, or by studying how infection impacts on epithelial cell proliferation, or on genes involved in epithelial cell differentiation or tumor development15. Using the infectious colitis model outlined in this protocol, bacterial numbers can also be assessed at extra-intestinal sites, such as the mesenteric lymph nodes, spleen and liver. Higher numbers in these organs may indicate that the mouse strain being tested is highly susceptible to the infection. Using the immunofluorescence staining technique described, a virtually endless number of mechanisms in response to infection can be explored. Once familiar with the techniques detailed here, the protocol can be modified to collect tissues to measure other endpoints, such as for RNA extraction and assessment of gene expression levels, to more thoroughly characterize responses to C. rodentium infection.
Disclosures
The authors have nothing to disclose.
Acknowledgements
This work was supported by operating grants to BAV from the Crohn’s and Colitis Foundation of Canada (CCFC) and the Canadian Institutes for Health Research (CIHR). GB was funded by a graduate studentship from CIHR. BAV is the Children with Intestinal and Liver Disorders (CHILD) Foundation Chair of Pediatric IBD Research and the Canada Research Chair in Pediatric Gastroenterology.
Materials
| Name of Reagent/Material | Company | Catalog Number | Comments |
| Luria Broth | ABM | G247 | Add 25 g of LB powder to 1L of water. Autoclave before using. |
| Square bottom plate with grid | Fisher | 08-757-11A | |
| Falcon culture tube | Sarstedt | 62.515.006 | |
| Bulb tipped gastric gavage needle | Fine Science Tools | 18060-20 | |
| 1 ml syringe | BD Biosciences | 309659 | |
| 4 kDa FITC-dextran | Sigma | FD-4 | |
| Citric acid | Sigma | C7129 | |
| Sodium citrate | Fisher | S279-500 | |
| Dextrose | Fisher | D16.1 | |
| Acid citrate dextrose | 20 mM ctiric acid, 110 mM sodium citrate, 5 mM dextrose | ||
| Black 96-well plate | Fisher | 07-200-762 | |
| Metal beads (5 mm) | Qiagen | 69989 | |
| 10% formalin | Fisher | 5F93-4 | |
| 5 ml vial | DiaMed | STK3205 | |
| Hematoxylin | Fisher | H345-23 | |
| Eosin | Fisher | E511-100 | |
| Xylene | Fisher | HC700-1GAL | |
| Tween 20 | Sigma | P5927 | |
| Coplin staining jar | VWR | 47751-792 | |
| Sodium citrate buffer | 10 mM sodium citrate, 0.05% Tween 20, pH 6.0 | ||
| Pap pen | Cedarlane | Mu22 | |
| Goat serum | Sigma | G902-3 | |
| Bovine Serum Albumin (BSA) | Fisher | BP1600-100 | |
| Triton X-100 | Sigma | T8532 | |
| Sodium azide | Sigma | SZ002 | |
| Blocking buffer | 2% goat serum, 1% BSA, 0.1% triton X-100, 0.05% Tween 20, 0.05% sodium azide, 0.01 M PBS, pH 7.2, mix & store at 4 °C. | ||
| Antibody dilution buffer | 0.1% triton X-100, 0.1% BSA, 0.05% sodium azide, 0.04% EDTA | ||
| Blocking buffer & Antibody dilution buffer for tir | Same recipes as above, but without addition of detergents (triton X-100 and tween 20) | ||
| Prolong Gold Antifade Reagent with DAPI | Invitrogen | P-36931 | |
| Coverslips | Fisher | 12.54SE | |
| Benchtop incubation shaker | Barnstead Lab Line | Max Q4000 | |
| Fluorometer | Perkin Elmer | Victor2D | |
| Refrigerated centrifuge | Beckman Coulter | Microfuge 22R | |
| Steamer | Black & Decker | ||
| Fluorescence microscope | Zeiss | Axio Image.Z1 |
References
- Nell, S., Seurbaum, S., Josenhans, C. The impact of microbiota on the pathogenesis of IBD: lessons from mouse infection models. Nature Reviews Microbiology. 8, 564-577 (2010).
- Cario, E. Toll-like Receptors in Inflammatory Bowel Diseases: A Decade Later. Inflammatory Bowel Diseases. 16 (9), 1583-1597 (2010).
- Eckmann, L. Animal Models of Inflammatory Bowel Disease. Annals New York Academy of Sciences. , 1027-38 (2006).
- Mundy, R., MacDonald, T. T., Dougan, G., Frankel, G., Wiles, S. Citrobacter rodentium of mice and man. Cellular Microbiology. 7, 1697-1706 (2005).
- Luperchio, S., Schauer, D. Molecular pathogenesis of Citrobacter rodentium and transmissible murine colonic hyperplasia. Microbes and Infection. 3 (4), 333-340 (2001).
- Bergstrom, K. S., Sham, H. P., Zarepour, M., Vallance, B. A. Innate host responses to enteric bacterial pathogens: a balancing act between resistance and tolerance. Cellular Microbiology. 14, 475-484 (2012).
- MacDonald, T. T., Frankel, G., Dougan, G., Goncalves, N. S., Simmons, C. Host defences to Citrobacter rodentium. International Journal of Medical Microbiology. 293, 87-93 (2003).
- Borenshtein, D., McBee, M. E., Schauer, D. B. Utility of the Citrobacter rodentium infection model in laboratory mice. Current Opinion in Gastroenterology. 24, 32-37 (2008).
- Gibson, D. L., Ma, C., Rosenberger, C. M., Bergstrom, K. S. B., Valdez, Y., Huang, J. T., Khan, M. A., Vallance, B. A. Toll-like receptor 2 plays a critical role in maintaining mucosal integrity during Citrobacter rodentium-induced colitis. Cellular Microbiology. 10, 388-403 (2008).
- Dennis, A., Kudo, T., et al. The p50 subunit of NF-κB is critical for in vivo clearance of the non-invasive enteric pathogen Citrobacter rodentium. Infection & Immunity. 76 (11), 4978-4988 (2008).
- Gibson, D. L., Ma, C., Bergstrom, K. S. B., Huang, J. T., Man, C., Vallance, B. A. MyD88 signalling plays a critical role in host defence by controlling pathogen burden and promoting epithelial cell homeostasis during Citrobacter rodentium-induced colitis. Cellular Microbiology. 10 (3), 618-631 (2008).
- Lebeis, S. L., Bommarius, B., Parkos, C. A., Sherman, M. A., Kalman, D. TLR signaling mediated by MyD88 is required for a protective innate immune response by neutrophils to Citrobacter rodentium. Journal of Immunology. 179 (1), 566-577 (2007).
- Bry, L., Brenner, M. B. Critical role of T cell dependent serum antibody, but not the gut-associated lymphoid tissue, for surviving acute mucosal infection with Citrobacter rodentium, an attaching and effacing pathogen. Journal of Immunology. 172 (1), 433-441 (2004).
- Simmons, C. P., Clare, S., et al. Central role for B lymphocytes and CD4+ T cells in immunity to infection by the attaching and effacing pathogen Citrobacter rodentium. Infection & Immunity. 71 (9), 5077-5086 (2003).
- Ahmed, I., Chandrakesan, P., Tawfik, O., Xia, L., Anant, S., Umar, S. Critical Roles of Notch and Wnt/β-Catenin Pathways in the Regulation of Hyperplasia and/or Colitis in Response to Bacterial Infection. Infection & Immunity. 80 (9), 3107-3121 (2012).

