Rapid Isolation of Single Cells from Mouse and Human Teeth
Summary
The current protocol presents a fast, efficient, and gentle method for isolating single cells suitable for single-cell RNA-seq analysis from a continuously growing mouse incisor, mouse molar, and human teeth.
Abstract
Mouse and human teeth represent challenging organs for quick and efficient cell isolation for single-cell transcriptomic or other applications. The dental pulp tissue, rich in the extracellular matrix, requires a long and tedious dissociation process that is typically beyond the reasonable time for single-cell transcriptomics. For avoiding artificial changes in gene expression, the time elapsed from euthanizing an animal until the analysis of single cells needs to be minimized. This work presents a fast protocol enabling to obtain single-cell suspension from mouse and human teeth in an excellent quality suitable for scRNA-seq (single-cell RNA-sequencing). This protocol is based on accelerated tissue isolation steps, enzymatic digestion, and subsequent preparation of final single-cell suspension. This enables fast and gentle processing of tissues and allows using more animal or human samples for obtaining cell suspensions with high viability and minimal transcriptional changes. It is anticipated that this protocol might guide researchers interested in performing the scRNA-seq not only on the mouse or human teeth but also on other extracellular matrix-rich tissues, including cartilage, dense connective tissue, and dermis.
Introduction
Single-cell RNA sequencing is a powerful tool for deciphering in vivo cell population structure, hierarchy, interactions, and homeostasis1,2. However, its results strongly depend on the first step of this advanced analysis – the preparation of a single-cell suspension of perfect quality out of the complex, well-organized tissue. This encompasses keeping cells alive and preventing unwanted, artificial changes in gene expression profiles of the cells3,4. Such changes might lead to the inaccurate characterization of population structure and misinterpretation of the collected data.
Specific protocols for the isolation out of a wide range of tissues have been developed5,6,7,8. They usually employ mechanical dissociation in combination with further incubation with various proteolytic enzymes. These typically include trypsin, collagenases, dispases, papain6,7,8,9, or commercially available enzyme mixtures such as Accutase, Tryple, etc.5. The most critical part affecting the transcriptome quality is enzymatic digestion. It was shown that prolonged incubation with enzymes at 37 °C influences the gene expression and causes the upregulation of many stress-related genes10,11,12,13. The other critical parameter of the isolation process is its overall length, as it has been shown that cell transcriptomes change after the tissue ischemia14. This protocol presents an efficient protocol for gentle isolation of single cells from mouse and human teeth, faster than other, previously utilized protocols for isolation of cells from complex tissues5,6,9,11,13,15,16.
This protocol presents how to quickly dissect soft tissue from the hard tooth and prepare a single-cell suspension suitable for scRNA-seq. This method employs only one centrifugation step and minimizes the effect of unwanted transcriptional changes by reducing the tissue handling and digestion time and keeping the tissue and cells at 4 °C most of the time. The procedure showcases the isolation of cells from mouse incisors, molar, and human wisdom teeth as an example, but principally should work for other teeth in various organisms. The complete protocol is schematically visualized in Figure 1. This protocol has been recently used to generate a dental cell type atlas obtained from mouse and human teeth1.
Protocol
All animal experiments were performed according to the International and local regulations and approved by the Ministry of Education, youth and sports, Czech Republic (MSMT-8360/2019-2; MSMT-9231/2020-2; MSMT-272/2020-3). This protocol was tested with both male and female wildtype C57BL/6 and CD-1 mice and with genetically modified Sox10::iCreERT2 mice17 (combined with various reporter systems) on a C57BL/6 background. Experiments with human samples were performed with the approval of the Committees for Ethics of the Medical Faculty, Masaryk University Brno & St. Anne´s Faculty Hospital in Brno, Czech republic.
1. Experimental set-up and preparation of solutions
- Instrument set-up
- Cool down the centrifuge to 4 °C.
- Heat the incubation chamber to 37 °C.
- Start the Fluorescence-activated cell sorting (FACS) machine and set all the temperatures of the sorter (including collection tube holder) to 4 °C. Perform the instrument quality control, set the drop delay.
- Set the preliminary gating strategy and perform the test sorting.
NOTE: When using FACS, use the 100 µm nozzle.
- Prepare the solutions (steps 1.2.1-1.2.3).
- Wash solution: Prepare fresh 2% FBS (Fetal Bovine Serum) in HBSS (Hanks' Balanced Salt Solution).
- Digestion mixture: Prepare fresh collagenase P (3 U/mL) fully dissolved in HBSS.
- Optional: Prepare fresh HBSS + BSA (0.04%) and chill analytical grade methanol in a -20 °C freezer for storing single-cell suspension at -80 °C.
NOTE: Step 1 needs to be performed before the start of the experiment. The composition of the utilized solutions is summarized in Supplementary Table 1.
2. Preparation of experimental animal/s and human tooth
- Prepare the experimental animals (mouse).
- Euthanize the mouse according to the local regulations; e.g. by anaesthetics overdose as described previously1.
CAUTION: Regulations for humane euthanizing of experimental animals varies locally. Always follow valid local regulations. - Immediately proceed to the tissue dissection step (step 3).
NOTE: If the tissue from the experimental animals cannot be dissected immediately (e.g., because of transfer from animal housing facility), place the experimental animals on ice and perform tissue dissection as soon as possible. To obtain more cells from mouse molar pulps, use younger (6 weeks and less) animals. With increasing age, the size of dental pulp decreases. Mouse incisors mostly keep their structure with increasing age so animals of various ages can be used for tissue dissection.
- Euthanize the mouse according to the local regulations; e.g. by anaesthetics overdose as described previously1.
- Prepare the human tooth
NOTE: Human teeth were extracted for a clinically relevant reason. Every diagnosis was treated individually, and an experienced dental surgeon always performed the tooth extraction.- Put the freshly extracted tooth immediately into a 50 mL tube with ice-cold HBSS and keep the tube on ice until further processing.
NOTE: Using retained wisdom teeth from patients until age 25-30 is recommended for the highest cell yield.
- Put the freshly extracted tooth immediately into a 50 mL tube with ice-cold HBSS and keep the tube on ice until further processing.
3. Tissue dissection
- Hold the experimental animal behind its head, looking on the ventral aspect of its head so that the tail points away.
- Using small, sharp scissors, quickly remove the skin from the mandible to expose the mandibular arch, the soft tissue between each half of the mandibles, and the adjacent facial muscles.
- Make a deep cut from each side of the mandible; firstly, through m. masseter along the buccal side of the mandible up to the temporomandibular joint, and then along the inner part of each half of the mandible through the base of the oral cavity (see Supplementary Figure 1).
- Cut all the muscles and ligaments along the mandible up to the temporomandibular joint from both outside and inside of the oral cavity.
NOTE: Avoid cutting bones. This might damage the most apical part of the incisor. - Grasp the mandible using bent tip tweezers and remove it. Then, split the dissected mandible into two halves with scissors by cutting through mandibular symphysis (see Supplementary Figure 1).
- Use an industrial low lint wipe to chafe the remaining soft tissue from each half of the mandible. After both parts of the mandible are cleaned, place them into a pre-prepared Petri dish with ice-cold HBSS.
NOTE: From this point forward, work on ice. Further dissection of mouse incisors and molars is performed under a stereomicroscope with a black background. - Mouse mandibular incisors
- For the dissection of mandibular incisors, remove the alveolar ridge with all three molars and transversally crack the mandibular arch in the place corresponding to the position between the first and second molar.
NOTE: A sharp scalpel blade no. 11 and tweezers are used to perform this step (see Table of Materials). - Carefully pull the incisor out of the rest of the dental socket.
NOTE: If successful, the extracted incisor will contain intact apical parts, including epithelial tissue with complete cervical loops. - If needed, remove the remaining fragments of bone still attached to the incisor with tweezers and a scalpel.
- Place the dissected incisors into fresh, ice-cold HBSS and dissect the tissue of interest: cervical loop, dental pulp, or other parts of the tooth.
- Place the dissected soft tissue into a droplet of fresh, ice-cold HBSS in the middle of a 10 cm Petri dish. Keep on ice.
- For the dissection of mandibular incisors, remove the alveolar ridge with all three molars and transversally crack the mandibular arch in the place corresponding to the position between the first and second molar.
- Mouse mandibular molars
- For dissecting mandibular molars, completely remove the alveolar ridge from the rest of the mandible using a scalpel blade.
- Move the dissected alveolar crest into fresh, ice-cold HBSS in a Petri dish and carefully remove all the remaining fragments of alveolar bones attached to the roots.
- Place the dissected molars without alveolar bone into fresh, ice-cold HBSS. To expose the pulp, start to remove parts of the dentin from the apical side using fine, sharp tip tweezers until you reach the pulp cavity.
- Once the pulp cavity is reached, carefully dissect dental pulp using a pair of sharp tip tweezers and place the soft tissue of the dental pulp into a droplet of fresh, ice-cold HBSS in the middle of a 10 cm Petri dish kept on ice.
NOTE: Since mouse molar pulps are extremely small, adapt magnification on the stereomicroscope accordingly.
- Human tooth
- Wash human tooth once again in ice-cold HBSS to remove the remaining blood.
- Place the tooth into three thick-walled sterile plastic bags and use a cast iron benchtop engineer's vise to crack the tooth. Use vise jaws with a flat surface to avoid penetrating the bags.
- Slowly tighten the vise until you hear the tooth cracking; remove it from the bags and place it into fresh, ice-cold HBSS in a Petri dish.
- Using two tweezers, take out the dental pulp, clean it from all the remnants of hard tissue and place it into one droplet of ice-cold HBSS in the middle of a 10 cm Petri dish kept on ice.
CAUTION: Human tissue might potentially be infectious. When working with human tissue, use protective equipment to avoid direct contact with the tissue.
4. Preparation of single-cell suspension
- Prepare a 15 mL tube with 2.5 mL of digestion mixture composed of Collagenase P (3 U/mL) fully dissolved in HBSS. Keep the solution on ice until use.
NOTE: Always use freshly prepared Collagenase P; do not freeze the aliquots. Collagenase P activity varies from batch-to-batch. Check the activity of your batch before diluting. Collagenase P is activated by calcium, whose amount is already sufficient in the lyophilized powder. Therefore, it is unnecessary to enrich the enzyme mix with calcium. Collagenase P is not inactivated by FBS but can be inactivated by chelating agents (e.g., Ethylenediaminetetraacetic – EDTA). Stopping the dissociation process is ensured by diluting the Collagenase P in Wash solution (2% FBS in HBSS). It has been shown that adding FBS increases cell viability and the final number of cells after sorting18. - Using a round-shaped scalpel blade no. 10, cut the tissue in all directions into the smallest possible pieces. Reaggregate the tissue pieces in the middle of the droplet and repeatedly cut the tissue aggregates using a round-shaped (No. 10) scalpel blade. Repeat this process several times until the material is sufficiently minced.
- Transfer the shredded tissue pieces using a 1 mL pipette tip into the prepared digestion mixture.
NOTE: In the case of a larger number of animals being processed at once, split the tissue into several tubes with Collagenase P. The maximum amount per tube are pulps obtained from approximately 10 mouse incisors. In the case of molars, where pulps are much smaller, the number of processed pulps can be increased adequately. When using human teeth, use a maximum of 2-3 pulps per tube, depending on the pulp size. - Place the tube into a 37 °C preheated incubator with a shaker. Tilt the tube inside the shaker to an angle of 60° and set the speed to 150-200 rpm to ensure constant suspension movements inside the tube.
- Vigorously triturate the suspension every 3-4 min with a 1 mL pipette tip to disintegrate all the clumps.
NOTE: With time, the clumps will become smaller and softer until they almost disappear. - In total, incubate for 15-20 min. At the end of the incubation, triturate for the last time, and then slowly add ice cold Wash solution to a final volume of 12 mL.
- Remove the remaining clumps or pieces of calcified tissue by filtering the suspension using a 50 µm cell strainer.
- Take 10-20 µL of the filtered cell suspension and count the cells during centrifugation using a cell counting chamber (Hemocytometer). Centrifuge for 5 min at 300 x g at 4 °C. Remove the supernatant using a 10 mL serological pipette.
NOTE: If the number of cells is limited that cannot be counted in diluted suspension, skip the cell counting and use all the cells for further processing. - Optional: Proceed for fixation and storage21.
- Re-suspend the pellets (up to 107 cells) in 100 uL of HBSS + BSA (0.04%).
- Add 400 µL of chilled methanol and mix slowly.
- Incubate the cell suspension for 15 min on ice.
- Store at -80 °C until processed (no longer than 1 month).
- Resuspend the pellet in the Wash solution. Aim for 700-1200 cells/µL of the Wash solution.
- Keep the tube on ice until further processing.
NOTE: If necessary, perform the fixation and storage at -80 °C. However, immediate processing of the cell suspension for scRNA-seq is recommended. Further steps will depend based on the single-cell RNA seq protocol. When the scRNA-seq protocol uses a microfluidic system (some sc-RNA-seq companies do), load the cells based on the manufacturer's guidelines either directly or after cell sorting. - For FACS, proceed to step 5.
5. Fluorescence-activated cell sorting (FACS)
- Before sorting, prepare the cell sorting instrument.
- Cool down the whole system to 4 °C, perform the instrument quality control, set the drop delay and the voltage of the deflection plates. Put collection tubes in the collection tube holder.
NOTE: To avoid additional centrifugation steps resulting in an inevitable cell loss, sort cells into a microtube (1.5 mL) with a small amount (25-50 µL) of the Wash solution.
- Cool down the whole system to 4 °C, perform the instrument quality control, set the drop delay and the voltage of the deflection plates. Put collection tubes in the collection tube holder.
- Optional: perform viability staining before FACS.
- Add Propidium iodide into cell suspension before FACS to a final concentration 0.5 µg/mL and incubate for 15 min at 4 °C.
- Load the sample into the cell sorter. Set a strict gating strategy to remove cell debris, doublets, and dead cells. Sort the cells into a prepared tube or multi-well plates. An example of a gating strategy is shown in Figure 2.
NOTE: To minimize manipulation steps and accelerate the protocol, applying a strict gating strategy is recommended (suggested in Figure 2) rather than using a viability staining. To separate the immune cells from the isolated population, CD45 antibody staining can be performed. To perform this, resuspend cell suspension in 100 µL of staining solution (PBS + 2% FBS + anti-CD45-APC conjugated antibody 1/100 dilution). Incubate for 15 min on ice protected from light and perform FACS directly.
Representative Results
Exemplary isolation of single cells was performed from two mandibular incisors from one 6-week-old C57BL/6 mouse male. Following this protocol, a single-cell suspension was prepared, and subsequently, single-cell sequencing was performed. The prepared single-cell suspension was analyzed and sorted using FACS (Figure 2). Firstly, the FSC-A (forward scatter, area) and SSC-A (side scatter, area) plotting was applied, and an appropriate gating strategy was used to select a population with expected size and granularity to filter our cell debris and cell doublets or aggregates (Figure 2A). This selected population (P1), counting 38% of all events, was further used, and FSC-A and FSC-H (forward scatter, height) parameters were applied to remove the remaining cell doublets (Figure 2B). The population without cell doublets (P2) counting 95% of P1 can be subsequently used for scRNA-seq. Alternatively, additional gating can be used to select the population of interest (e.g., expression of fluorescent proteins or live/dead staining). To check the number of live/dead cells in the final suspension, the PI (propidium iodide) staining was performed (Figure 2C). The P3 population containing PI– (living) cells was 98.4% out of the parent P2 population and 35.5% out of the total events. The total number of filtered out, dead (PI+) cells was 1887.
The final number of cells obtained without viability staining suitable for RNA-seq (P2) counted 118,199 cells from two mouse incisor pulps. This means the number of almost 60,000 living cells from one mandibular incisor.
To clarify the number of immune cells in the final single-cell suspension, two approaches were used. Firstly, the CD45 antibody staining and subsequent FACS analysis were used. As a complementary method, the total number of immune cells (CD45+) in scRNA-seq data was analysed. FACS analysis showed 14.44% of CD45+ cells (13.20% alive and 1.24% dead) (Figure 3A). Analysis of scRNA-seq data showed 10.90% of CD45-expressing cells (Figure 3B). The decrease of CD45+ cells in scRNA-seq data can be caused by additional thresholding during scRNA-seq analysis.
These representative data on the example of mouse incisor show that the given protocol in combination with strict gating strategy is efficient in obtaining a high number of cells out of a single mouse tooth without the necessity of additional use of viability staining. The ratio of immune (CD45+) cells was minor (13.2%). Moreover, it was previously shown that the immune cells are essential in maintaining tooth homeostasis, so removing them from scRNA-seq analysis during the FACS would be counterproductive in some applications.
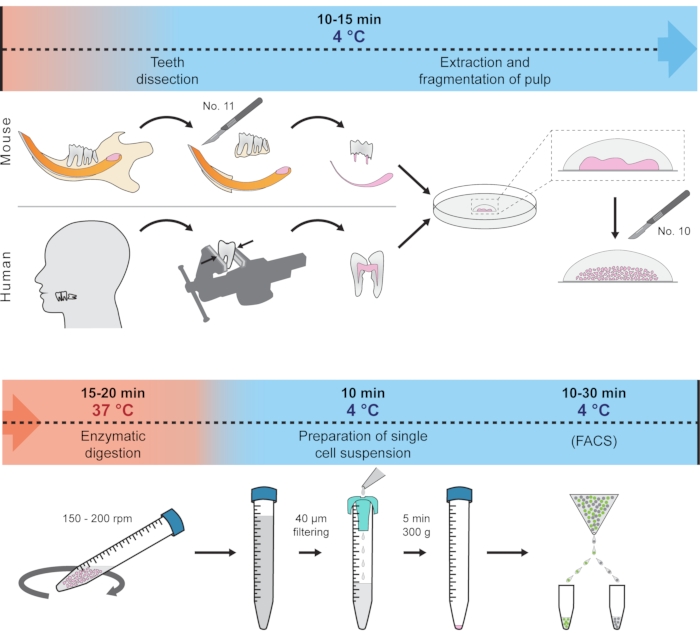
Figure 1: Schematic representation of the protocol. Different steps, including temperature conditions and expected time, are represented to prepare single-cell suspension from mouse and human teeth. Please click here to view a larger version of this figure.
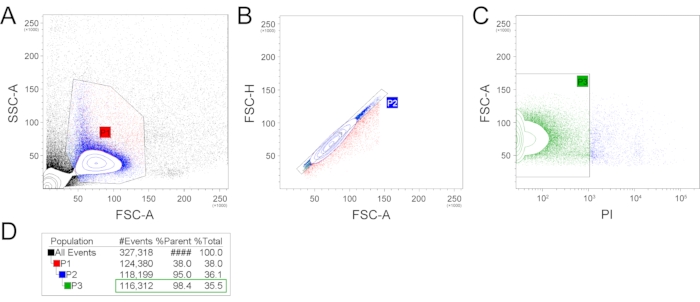
Figure 2: Example of the gating strategy. FSC-A and SSC-A gating was used to produce the P1 gate, reflecting the cell population with expected size and granularity and filtering out the cell and extracellular matrix debris and most large events (A). Subsequently, the P1 population was plotted in FSC-H and FCS-A plot, which filtered out cell doublets (B). This P2 population was then analyzed for the presence of dead cells by propidium iodide (C). The number of events/cells per gate are represented in (D). (FSC-A – forward scatter, area; SSC-A – side scatter, area; FSC-H – forward scatter, height; PI – propidium iodide). Please click here to view a larger version of this figure.

Figure 3: Quantification of the immune cells. Quantification of the immune cells was performed by FACS analysis of cells stained with anti-CD45 antibody and Live/Dead analysis using propidium iodide staining (A). Further quantification of immune cells was performed during scRNA-seq analysis (B). (CD45-APC – anti-CD45 allophycocyanin conjugated antibody; PI – propidium iodide; t-SNE – t-distributed stochastic neighbor embedding). Please click here to view a larger version of this figure.
Supplementary Figure 1: Overview of the mandible dissection process. Dashed lines illustrate suggested cuts. TMJ – temporomandibular joint, m. masseter – musculus masseter. Please click here to download this File.
Supplementary Table 1: The compositions of the solutions used in the study. Please click here to download this Table.
Discussion
Studying teeth and bones on the cellular or molecular level is generally challenging since cells forming these tissues are surrounded by different kinds of hard matrices19. One of the main goals for performing single-cell RNA-seq on dental tissue is the need to obtain cells of interest fast and without any artificial changes in their transcriptomes. To accomplish this, a highly efficient protocol suitable for isolating cells from mouse and human tooth pulps was developed, which allows for quick generation of single-cell suspensions for all transcriptomic applications. This was ensured by fast tissue isolation, minimizing the steps of tissue and cell manipulations, and streamlining the mechanical and enzymatic digestion.
The most critical steps of this protocol are fast tissue processing and adequate single-cell suspension preparation8,9. A manual approach is used to obtain dental pulps without utilizing a dental drill or other heat-generating devices. Overheating may cause an artificial expression of heat shock proteins and other genes, ultimately leading to the analyzed gene expression patterns being unrepresentative of the original tissue20. Manual tissue harvesting may be a challenging step that will likely need some training beforehand. The pulp is then cut into small pieces and enzymatically digested at 37 °C. Except for the 15-20 min of enzymatic digestion, the whole protocol is performed at 4 °C. The tissue processing and especially the enzymatic digestion were minimized to the shortest possible time since more prolonged incubation at 37 °C can cause changes in gene expression patterns10. Mechanical removing of the dentin is recommended before enzymatic digestion. Dentin and the pulp-attached predentin contain a large amount of collagen, and its excessive presence might decrease the effectiveness of the digesting solution. After being removed from the body (or death of organisms), it was shown that cells start to modify their gene expression patterns quickly12. Therefore, cell isolation and processing should be carried out as fast as possible. The current protocol reduces the processing time to 35-45 min from isolating the tissue (euthanizing animal) to preparing single-cell suspension.
One alternative modification of this technique is cell preservation for later use. This is achieved by methanol fixation. Methanol-fixed cell suspension can be stored for up to 1 month at -80 °C, as described in the protocol21. However, whenever possible, perform scRNA-seq directly, since it was shown that the single-cell data from methanol-fixed single-cell suspensions might suffer from increased expression of stress-related genes and contamination with ambient RNA22. This step might need additional modification according to the manufacturer's protocols.
Before the first application of this protocol, performing several validation steps are recommended to test the technique. From our experience, we suggest testing the aforementioned critical steps of the protocol. Additionally, we suggest testing the effectiveness of the collagenase P solution and testing the handling of the tissue dissociation step. Specifically, around the first 5 min after the initiation of collagenase P incubation, the pieces of tissue should aggregate together. This is a common situation. Aggregates are disintegrated every 3-4 min using a 1 mL pipette, and with increasing time, they should become smaller until barely visible.
Furthermore, it is recommended to perform cell counting in a cell counting chamber before centrifugation and before and after filtering to detect possible cell losses due to suboptimal supernatant removal. If the final single-cell suspension needs to be purified, FACS can be used. Cell sorting enables not only to remove debris or dead cells but importantly enables to enrich final suspension with fluorescently labeled cells13,19. To avoid shear stress or clogging of the cell sorter, a wide nozzle (85 µm or 100 µm) is used. This will further improve the viability of the sorted cells.
This technique was designed and tested on both mouse and human teeth. The major limiting factor is the small number of cells in the reduced dental pulps of the teeth of older mice (molars) and humans. Suppose a larger number of cells need to be obtained or cells from the teeth of older patients are to be acquired. One possible solution is to process a higher number of teeth and merge them into a single batch, subsequently processed as one sample.
Living cells of human dental pulp were firstly isolated more than twenty years ago using an enzyme mixture of collagenase I and dispase23. Since then, isolations of dental pulp cells became widely utilized, and several techniques have been used5,6,7,8. The critical significance of the method presented here is the adaption of all isolation steps to make the isolation fast and gentle to ensure the high quality of the final cell suspension for scRNA-seq. Higher cell yield can be obtained by more prolonged incubation with enzymes. This protocol provides an efficient solution for quickly obtaining single cells from mouse and human teeth of suitable quality for single-cell RNA-sequencing. This technique is expected to be widely used for other tissues or organisms with just slight technical modifications.
Disclosures
The authors have nothing to disclose.
Acknowledgements
J.K. was supported by the Grant Agency of Masaryk University (MUNI/H/1615/2018) and by funds from the Faculty of Medicine MU to junior researcher. J.L. was supported by the Grant Agency of Masaryk University, (MUNI/IGA/1532/2020) and is a Brno Ph.D. Talent Scholarship Holder – Funded by the Brno City Municipality. T.B. was supported by the Austrian Science Fund (Lise Meitner grant: M2688-B28). We thank to Lydie Izakovicova Holla and Veronika Kovar Matejova for their help with the obtaining of human teeth. Finally, we thank Radek Fedr and Karel Soucek for their kind assistance with FACS sorting.
Materials
| APC anti-mouse CD45 Antibody | BioLegend | 103112 | |
| Bovine Serum Albumin Fraction V | Roche | 10735078001 | |
| CellTrics 50 µm, sterile | Sysmex | 04-004-2327 | |
| Collagenase P (COLLP-PRO) | Roche | 11213857001 | |
| CUTFIX Scalpel Blades, Fig. 10 | AESCULAP | 16600495 | |
| CUTFIX Scalpel Blades, Fig. 11 | AESCULAP | 16600509 | |
| Fetal Bovine Serum (South America), Ultra low Endotoxin | Biosera | FB-1101/500 | |
| Hank's balanced salt solution (HBSS) without Ca2+ and Mg2+ | SIGMA | H6648 | |
| Industrial low lint wipes, MAX60 | Dirteeze | MAX60B176 | |
| Industrial strong tweezers, style 660, bent serrated pointed tips, 150mm | Value-Tec | 50-014366 | |
| Methyl alcohol A.G. | Penta | 21210-11000 | |
| Propidium Iodide Solution | BioLegend | 421301 | |
| Scalpel handle | CM Instrumente | AG-013-10 | |
| Serological pipettes 10mL, individually wrapped, 200 pcs. | CAPP | SP-10-C | |
| Stereo microscope | Leica | EZ4 | |
| Surgical scissors (9cm) | CM Instrumente | AI-430-09Y | |
| Tissue culture dish Ø 100 mm | TPP | 93100 |
References
- Krivanek, J., et al. Dental cell type atlas reveals stem and differentiated cell types in mouse and human teeth. Nature Communications. 11 (1), 4816 (2020).
- Soldatov, R., et al. Spatiotemporal structure of cell fate decisions in murine neural crest. Science. 364 (6444), (2019).
- Machado, L., Relaix, F., Mourikis, P. Stress relief: emerging methods to mitigate dissociation-induced artefacts. Trends in Cell Biology. , (2021).
- Nguyen, Q. H., Pervolarakis, N., Nee, K., Kessenbrock, K. Experimental considerations for single-cell RNA sequencing approaches. Frontiers in Cell and Developmental Biology. 6, (2018).
- Chiba, Y., et al. Single-cell RNA-sequencing from mouse incisor reveals dental epithelial cell-type specific genes. Frontiers in Cell and Developmental Biology. 8, (2020).
- Debnath, S., et al. Discovery of a periosteal stem cell mediating intramembranous bone formation. Nature. 562 (7725), 133-139 (2018).
- Kanton, S., Treutlein, B., Camp, J. G. Chapter 10 – Single-cell genomic analysis of human cerebral organoids. Methods in Cell Biology. 159, 229-256 (2020).
- Price, F. D., et al. Inhibition of JAK-STAT signaling stimulates adult satellite cell function. Nature Medicine. 20 (10), 1174-1181 (2014).
- vanden Brink, S. C., et al. Single-cell sequencing reveals dissociation-induced gene expression in tissue subpopulations. Nature Methods. 14 (10), 935-936 (2017).
- O’Flanagan, C. H., et al. Dissociation of solid tumor tissues with cold active protease for single-cell RNA-seq minimizes conserved collagenase-associated stress responses. Genome Biology. 20 (1), 210 (2019).
- van Velthoven, C. T. J., de Morree, A., Egner, I. M., Brett, J. O., Rando, T. A. Transcriptional profiling of quiescent muscle stem cells in vivo. Cell Reports. 21 (7), 1994-2004 (2017).
- Miyawaki-Kuwakado, A., et al. Transcriptome analysis of gene expression changes upon enzymatic dissociation in skeletal myoblasts. Genes to Cells. 26 (7), 530-540 (2021).
- Mattei, D., et al. Enzymatic dissociation induces transcriptional and proteotype bias in brain cell populations. International Journal of Molecular Sciences. 21 (21), 7944 (2020).
- Ferreira, P. G., et al. The effects of death and post-mortem cold ischemia on human tissue transcriptomes. Nature Communications. 9 (1), 490 (2018).
- He, W., Ye, J., Xu, H., Lin, Y., Zheng, Y. Differential expression of α6 and β1 integrins reveals epidermal heterogeneity at single-cell resolution. Journal of Cellular Biochemistry. 121 (3), 2664-2676 (2020).
- Machado, L., et al. In situ fixation redefines quiescence and early activation of skeletal muscle stem cells. Cell Reports. 21 (7), 1982-1993 (2017).
- Laranjeira, C., et al. Glial cells in the mouse enteric nervous system can undergo neurogenesis in response to injury. The Journal of Clinical Investigation. 121 (9), 3412-3424 (2011).
- Song, X., et al. Improved strategy for jet-in-air cell sorting with high purity, yield, viability and genome stability. FEBS Open Bio. 11 (9), 2453-2467 (2021).
- Greenblatt, M. B., Ono, N., Ayturk, U. M., Debnath, S., Lalani, S. The unmixing problem: A guide to applying single-cell RNA sequencing to bone. Journal of Bone and Mineral Research. 34 (7), 1207-1219 (2019).
- Charlebois, D. A., Hauser, K., Marshall, S., Balázsi, G. Multiscale effects of heating and cooling on genes and gene networks. Proceedings of the National Academy of Sciences. 115 (45), 10797-10806 (2018).
- Chen, J., et al. PBMC fixation and processing for Chromium single-cell RNA sequencing. Journal of Translational Medicine. 16 (1), 198 (2018).
- Denisenko, E., et al. Systematic assessment of tissue dissociation and storage biases in single-cell and single-nucleus RNA-seq workflows. Genome Biology. 21 (1), 130 (2020).
- Gronthos, S., Mankani, M., Brahim, J., Robey, P. G., Shi, S. Postnatal human dental pulp stem cells (DPSCs) in vitro and in vivo. Proceedings of the National Academy of Sciences of the United States of America. 97 (25), 13625-13630 (2000).

