Protocols for C-Brick DNA Standard Assembly Using Cpf1
Summary
CRISPR-associated protein Cpf1 can be guided by a specially designed CRISPR RNA (crRNA) to cleave double-stranded DNA at desired sites, generating sticky ends. Based on this characteristic, a DNA assembly standard (C-Brick) was established, and a protocol detailing its use is described here.
Abstract
CRISPR-associated protein Cpf1 cleaves double-stranded DNA under the guidance of CRISPR RNA (crRNA), generating sticky ends. Because of this characteristic, Cpf1 has been used for the establishment of a DNA assembly standard called C-Brick, which has the advantage of long recognition sites and short scars. On a standard C-Brick vector, there are four Cpf1 recognition sites – the prefix (T1 and T2 sites) and the suffix (T3 and T4 sites) – flanking biological DNA parts. The cleavage of T2 and T3 sites produces complementary sticky ends, which allow for the assembly of DNA parts with T2 and T3 sites. Meanwhile, a short "GGATCC" scar is generated between parts after assembly. As the newly formed plasmid once again contains the four Cpf1 cleavage sites, the method allows for the iterative assembly of DNA parts, which is similar to those of BioBrick and BglBrick standards. A procedure outlining the use of the C-Brick standard to assemble DNA parts is described here. The C-Brick standard can be widely used by scientists, graduate and undergraduate students, and even amateurs.
Introduction
The standardization of DNA biological parts is important for the development of synthetic biology1. The development of a DNA assembly procedure can replace ad hoc experimental designs and remove many of the unexpected outcomes that arise during the assembly of genetic components into larger systems. The BioBrick standard (BBF RFC 10) was one of the earliest proposed DNA assembly standards. It uses the prefix sequence (containing EcoRI and XbaI cutting sites) and the suffix sequence (containing SpeI and PstI cutting sites)2,3. Because XbaI and SpeI have complementary cohesive ends, BioBrick DNA parts that are cut with XbaI and SpeI can be joined together, generating a new BioBrick for further iterative assembly.
Some defects have been identified with the use of the BioBrick standard4. For example, it produces an 8-bp scar between the DNA parts, which does not allow for the construction of in-fusion proteins. Besides, the four abovementioned types of 6-bp restriction sites must be removed from the DNA parts, which is very inconvenient. The BglBrick standard was established to solve the first problem5. It creates a 6-bp "GGATCT" scar, producing Gly-Ser and allowing for the fusion of multiple proteins or protein domains. iBrick was developed to deal with the second problem6. It uses homing endonucleases (HEs) that recognize long DNA sequences. As the HE recognition sites rarely exist in natural DNA sequences, the iBrick standard can be used for the direct construction of iBrick parts without modifying their DNA sequences. However, the iBrick standard leaves a 21 bp scar between the DNA parts, which might be the reason for its unpopularity.
In recent years, the clustered regularly interspaced short palindromic repeats (CRISPR) system has developed rapidly7,8. Among the CRISPR-associated (Cas) proteins, Cas9 endonuclease from Streptococcus pyogenes is now widely used. It mostly introduces double-stranded DNA breaks (DSBs) with blunt ends9.
In 2015, Zhang and coworkers characterized Cpf1 (CRISPR from Prevotella and Francisella 1) for the first time. It belongs to the class 2 type V CRISPR-Cas system and is a CRISRP RNA (crRNA)-guided endonuclease10. Unlike Cas9, Cpf1 introduces a DSB with a 4 or 5 nt 5' overhang10. Based on this characteristic, Cpf1 was used to develop a DNA assembly standard, C-Brick4. On a C-Brick standard vector, four Cpf1 target sites of prefixed T1/T2 and suffixed T3/T4 flank the biological parts; this is similar to the BioBrick standard. As the cleavage of T2 and T3 sites produces complementary sticky ends, it is possible to perform the iterative assembly of DNA parts while generating a "GGATCC" scar between the parts. Notably, the C-Brick standard has two main advantages: recognizing long target sequences and leaving short scars. The 6 bp "GGATCC" scar generated by C-Brick encodes Gly-Ser, which allows for the construction of fusion proteins. Moreover, the C-Brick standard is also partially compatible with the BglBrick and BioBrick standards.
Protocol
1. Preparation of crRNA
- Preparation of crRNA templates
- Re-suspend individual oligonucleotides (Table 1) in RNase-free water to a concentration of 10 µM.
- Add 22.5 µL of top-strand oligonucleotide (T7-F in Table 1), 22.5 µL of bottom-strand oligonucleotide (Table 1), and 5 µL of 10x annealing buffer to a 0.2 mL PCR tube. Ensure that the total volume is 50 µL.
NOTE: Six different bottom-strand oligonucleotides are shown in Table 1, each of which should be individually paired with the top-strand T7-F. The lowercase letters in Table 1 represent the sequence to be transcribed into the guide sequence of crRNA. 10x Taq PCR buffer can be used instead of the 10x annealing buffer. - Place the PCR tube into a thermocycler.
- Run the annealing program: initial denaturation at 95 °C for 5 min and then a cooldown from 95-20 °C, with a 1 °C decrease per min on the thermocycler.
NOTE: Immediately use the samples for step 1.2.1.
- Transcription and purification of crRNA
- Add 38 µL of RNase-free water, 8 µL of the template from step 1.1.4, 20 µL of 5x T7 transcription buffer, 20 µL of NTP mixture (10 µM), 4 µL of recombinant RNase inhibitor (RRI), and 10 µL of T7 RNA polymerase to a 1.5 mL microcentrifuge tube. Ensure that the total volume is 100 µL.
- Put the microcentrifuge tube in a 37 °C water bath overnight (for about 16 h).
- Use an RNA cleanup and concentration kit to purify the transcribed RNA; follow the manufacturer's protocol.
- Quantitate the RNAs with UV-Vis spectrophotometers and dilute the RNA to a concentration of 10 µM. Use the samples immediately or store them at -80 °C.
NOTE: The crRNA sequences are listed in Table 2.
2. Construction of C-Brick Parts ( i.e., the Insertion of Biological Parts into a C-Brick Standard Vector)
NOTE: This step has three sub-steps. For short biological parts (e.g., promoters and terminators), it is best to use the direct PCR method to insert the biological parts into a C-Brick standard vector4. The whole vector sequence is shown in the supplementary data, and the prefix and suffix sequence is shown in Figure 1 (step 2.1, below). For biological parts that can be easily obtained via PCR (e.g., with templates of genomic DNA, plasmids, or de novo synthesized DNA sequences), it is best to use the seamless assembly method to insert the biological parts into a C-Brick standard vector (step 2.2, below). For those parts obtained from BioBrick and BglBrick standards, restriction enzyme-mediated digestion and T4 DNA ligase-mediated ligation can be used to insert the biological parts into a C-Brick standard vector (step 2.3, below).
- Construction via direct PCR amplification
- Design and order oligonucleotides for PCR amplification.
NOTE: The 5'-end of the oligonucleotide contains the sequence of the DNA part, and 3'-end of the oligonucleotide contains the vector sequence (i.e., "GGATCCACTAGTCTCTAGCTCG" at the 3'-end of the forward strand and "GGATCCTTTCTCCTCTTTCTAG" in the reverse strand). Either no overhang or a ~20-bp overhang can be designed at the 5'-end of the oligonucleotide. Specific examples can be found in a previous study4. - Re-suspend individual oligonucleotides in ultra-pure water to a concentration of 10 µM.
- Set up PCR reactions on ice in a 0.2 mL PCR tube: add 18 µL of ultra-pure water, 25 µL of 2x PCR buffer, 1 µL of dNTPs (10 µM), 2.5 µL each of the forward and reverse primers (10 µM) from step 2.1.2, 0.5 µL of C-Brick standard vector (50 ng) as a template, and 0.5 µL of high-fidelity DNA polymerase. Ensure that the total volume is 50 µL.
- Place the tube directly into a thermocycler and start the thermocycler program. Use the samples immediately or freeze and store them at -20 °C.
- Thermocycler program: one cycle for 3 min at 98 °C; thirty cycles for 10 s at 98 °C, 20 s at 55 °C, and 2 min at 72 °C; and one cycle for 10 min at 72 °C. Keep the sample at 16 °C.
- Add 9 µL of the mixture from step 2.1.4 to a 1.5 mL microcentrifuge tube.
- Add 1 µL of DpnI and incubate for 1 h in a 37 °C water bath.
NOTE: If the primers have no overlapping sequences, add 0.5 µL of T4 polynucleotide kinase (PNK) and 0.5 µL of T4 DNA ligase and incubate in a 22 °C water bath for 1 h. Otherwise, if the primers contain a ~20 nt overlapping sequence, directly transform the DpnI-treated products into E. coli (DH10B)-competent cells (step 2.4). The linearized PCR product will be circularized by the recombination system in the host. - Use the samples immediately or store them at -20 °C.
- Design and order oligonucleotides for PCR amplification.
- Construction via seamless assembly
- Design and order oligonucleotides for PCR amplification.
NOTE: The 3'-end of the oligonucleotide contains the terminal sequence of the DNA part, and the 5'-end of the oligonucleotide contains the terminal of the linearized C-Brick standard vector sequence (i.e., "CTAGAAAGAGGAGAAAGGATCC" for the 5'-end of the forward strand and "CGAGCTAGAGACTAGTGGATCC" for the 5'-end of the reverse strand). Specific examples can be found in a previous study4. - Re-suspend the individual oligonucleotides in ultra-pure water to a concentration of 10 µM.
- Set up PCR reactions on ice in a 0.2 mL PCR tube: add 18 µL of ultra-pure water, 25 µL of 2x PCR buffer, 1 µL of dNTPs (10 µM), 2.5 µL each of the forward and reverse primers (10 µM) from step 2.2.2, 0.5 µL of template (20-500 ng), and 0.5 µL of high-fidelity DNA polymerase. Ensure that the total volume is 50 µL.
- Place the tube directly into the thermocycler and run the thermocycler program. Use the samples immediately or freeze and store them at -20 °C.
NOTE: Set the thermocycler program to: one cycle for 3 min at 98 °C; 30 cycles for 10 s at 98 °C, 20 s at 55 °C (according to the annealing temperature of the primer), and 1 min (according to the length of the biological parts) at 72 °C; and one cycle for 10 min at 72 °C. Keep the sample at 16 °C. - Purify the PCR product using a PCR cleanup system kit; follow the manufacturer's protocol.
- Digest the C-Brick standard vector with BamHI: add 34 µL of ultra-pure water, 10 µL of plasmid (1 µg), 5 µL of 10x buffer, and 1 µL of BamHI to the microcentrifuge tube. Incubate for up to 2 h at 37 °C in a water bath.
- Purify the above product using a gel cleanup system kit; follow the manufacturer's protocol.
- Add 2 µL of linearized vector (50 ng) from step 2.2.7, 5 µL of fragment (200 ng) from step 2.2.4, 2 µL of 5x seamless assembly buffer, and 1 µL of seamless assembly enzyme from a seamless assembly kit to a 1.5-mL microcentrifuge tube.Ensure that the total volume is 10 µL.
- Place this in a 37 °C water bath for 30 min. Use the samples immediately or freeze and store them at -20 °C.
- Design and order oligonucleotides for PCR amplification.
- Construction via restriction enzyme-mediated digestion and T4 DNA ligase-mediated ligation.
NOTE: For those parts obtained from the BioBrick (step 2.3.4-2.3.7) and BglBrick (step 2.3.1-2.3.3) standards, restriction enzyme-mediated digestion and T4 DNA ligase-mediated ligation can be used to insert the biological parts into a C-Brick standard vector.- Digest the BglBrick standard parts with BamHI and BglII: add 34 µL of ultra-pure water, 10 µL of plasmid (2 µg), 5 µL of 10x buffer 3, 0.5 µL of BamHI, and 0.5 µL of BglII to the microcentrifuge tube. Incubate for 2 h in a 37 °C water bath.
- Run a 1% agarose gel electrophoresis and purify the fragment using a gel cleanup system kit.
- Ligate the fragment and the C-Brick standard vector: add 2 µL of linearized vector (50 ng) from step 2.1.12 and 6 µL of fragment (200 ng) from step 2.3.3 to a 1.5 mL microcentrifuge tube. Add 1 µL of 10 x T4 DNA ligase buffer and 1 µL of T4 DNA ligase. Incubate for 2 h at 22 °C. Use the samples immediately or freeze and store them at -20 °C.
- Digest the BioBrick standard parts with XbaI and SpeI: add 34 µL of ultra-pure water, 10 µL of plasmid (2 µg), 5 µL of 10x buffer, 0.5 µL of XbaI, and 0.5 µL of SpeI to the microcentrifuge tube. Incubate for 2 h in a 37 °C water bath.
- Digest the C-Brick standard vector with XbaI and SpeI: add 34 µL of ultra-pure water, 10 µL of plasmid (1 µg), 5 µL of 10x buffer, 0.5 µL of XbaI, and 0.5 µL of SpeI to the microcentrifuge tube. Incubate for 2 h in a 37 °C water bath.
- Run a 1% agarose gel electrophoresis and purify the fragment from step 2.3.4 and the linearized vector from step 2.3.5 with a gel cleanup system kit; following the manufacturer's protocol.
- Ligate the fragment and the C-Brick vector: add 2 µL of linearized vector (50 ng) from step 2.3.5 and 6 µL of fragment (200 ng) from step 2.3.4 to a 1.5 mL microcentrifuge tube. Add 1 µL of 10x T4 DNA ligase buffer and 1 µL of T4 DNA ligase. Incubate for 2 h at 22 °C. Use the samples immediately or freeze and store them at -20 °C.
- Add 10 µL of the products from step 2.1.8, 2.2.9, 2.3.3, or 2.3.7 for chemical transformation into the competent cells of E. coli (DH10B).
- Pick-up several clones to a 5-mL liquid Luria-Bertani (LB) tube and incubate at 37 °C on a shaker (220 rpm) overnight.
- Extract the plasmid using a plasmid preparation kit; follow the manufacturer's protocol.
- Identify the correct clones by Sanger sequencing11.
3. C-Brick Assembly (Figure 2)
- Digestion of the C-Brick vector with Cpf1
- Add 22.5 µL of RNase-free water, 4 µL of 10x Cpf1 buffer, 10 µL of the plasmid from step 2.6 (1 µg), 0.5 µL of RRI, and 1 µL of Cpf1 (5 µM) to a 1.5-mL microcentrifuge tube.
NOTE: Cpf1 can be purchased commercially or purified following the protocol in a previous study4. - To insert a foreign DNA fragment into the T1 and T2 sites, add 1 µL of crRNA-T2 (10 µM) and incubate in a water bath at 37 °C for 30 min. Add 1 µL of crRNA-T1 (10 µM) and Cpf1 (5 µM) for another 30 min.
NOTE: Alternatively, to insert a foreign DNA fragment into the T3 and T4 sites, add 1 µL of crRNA-T3 (10 µM) and incubate in a water bath at 37 °C for 30 min. Add 1 µL of crRNA-T4 (10 µM) for another 30 min. - Add 1 µL of thermosensitive alkaline phosphatase and incubate the tube in a 37 °C water bath for another 1 h.
- Run an agarose gel electrophoresis and purify the vector using a gel cleanup system kit. Use the samples immediately or freeze and store them at -20 °C.
- Add 22.5 µL of RNase-free water, 4 µL of 10x Cpf1 buffer, 10 µL of the plasmid from step 2.6 (1 µg), 0.5 µL of RRI, and 1 µL of Cpf1 (5 µM) to a 1.5-mL microcentrifuge tube.
- Digestion of the DNA fragment with Cpf1
- Add 21.5 µL of RNase-free water, 4 µL of 10x Cpf1 buffer, 10 µL of plasmid from step 2.6 (2 µg), 0.5 µL of RRI, and 2 µL of Cpf1 (5 µM) to a 1.5-mL microcentrifuge tube.
- To insert a foreign DNA fragment into the T1 and T2 sites, add 1 µL of crRNA-T1 (10 µM) and 1 µL of crRNA-T3 (10 µM) and incubate in a water bath at 37 °C for 2 h.
NOTE: Alternatively, add 1 µL of crRNA-T2 (10 µM) and 1 µL of crRNA-T4 (10 µM) and incubate in a water bath at 37 °C for 2 h. - Run an agarose gel electrophoresis and purify the fragment using a gel cleanup system kit. Use the samples immediately or freeze and store them at -20 °C.
- C-Brick assembly and further verification
- Ligate the foreign DNA fragment and C-Brick vector: add 2 µL of the linearized vector (50 ng) from step 3.1.4 and 6 µL of the DNA fragment (200 ng) from step 3.2.3 to a 1.5 mL microcentrifuge tube. Add 1 µL of 10x T4 DNA ligase buffer and 1 µL of T4 DNA ligase. Incubate for 2 h at 22 °C. Use the samples immediately or freeze and store them at -20 °C.
- Add 10 µL of the ligation products (step 3.3.1) for chemical transformation into E. coli (DH10B)-competent cells.
- Add several clones to a 5 mL liquid LB tube and incubate overnight on a shaker at 220 rpm and 37 °C.
- Extract the plasmid using a plasmid preparation kit; follow the manufacturer's protocol.
- Identify correct clones by Sanger sequencing11.
- Perform a new round of C-Brick assembly. Correct clones from step 3.3.4 can be used as a new C-Brick vector or DNA part for further iterative C-Brick assembly, following the same protocol from steps 3.1-3.3.
NOTE: T2' and T3' sites are designed as the backup of T2 and T3 sites (Figure 1a). For plasmids obtained from step 2.3.7, only T2' and T3' can be used for the C-Brick standard assembly.
Representative Results
This protocol demonstrated the assembly of three chromoprotein cassettes (cjBlue (BBa_K592011), eforRed (BBa_K592012), and amilGFP (BBa_K592010)). First, the coding sequences of the three abovementioned genes and terminators were individually cloned into a C-Brick standard vector. Short DNA parts, promoter and terminator, were introduced into the C-Brick vector through PCR amplification using primers containing the short DNA parts on the 5' terminal. This was followed by the self-ligation method. Three chromoprotein-encoding sequences were first de novo synthesized and then cloned into the C-Brick standard vector through seamless assembly. The primers were described in a previous study4.
After that, cjBlue, eforRed, and amilGFP sequences were ligated with terminator and promoter sequences in the same procedure shown in Figure 2. Correct constructs were transformed into E. coli, demonstrating beautiful colors (Figure 3a). Subsequently, two color-expression cassettes were further assembled with the C-Brick standard to create more colors (i.e., the red color from amilGFP plus eforRed, the green color from amilGFP plus cjBlue, and the light purple color from eforRed plus cjBlue; Figure 3b).
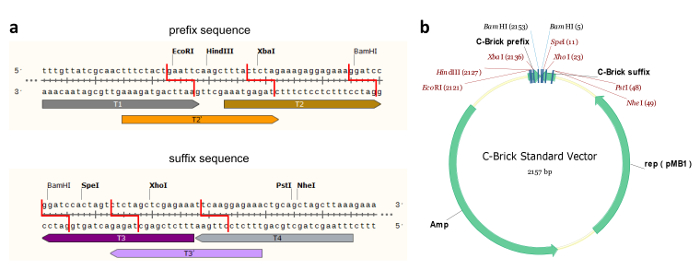
Figure 1: C-Brick Standard Interface DNA Sequence and C-Brick Standard Vector. (a) The sequence of the C-brick standard interface-T1, T2, T3, T4, T2', T3'-can be recognized by its corresponding crRNA and then introduced into a 5' overhang using Cpf1. The cleavage of T2 and T3 (or T2' and T3') sites produces complementary overhangs. With the eight restriction sites designed, the C-Brick standard is partially compatible with BioBrick and BglBrick. (b) Plasmid map for the C-Brick standard vector with restriction digestion sites. Please click here to view a larger version of this figure.
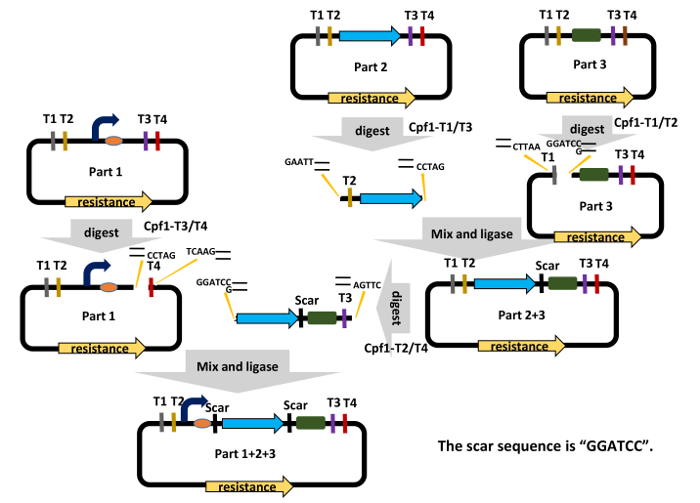
Figure 2: The Workflow for DNA Assembly in the C-Brick Standard. Cpf1-digested T2 and T3 target sites produce complementary cohesive ends of "GGATC" and "GATCC," respectively, which can be ligated to generate a short "GGATCC" scar. Detailed procedures can be found in step 3 in the text ("C-Brick assembly"). Please click here to view a larger version of this figure.
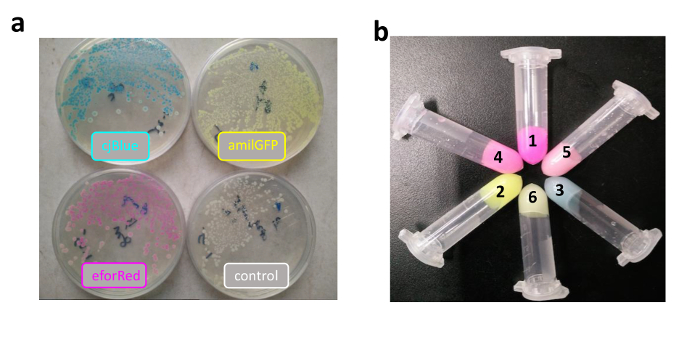
Figure 3: Colorful Bacterial Pigments Produced by E. coli Harboring Constructs Assembled in the C-Brick Standard. (a) Three chromoproteins were expressed in E. coli grown on a plate. Bacteria with no chromoproteins (negative control) are shown on the 4th plate. (b) Three chromoproteins and three assembled dual chromoproteins were expressed in E. coli grown in liquid LB medium. From 1 to 6, the constructs expressed eforRed (1), amilGFP (2), cjBlue (3), amilGFP plus eforRed (4), amilGFP plus cjBlue (5), and eforRed plus cjBlue (6). The colorful bacteria on a single plate or in a single microcentrifuge tube were cultured from a single sequence-verified clone. Please click here to view a larger version of this figure.
| name | sequence | ||
| T7-F | GAAATTAATACGACTCACTATAGGG | ||
| T7-T1-R | gaattcagtagaaagttgcgataaATCTACAACAGTAGAAATTCCCTATAGTGAGTCGTATTAATTTC | ||
| T7-T2'-R | ctagagtaaagcttgaattcagtaATCTACAACAGTAGAAATTCCCTATAGTGAGTCGTATTAATTTC | ||
| T7-T2-R | ggatcctttctcctctttctagagATCTACAACAGTAGAAATTCCCTATAGTGAGTCGTATTAATTTC | ||
| T7-T3-R | ggatccactagtctctagctcgaATCTACAACAGTAGAAATTCCCTATAGTGAGTCGTATTAATTTC | ||
| T7-T3'-R | ctctagctcgagaaattcaaggaATCTACAACAGTAGAAATTCCCTATAGTGAGTCGTATTAATTTC | ||
| T7-T4-R | ttcaagggaaactgcagctagctATCTACAACAGTAGAAATTCCCTATAGTGAGTCGTATTAATTTC | ||
Table 1: Oligos for the Preparation of the crRNA Transcription Templates used in this Study. T7-F was the top-strand oligonucleotide, and the others were the bottom-strand oligonucleotides. The lowercase bases in the sequence will be transcribed into the guide sequence in crRNA.
| names | RNA sequences (5'-3') | ||
| crRNA-T1 | GGGAAUUUCUACUGUUGUAGAUUUAUCGCAACUUUCUACUGAAUUC | ||
| crRNA-T2 | GGGAAUUUCUACUGUUGUAGAUCUCUAGAAAGAGGAGAAAGGAUCC | ||
| crRNA-T3 | GGGAAUUUCUACUGUUGUAGAUUCGAGCUAGAGACUAGUGGAUCC | ||
| crRNA-T4 | GGGAAUUUCUACUGUUGUAGAUAGCUAGCUGCAGUUUCUCCUUGAA | ||
| crRNA-T2' | GGGAAUUUCUACUGUUGUAGAUUACUGAAUUCAAGCUUUACUCUAG | ||
| crRNA-T3' | GGGAAUUUCUACUGUUGUAGAUUCCUUGAAUUUCUCGAGCUAGAG | ||
Table 2: The crRNA Sequences used in this Study. The crRNAs were produced through the in vitro transcription in step 1 in the text ("Preparation of crRNA").
Discussion
This protocol describes a procedure for the DNA assembly standard C-Brick. The most important step in this protocol is the linearization of the C-Brick standard vector; incomplete cleavage of the vector could seriously affect the success rate. Additionally, although Cpf1 mainly cleaves target DNA sequences in the "18-23" cleavage pattern, inaccurate cleavage near the two bases was also detected4, which can cause a small number of mutations after DNA assembly. Therefore, Sanger sequencing is needed to verify the assembled construct.
The efficiency of the C-Brick assembly is lower than traditional restriction enzyme and ligation methods that have been described in a previous study4. The major reason affecting the assembly efficiency may be the inaccurate cleavage characteristics of Cpf1. In the future, improvements to the cleavage accuracy can be extremely useful for the standard and are currently being explored.
C-Brick is also partially compatible with the BglBrick and BioBrick standards. For example, BglBrick parts can be cut with BglII and BamHI and then inserted into the BamHI site of a C-Brick standard vector, generating a C-Brick part. However, the insertional direction of the BglBrick part should be verified, and there would be a "GGATCT" scar after assembly. When a C-Brick DNA part is obtained from a BioBrick part (i.e., with XbaI and SpeI digestion), both T2 and T3 sites will be destroyed, and two backup Cpf1 target sequences (T2' and T3') can be used.
As the Cpf1 cleavage requires sgRNA, which is extremely sensitive to RNase, all materials should be RNase-free. Currently, all C-Brick standard vectors, RNase-free Cpf1 nucleases, and crRNAs can be ordered commercially12. Therefore, the procedures for C-Brick are actually as simple as those of the BioBrick standard.
In the past few years, several useful DNA assembly methods have been developed and are widely used, including Golden Gate Assembly13, Gibson Assembly14, and others15. However, as DNA assembly standards have the potential to provide cost-effective, reliable, high-throughput, and automatic assembly reactions5, standards may facilitate the construction and testing of newly designed or re-designed genes, genetic circuits, and pathways15,16,17, especially in the field of synthetic biology. Among the currently published DNA assembly standards, the C-Brick standard has both obvious advantages and disadvantages4 (i.e., the ligation efficiency). If the ligation efficiency is improved, the standard can be widely adopted in the future.
Disclosures
The authors have nothing to disclose.
Acknowledgements
We thank Shanghai Tolo Biotech for their technical assistance during the development of the C-Brick standard. This work was supported by grants from the Strategic Priority Research Program of the Chinese Academy of Sciences (Grant No. XDB19040200).
Materials
| Comercial Oligonucleotide | Sangon Biotech | ||
| 10x Taq PCR Buffer | Transgen | #J40928 | |
| Ultra Pure Distilled Water | Invitrogen | 10977-015 | |
| 5x RNA Transcription Buffer | Thermo Scientific | K0441 | |
| T7 RNA polymerase | Thermo Scientific | #EP0111 | |
| NTP mixture | Sangon | #ND0056 | |
| RRI(Recombinant RNase Inhibitor) | Takara | 2313A | |
| RNA Clean & Concentrator-5 | Zymo Research | R1015 | |
| UV-Vis Spectrometer | Thermo Scientific | Nano-Drop 2000c | |
| 2x Phanta Max Buffer | Vazyme | PB505 | PCR buffer |
| dNTPs | Transgen | AD101 | |
| Phanta Max Super-Fidelity DNA Polymerase | Vazyme | P505-d1 | |
| Ezmax for One-step Cloning | Tolobio | 24303-1 | seamless assembly kit |
| 5x Buffer for Ezmax One-step Cloning | Tolobio | 32006 | |
| BamHI | NEB | #R0136L | |
| BamHI-HF | NEB | #R3136L | |
| BglII | NEB | #R0144L | |
| XbaI | NEB | #R0145L | |
| SpeI | NEB | #R3133L | |
| 10x Buffer 3 | NEB | #B7003S | |
| 10x CutSmart Buffer | NEB | B7204S | |
| 10x T4 DNA ligase Buffer | Tolobio | 32002 | |
| T4 PNK | Tolobio | 32206 | |
| T4 DNA ligase | Tolobio | 32210 | |
| DpnI | NEB | #R01762 | |
| SV Gel and PCR clean-up system | Promega | A9282 | |
| Plasmid Mini Kit I | Omega | D6943-02 | plasmid preparation kit |
| thermosensitive alkaline phosphatase | Thermo Scientific | #EF0651 | FastAP |
| 10x Cpf1 buffer | Tolobio | 32008 | |
| Cpf1 | Tolobio | 32105 | FnCpf1 |
| thermocycler | Applied Biosystems | veriti 96 well | |
| C-Brick standard vector | Tolobio | 98101 | |
| E. coli [DH10B] | Invitrogen | 18297010 | |
| Luria-Bertani media (tryptone) | Oxoid | LP0042 | |
| Luria-Bertani media (yeast extract) | Oxoid | LP0021 | |
| Luria-Bertani media (NaCl) | Sangon Biotech | B126BA0007 |
References
- Canton, B., Labno, A., Endy, D. Refinement and standardization of synthetic biological parts and devices. Nat Biotechnol. 26 (7), 787-793 (2008).
- Shetty, R. P., Endy, D., Knight, T. F. Engineering BioBrick vectors from BioBrick parts. J Biol Eng. 2, (2008).
- Knight, T. . Idempotent Vector Design for Standard Assembly of Biobricks. , 1-11 (2003).
- Li, S. Y., Zhao, G. P., Wang, J. C-Brick: A New Standard for Assembly of Biological Parts Using Cpf1. ACS Synth Biol. 5 (12), 1383-1388 (2016).
- Anderson, J. C., et al. BglBricks: A flexible standard for biological part assembly. J Biol Eng. 4 (1), (2010).
- Liu, J. K., Chen, W. H., Ren, S. X., Zhao, G. P., Wang, J. iBrick: a new standard for iterative assembly of biological parts with homing endonucleases. PLoS One. 9 (10), e110852 (2014).
- Hsu, P. D., Lander, E. S., Zhang, F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 157 (6), 1262-1278 (2014).
- Wright, A. V., Nunez, J. K., Doudna, J. A. Biology and Applications of CRISPR Systems: Harnessing Nature’s Toolbox for Genome Engineering. Cell. 164 (1-2), 29-44 (2016).
- Jinek, M., et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 337 (6096), 816-821 (2012).
- Zetsche, B., et al. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell. 163 (3), 759-771 (2015).
- Sanger, F., Nicklen, S., Coulson, A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 74 (12), 5463-5467 (1977).
- Engler, C., Kandzia, R., Marillonnet, S. A one pot, one step, precision cloning method. PLoS One. 3 (11), e3647 (2008).
- Gibson, D. G., et al. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods. 6 (5), 343-345 (2009).
- Casini, A., Storch, M., Baldwin, G. S., Ellis, T. Bricks and blueprints: methods and standards for DNA assembly. Nat Rev Mol Cell Biol. 16 (9), 568-576 (2015).
- Cameron, D. E., Bashor, C. J., Collins, J. J. A brief history of synthetic biology. Nat Rev Microbiol. 12 (5), 381-390 (2014).
- Keasling, J. D. Synthetic biology for synthetic chemistry. ACS Chem Biol. 3 (1), 64-76 (2008).

