Imaging of Podocytic Proteins Nephrin, Actin, and Podocin with Expansion Microscopy
Summary
The presented method enables visualization of fluorescently labeled cellular proteins with expansion microscopy leading to a resolution of 70 nm on a conventional microscope.
Abstract
Disruption of the glomerular filter composed of the glomerular endothelium, glomerular basement membrane and podocytes, results in albuminuria. Podocyte foot processes contain actin bundles that bind to cytoskeletal adaptor proteins such as podocin. Those adaptor proteins, such as podocin, link the backbone of the glomerular slit diaphragm, such as nephrin, to the actin cytoskeleton. Studying the localization and function of these and other podocytic proteins is essential for the understanding of the glomerular filter's role in health and disease. The presented protocol enables the user to visualize actin, podocin, and nephrin in cells with super resolution imaging on a conventional microscope. First, cells are stained with a conventional immunofluorescence technique. All proteins within the sample are then covalently anchored to a swellable hydrogel. Through digestion with proteinase K, structural proteins are cleaved allowing isotropical swelling of the gel in the last step. Dialysis of the sample in water results in a 4-4.5-fold expansion of the sample and the sample can be imaged via a conventional fluorescence microscope, rendering a potential resolution of 70 nm.
Introduction
Albuminuria is a surrogate parameter of cardiovascular risk and results from disruption of the glomerular filter1. The glomerular filter is composed of the fenestrated endothelium, the glomerular basement membrane and the slit diaphragm formed by podocytes. Primary and secondary foot processes of podocytes wrap around the capillary wall of the glomerulum2. The delicate structure of foot processes is maintained by cortical actin bundles which also serve as anchors for multiple slit diaphragm proteins and other adaptor proteins2. The slit diaphragm's backbone protein is called nephrin and interacts in a homophilic manner with nephrin molecules of opposing podocytes. Via diverse adaptor proteins, nephrin is linked to the actin cytoskeleton2,3. Mutations in the nephrin-encoding gene NPHS1 lead to nephrotic syndrome of the Finnish type4.
One of nephrin's interacting proteins is podocin, a hairpin-like protein of the stomatin family3. Podocin recruits nephrin to lipid rafts and links it to the actin cytoskeleton5. Podocin is encoded by the NPHS2 gene. Mutations in NPHS2 lead to steroid-resistant nephrotic syndrome6.
To visualize and co-localize actin adaptor proteins, immunofluorescence techniques may be used. Unfortunately, the diffraction barrier of the light limits the resolution of conventional fluorescence microscopes to 200-350 nm7. Novel microscopy techniques, e.g., stimulated emission depletion (STED)8, photo-activated localization microscopy (PALM)9, stochastic optical reconstruction microscopy (STORM or dSTORM) or ground state deletion microscopy followed by individual molecule return (GSDIM)9,10,11, enable a resolution up to approximately 10 nm. However, these super resolution techniques require highly expensive microscopes, well-trained personnel and are therefore not available in many laboratories.
Expansion microscopy (ExM) is a novel and simple technique that enables super resolution imaging with conventional microscopes and is potentially available to a large research community12. In protein retention expansion microscopy (proExM), the sample of interest (cells or tissue) is fixed and stained with fluorophores13. Proteins within the sample are then covalently anchored by a small molecule (6-((Acryloyl)amino)hexanoic acid, succinimidyl ester, AcX) into a swellable hydrogel13. Through enzymatic digestion with proteinase K (ProK), proteins and fluorophores maintain their relative position within the gel after expansion13. After swelling of the gel, the sample expands up to 4.5-fold (90-fold volumetric expansion) leading to an effective lateral resolution of approximately 60-70 nm (300 nm/4.5). Modifications of this technique can even allow for a 10-fold expansion (1,000-fold volumetric expansion), rendering a resolution of 20-30 nm on conventional microscopes14,15,16.
Glomerular structures of mouse and human kidneys have been visualized via ExM17. Within this paper, we present a detailed proExM protocol to visualize super resolution images of F-actin and the actin-adapter protein podocin within cells using a conventional fluorescence microscope.
Protocol
1. Splitting and seeding of cells
- Warm up sterile Dulbecco's Modified Eagle's Medium (DMEM) including 10% fetal calf serum (FCS), sterile Phosphate buffered saline (PBS) and sterile trypsin to 37 °C. Activate the clean bench.
- Prepare a 6-well plate by adding one sterile glass cover slip (10 mm) to each well using sterile forceps.
- Put a 10 cm cell culture dish with Cos7 cells under the clean bench. Under the clean bench, aspirate the cells' medium using a vacuum device.
- Hold the angulated cell culture dish in one hand and add 10 mL of sterile PBS to the side of the cell culture dish to avoid rinsing off cells. Put the cell culture dish down so that PBS rinses the complete cell culture dish.
- Remove the PBS and add 1 mL of trypsin to the middle of the cell culture dish and incubate for 5 min at 37 °C.
- Stop the trypsinization reaction by adding 10 mL of DMEM including 10% FCS and pipette the cell solution up and down with a pipettor to manually separate the cells.
- Analyze the cell number per milliliter using a counting chamber.
- Seed 68,000 cells in 2 mL medium per well onto the glass cover slips in the 6-well plate. Distribute the cells within the 6-well plate by cautiously shaking the plate horizontally and vertically.
NOTE: Cells should lie scattered. - Incubate the cells overnight in a 37 °C incubator with 5% CO2.
2. Transfection of cells
- Prepare two sterile 1.5 mL tubes (one for diluted DNA (A) and one for diluted reagent for cationic lipid transfection (B)). Pipette 0.75 µg of nephrin and 0.75 µg of podocin cDNA expression plasmid per well into one 1.5 mL tube and dilute it into 100 µL of reduced serum medium per well. Add 3 µL of cationic lipid transfection reagent per well to the second 1.5 mL tube and dilute with 100 µL of reduced serum medium. Incubate both reactions for 5 min at room temperature.
- Combine both reactions (A and B) to a DNA-lipid complex and incubate for 20 min at room temperature. Cautiously add 200 µL of the DNA-lipid complex to each well.
- Incubate the transfected cells at 37 °C with 5% CO2 for 48 h without changing the medium.
3. Immunolabeling of cellular structures
- Prepare a fixing solution (4% (w/v) paraformaldehyde in PBS, 1 mL/well), the permeabilization solution (0.5% (w/v) Triton X-100 in PBS, 1 mL/well), and a blocking solution (5% (v/v) bovine serum albumin in PBS, 1 mL/well).
- Remove the medium with a vacuum device. Add 2 mL of PBS to each well to remove extra medium. Aspirate the PBS completely.
NOTE: To avoid washing away cells with PBS, make sure to pipette PBS not onto the glass cover slip directly. - Fix cells with 4% (w/v) paraformaldehyde (PFA) dissolved in PBS for 10 min at room temperature.
NOTE: Alternatively, fix cells in 3% (v/v) glyoxal-ethanol18. - Discard PFA and wash cells twice in PBS (2 mL each) by avoiding direct pipetting onto the glass cover slips. Permeabilize the fixed cells with Triton X-100 0.5% (w/v) in PBS for 10 min at room temperature.
- Remove the permeabilization solution and wash twice with PBS as indicated in step 3.2.
- Block the cells by adding 1 mL of 5% (v/v) BSA in PBS for 1 h at room temperature. Incubate the cells with 200 µL of the primary antibody (anti-podocin antibody 1:200 in 1% (v/v) BSA in PBS) overnight at 4 °C.
NOTE: Alternatively, incubate the primary antibody for 1 h at room temperature. - Remove the primary antibody and wash three times with PBS as in step 3.2. Add the secondary antibody (goat anti-rabbit Alexa 488 1:1000 in 1% (v/v) BSA in PBS) for 1 h at room temperature. Keep the cells in the dark using a box.
- Discard the secondary antibody and wash with PBS three times.
- Remove PBS and incubate the cells with 200 µL of anti-nephrin antibody 1:100 in 1% (v/v) BSA in PBS for 1 h at room temperature. Wash with PBS three times. Keep the cells in the dark using a box.
- Remove PBS and incubate with the secondary antibody donkey anti-guinea pig 633, 1:200, for 1 h at room temperature. Wash with PBS three times. Keep the cells in the dark using a box.
4. Expansion microscopy
- Preparation
- To form the spacers for the gelation chamber, cut glass cover slips #1.0 and #1.5 into 5 mm stripes (four each per glass slide) using a diamond knife. Position the #1.5 mm cover slip stripes so that they form a square of 2.5 cm length on a glass slide and place them into a staining chamber (Figure 2A1–C1). Pipette a droplet of ddH2O into the corners of the square to adhere the glass cover slip stripes to each other and to the glass slide (Figure 2A2–C2).
NOTE: Avoid complete drying of ddH2O as the adhesion force will get lost. If needed, apply more droplets of ddH2O. Wait for approximately 20 min so that #1.5 mm cover slips are stably attached before starting with step 4.1.2. - Place four #1.0 glass cover slip stripes per cover slip onto the #1.5 mm cover slips. Adhere the stripes by pipetting a droplet on each #1.5 mm cover slip stripe (Figure 2A3–C3).
NOTE: The gel's thickness should be at least 0.15 mm to facilitate handling in the progress of the protocol. However, to avoid excessive gel on top of the cells, keep the spacers height close to the cell substrate. By using #1.5 and #1.0 cover slip stripes as spacers, the spacers height will be approximately 0.3 mm. The cells adhere to the cover glass (height 0.12 mm). Therefore, the gel will be thick enough to handle but excessive gel on top of the cells is avoided using #1.0 and #1.5 cover glass stripes as spacers. - For the gelation chamber lid, wrap a cover glass (#1.5) with paraffin film. Avoid any folds or dirt on the paraffin film (Figure 2A5-C5).
- To form the spacers for the gelation chamber, cut glass cover slips #1.0 and #1.5 into 5 mm stripes (four each per glass slide) using a diamond knife. Position the #1.5 mm cover slip stripes so that they form a square of 2.5 cm length on a glass slide and place them into a staining chamber (Figure 2A1–C1). Pipette a droplet of ddH2O into the corners of the square to adhere the glass cover slip stripes to each other and to the glass slide (Figure 2A2–C2).
- Anchoring and polymerization (gelation)
- Prepare the anchoring buffer (see Table 1). For anchoring treatment, replace PBS with 250 µL of anchoring buffer per well directly onto the glass cover slip and incubate for 3 h at room temperature. Keep substrate in the dark using a box.
NOTE: Alternatively, incubate overnight at room temperature. Prepare fresh anchoring buffer for every experiment and wait for 10-15 min until it is dissolved properly. 6-((Acryloyl)amino)hexanoic acid, succinimidyl ester (AcX) should be replaced every 4-5 months to ensure adequate anchoring. - Remove the anchoring buffer and wash once with 1.5 mL PBS per well.
- To stain actin fibres, thaw an ExM compatible phalloidin-solution and incubate the phalloidin (5 µL of phalloidin diluted in 195 µL of 1% (v/v) BSA in PBS/well) for 45 min at room temperature. Keep samples in the dark using a box.
- In the meantime, dissolve sodium acrylate in ddH2O using a stirring device. On ice, prepare the monomer solution (see Table 1).
NOTE: Dissolved sodium acrylate should be a clear and colorless solution. If the solution is yellow, replace with new sodium acrylate. - Prepare the gelling solution on ice (see Table 1). Pipette ammoniumperoxidsulfate (APS) into the gelling solution right before the gelling solution is applied to the gelation chamber.
- Remove phalloidin from the cells and wash with 1.5 mL of PBS twice at room temperature. Leave 1.5 mL PBS within the well to facilitate removal of the sample on the glass cover slip.
- Place the cells on the cover glass into the gelling chamber using forceps and a cannula to lift the cover glass slip from the 6-well plate.
NOTE: The cells should be on the top of the glass cover slip. The glass cover slip should not touch the spacers. - Add APS to the gelling solution and vortex shortly. Pipette 200 µL of gelling solution on the sample (Figure 2A4-C4). Cautiously close the gelation chamber by avoiding air bubbles within the gel (Figure 2A5–C5).
- Incubate the gelling chamber for at least 1 h at 37 °C to polymerize the gel in the wet staining chamber.
- Prepare the anchoring buffer (see Table 1). For anchoring treatment, replace PBS with 250 µL of anchoring buffer per well directly onto the glass cover slip and incubate for 3 h at room temperature. Keep substrate in the dark using a box.
- Homogenization (digestion)
- Take the staining chamber out of the incubator. To open the gelation chamber lid, introduce a razor blade between the lid and the spacer. Remove the lid cautiously. Remove the spacers with the razor blade and eliminate all extra gel by cutting it with the razor blade.
- Put the slide with the gel and cover glass into a dish filled with PBS. By shaking gently, remove the detached cover glass from the gel. To remove the gel from the dish easily, put the slide below the gel to attach the gel to the slide.
- With the gel on the slide, divide the gel in small pieces (quarter of the gel is divided into two to three pieces) using the razor blade. Gently push one piece of gel into a well of a 6-well plate with glass bottom and enfold it by a paintbrush. Keep the gel moisturized with a little amount of PBS using the paintbrush to avoid dehydration of the gel.
NOTE: The cells face downwards. - Using an inverted microscope, take overview images with low numerical aperture to determine the expansion factor after expansion.
- Prepare the digestion buffer (see Table 1). Dilute Proteinase K to 4 U/mL in digestion buffer to receive the digestion solution.
NOTE: Digestion buffer without Proteinase K can be stored at 4 °C for 1-2 weeks. - Add 500 µL of the digestion solution to each well and immerse the gel within the solution. Let it digest overnight at room temperature and close the lid keeping the samples in the dark.
NOTE: Alternatively, let it digest at 37 °C for 1 h.
- Expansion
- Remove the digestion solution with a pipette and discard it. Add 1 mL of ddH2O. Incubate the immersed gel for 10 min at room temperature.
- Remove the water and add 1 mL of fresh ddH2O. Wait for 10 min and continue exchanging water every 10 min until a plateau of expansion is reached.
NOTE: Sample expansion up to 4.5-fold is achievable. The gel becomes optically clear.
- Imaging
- Remove the water from the gel and directly start microscopy. Using an inverted microscope, primarily use an air objective (low magnification) to find imaged cells in the pre-expansion state (step 4.3.4).
- Switch to a 40x (oil/water) and 63x objective for better resolution. Excite with the wavelength of interest and take the image via the camera.
- Validation
- Take an overview image of the sample. Find and match the same structures within the sample that were imaged in step 4.3.4. Use the channel with the best signal-to-noise ratio for validation (Figure 3 A-B).
NOTE: Adjust the imaging parameters to achieve similar brightness as in the image acquired in the non-expanded state (step 4.3.4). - Overlay pre- and post-expansion images by rotating and shifting them with ImageJ. Use the distance measuring tool in ImageJ to measure the distances between clearly identifiable structures. Measure at least 10 different structures.
NOTE: Alternatively, use a Python script to measure distances as described in14. - Calculate the expansion factor by dividing post-/pre-expansion measurements.
- To determine distortions, take images with a higher numerical aperture (Figure 4A–B). Overlay these images and analyze them with ImageJ or as described in15.
NOTE: Determination of distortions should be performed routinely but not necessarily on every sample.
- Take an overview image of the sample. Find and match the same structures within the sample that were imaged in step 4.3.4. Use the channel with the best signal-to-noise ratio for validation (Figure 3 A-B).
Representative Results
The concept and timing of this proExM protocol is depicted in Figure 1. On day 5, transfected cells are fixed and stained with fluorescent antibodies targeting the protein of interest (Figure 1A,B). On day 6, treatment with AcX leads to formation of amine groups on all proteins (including fluorophores) (Figure 1A,B)12. Upon polymerization of the hydrogel, these amine groups bind covalently to the hydrogel (day 6). After polymerization of the gel, homogenization (digestion) is performed with proteinase K resulting in the destruction of structural proteins of the cell (day 6, Figure 1A,B). Fluorescently labeled antibodies remain mostly preserved after digestion. Due to the disruption of structural proteins, water dialysis of the hydrogel results in isotropic expansion of the cell within the hydrogel on day 7 (Figure 1A,B). Imaging of the sample is performed with a conventional fluorescence microscope (Figure 1A). Data validation to determine the expansion factor and to exclude distortions should be performed (Figure 1A).
To perform expansion of the cell isotropically, the gelation step is essential. Figure 2 shows the lateral and top view of a gelation chamber. Glass cover slips build the spacers of the gelation chamber (Figure 2A1-3/C1-3). The cover glass with the fixed and stained cells is positioned with the cells upward onto a glass slide (Figure 2A4-C4). The lid of the gelation chamber is wrapped with parafilm and is closed bubble-free (Figure 2A5-C5).
This ExM protocol enables expansion of up to four-fold. To determine the expansion factor, it is essential to image cells before and after expansion (Figure 3A + B). Insufficient anchoring and homogenization may lead to distortions and ruptures of cells. Figure 4A + B shows representative examples of ruptured cells in different magnification images.
This method can be used to investigate the co-localization of F-actin and actin adaptor proteins, e.g., podocin and nephrin (Figure 5). Podocin is depicted in green while actin is labeled in blue (Figure 5). Nephrin is marked in green. White areas indicate co-localization.
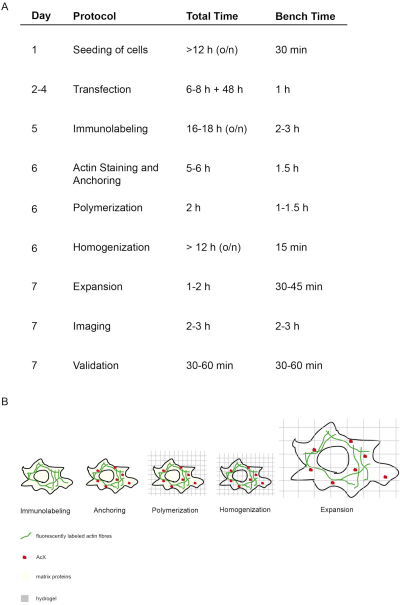
Figure 1: Concept and timing of this ExM protocol. (A) In the "protocol" column, each step of the protocol is outlined. (A + B) After seeding and transfecting cells, immunofluorescent labeling is performed (Immunolabeling). (A + B) The small molecule AcX (red dot) binds to all proteins and anchors them to the hydrogel (Anchoring). (A + B) Via polymerization all proteins including fluorophores are covalently bound via AcX to the hydrogel (Polymerization). (A + B) Homogenization leads to digestion of structural matrix proteins. (A + B) Expansion is achieved by dialysis in water. (A) Imaging and validation of imaging finalizes the experiment. (A) The entire protocol requires 7 days (column "day") with many incubation steps (total time per day column "total time"), but actual bench time is much less as indicated in the respective column "bench time". Modified from14. Please click here to view a larger version of this figure.
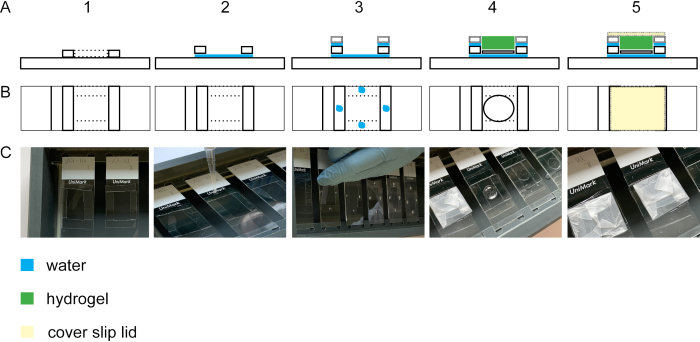
Figure 2: Building the gelation chamber. Side view (A1) and top view (B1 + C1) of a glass slide with four #1.5 cover stripes. By adding a droplet of water between the glass slide and the cover slip stripes, the stripes will adhere to the glass slide (side view A2, top view B2, C2). Droplets of water on the #1.5 cover stripes lead to adhesion of #1.0 cover stripes laid on top of the #1.5 cover stripes (side view A3, top view B3, C3). The sample on the cover slip is placed in the middle of the rectangle using forceps. The gel is pipetted on top (side view A4, top view B4, C4). (A5) Side view and top view (B5 and C5) of the assembled gelation chamber including the closed lid which is built from a cover slip wrapped in parafilm. Please click here to view a larger version of this figure.
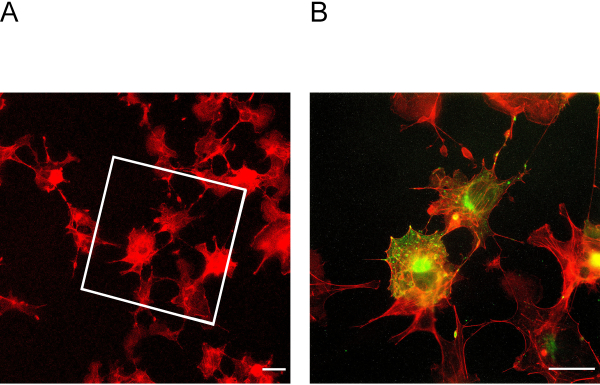
Figure 3: Cells before and after expansion. (A) Cells before expansion stained for actin. The box indicates in which area the expanded cells in Figure 3B lie. (B) Cells after expansion stained for actin in red and podocin in green. Podocin co-localizes with actin in the cell periphery. Scale bar = 5 µm, expansion factor = 2. Please click here to view a larger version of this figure.

Figure 4: Distortions and ruptures of cells. (A + B) Representative microscopic images of cos7 cells immuno-stained for actin (red). Cells were fixed, stained, anchored, digested and expanded. (A) Ruptures of cells. Arrows indicated ruptured areas. Scale bar = 5 µm, expansion factor = 4. (B) Ruptures and distortion of cells. White arrows indicated ruptured areas. Scale bar = 5 µm, expansion factor = 4. Please click here to view a larger version of this figure.
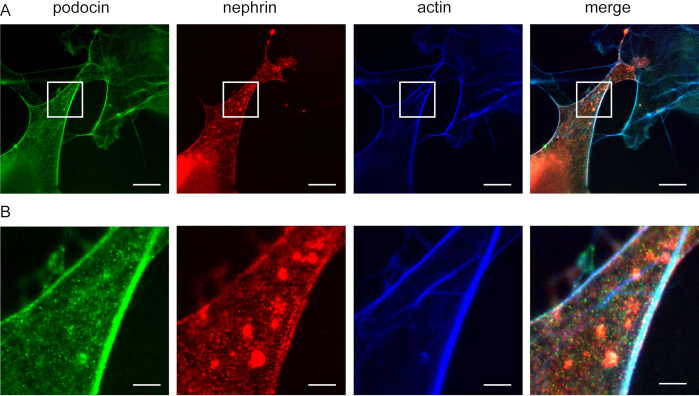
Figure 5: Podocin co-localizes with nephrin and actin. Cos7 cells immunofluorescently labeled for podocin, actin, and nephrin. (A) Cos7 cells stained for podocin (green), actin (blue), and nephrin (red) with ExM. Podocin co-localizes with actin and nephrin. Scale bar = 200 nm, expansion factor = 4. (B) Magnification of the indicated area in (A), Scale bar = 40 nm. Please click here to view a larger version of this figure.
| Solutions for ExM | |||
| Anchoring buffer | final concentration | ||
| NaHCO3 | 150 mM | ||
| Acryloyl-X, SE (AcX) | 0.1 mg/ml | ||
| Monomer solution | Stock solution concentration g/100ml | amount (ml) | final concentration (g/100 ml) |
| sodium acrylate | 38 | 2.25 | 8.6 |
| acrylamide | 50 | 0.5 | 2.5 |
| N,N’-Methylenebisacrylamide | 2 | 0.5 | 0.10 |
| sodium chloride | 29.2 | 4 | 11.7 |
| PBS | 10x | 1 | 1x |
| water | 1.15 | ||
| total | 9.4 | ||
| Gelling solution | Stock solution concentration | amount (µl) | final concentration (mg/ml) |
| monomer solution | NA | 190 | NA |
| APS | 10% | 4 | 2 |
| TEMED | 10% | 4 | 2 |
| water | NA | 2 | NA |
| total | 200 | ||
| Digestion solution | Stock solution concentration | amount (µl) | final concentration |
| Tris Cl, pH 8.0 | 1 M | 1000 | 50 mM |
| EDTA pH 8.0 | 0.5 M | 40 | 1 mM |
| Triton X-100 | 10% | 1000 | 0.5 % |
| Guanidin HCL | 8M | 2000 | 0.8 M |
| water | ad 20 ml | ||
| proteinase K | 800 U/ml | 100 | 4 U/ml |
Table 1: Solutions for ExM.
Discussion
The presented method enables the investigator to visualize cellular proteins, e.g., podocin, nephrin, and cytoskeletal components, e.g., F-actin. Within this protocol, transfected cos7 cells are used as a model to study interaction of slit diaphragm proteins with F-actin. Unfortunately, immortalized podocyte cell lines do not express sufficient endogenous amounts of slit diaphragm proteins19.
With this method, cellular proteins can be visualized with nanoscale resolution using a conventional fluorescence microscope. The most critical steps within the protocol are: 1) sufficient anchoring of protein amine groups to the hydrogel with AcX, 2) adequate polymerization of the hydrogel, 3) optimal timing for digestion and 4) selection of compatible fluorophores.
Anchoring of cellular proteins to the hydrogel is essential for this method in order to preserve the protein's position within the hydrogel during expansion. AcX is a small molecule that binds to amine groups of proteins within cells and tissues. AcX creates a carbon-carbon double band with proteins, enabling incorporation of the proteins into the hydrogel in the polymerization step20. AcX also integrates antibodies so that labeling with immunofluorescence antibodies can be performed before AcX treatment. Insufficient anchoring may lead to ruptures and distortions of cells. Due to modification of amine groups by fixatives, one needs to optimize the fixative or the time of fixation. In addition, insufficient storage or non-optimized anchoring conditions may result in ruptures and distortions. Based on our experience, AcX loses its optimal effect when used for more than 3-4 months.
Polymerization of the gel is temperature dependent. We, therefore, recommend keeping the polymerization solutions on ice before pipetting it into the gelation chamber. In addition, the handling time of the gelation step should be kept short (less than 5 min) in order to avoid premature gel formation. Thorough mixing of the polymerization solution prevents uneven polymerization. Air bubbles will affect the expansion process when touching the sample and can be prevented by adding more polymerization solution.
After incorporation of the cellular proteins within the hydrogel, the mechanical homogenization step (or digestion) is needed to ensure expansion. Different methods, e.g., heat and detergent or enzymatic digestion, exist and need to be customized to the investigated sample12,14,20. Within this protocol, the protease Proteinase K is used for enzymatic digestion. Proteinase K is applied at a dosage sufficient to destroy structural proteins whilst preserving most other proteins including fluorescent antibodies12. If digestion is incomplete, the sample expansion is insufficient. In addition, the sample can tear during the expansion process (Figure 3). If an inadequate sample expansion has occurred, water replacement is recommended. Alternatively, the time for the enzymatic digestion can be adjusted or a new aliquot of the Proteinase K opened.
If the sample is over-digested, fluorescence signals will be diminished. In this case, the digestion time should be reduced. In ExM in general, the fluorescence signal intensity per unit of volume is reduced due to the volumetric expansion of the sample14. Therefore, longer exposure times during imaging need to be considered.
It is essential to select ExM compatible fluorophores. Cyanine dyes are degraded during the polymerization step13. Fluorescence proteins based on bacteriophytochromes are also largely destroyed13. However, most GFP-like proteins will be preserved13. In addition, streptavidin can also be applied pre-expansion, labeling post-translational modifications such as S-nitrolysation via a small molecule tag13.
Phalloidin, a small labeling molecule to target the actin cytoskeleton, is not compatible with ExM21. To overcome insufficient anchoring of phalloidin, trivalent anchoring (TRITON) has been introduced21. This approach offers simultaneous targeting, labeling and grafting of biomolecules21.
This method can be modified to stain RNA molecules (ExFish)22. In iterative expansion microscopy (iExM) or X10 microscopy, the resolution of 60-70 nm can be extended to approximately 25 nm by applying a second swellable gel within the first expanded hydrogel or conducting a single expansion step using a different hydrogel15,16. Ultrastructure expansion microscopy (U-ExM) enables super resolution of proteins preserving their attribution to an ultrastructural element (e.g., mitochrondria, microtubules)23. A combination of ExFish (RNA and DNA) and proExM methods have previously been performed as well22,24. The presented protocol uses transfected cos7 cells as a model to investigate slit diaphragm proteins. We expect that other resident cultured kidney cells, e.g., HEK293T cells, can be similarly used for this protocol. Depending on the cell line, adjustments may need to be made for the different culturing and transfection conditions.
ExM enhances resolution of immuno-stained samples by about 4-fold reaching a lateral spatial resolution of 70 nm13. Compared to other super-resolution techniques, ExM is performed on a conventional fluorescence microscope13,14. Therefore, no expensive equipment or specifically trained personnel is necessary to conduct the ExM method14. Even though not all fluorophores are compatible with ExM, there are generally many available antibodies with optimized fluorophores with photo-physical properties needed for super resolution microscopy14. The main disadvantage of this method is that ExM is incompatible with live samples12,14.
In the future, improving the hydrogel's chemical composition may lead to even higher spatial resolution12. The combination of different protocols may also enable visualization of proteins, RNA, DNA, or lipids in complexes within the same sample with such high resolution12.
Disclosures
The authors have nothing to disclose.
Acknowledgements
The authors would like to thank Blanka Duvnjak and Nikola Kuhr for their excellent technical assistance.
Materials
| Acrylamide >99% | Sigma-Aldrich | A3553-100G | |
| 6-((Acryloyl)amino)hexanoic acid, succinimidyl ester, Acryloyl-X, SE | invitrogen | A-20770 | store up to 4 months |
| APS | Sigma-Aldrich | A3678-25G | |
| Deckgläser (cover glasses) | Engelbrecht | K12432 | 24x32mm #1.0 |
| Diamont cutter | VWR | 201-0392 | for cutting the cover slips |
| Guanidine HCl | Sigma-Aldrich | G3272-100G | 8M Stock can be kept at RT |
| Marten hair paintbrush | Leon Hardy | 3 (770) | |
| "Menzel" Deckgläser (cover glasses) | Thermo Fischer | 15654786 | 24x24mm #1.5 |
| N,N`-Methylenbisacrylamide | Sigma-Aldrich | M7256-25G | |
| Objektträger UniMark | Marienfeld | 703010 | |
| Proteinase K | New England Biolabs | P8107S | |
| Sodium Acrylate | Sigma-Aldrich | 408220 | check purity |
| Sodium Bicarbonate | Sigma-Aldrich | S5761 | |
| Staining chamber | produced at the university's workshop | ||
| TEMED | ROTH | 2367.1 | |
| 6-Well glass bottom plates | Cellvis | P06-1.5H-N | |
| Antibodies | |||
| Actin-ExM 546 | chrometra | non-available | 1:40 |
| Anti Podocin produced in rabbit | Sigma | P-0372-200UL | 1:200 |
| Donkey anti guinea-pig CF633 | Sigma | SAB4600129-50UL | 1:200 |
| Goat anti rabbit 488 | Life Technologies | A11034 | 1:1000 |
| Guinea pig anti nephrin | Origene | BP5030 | 1:100 |
| Software | |||
| FIJI | |||
| Visiview | |||
| microscope | |||
| AXIO Observer Z1 | Zeiss | non-available |
References
- Matsushita, K., et al. Association of estimated glomerular filtration rate and albuminuria with all-cause and cardiovascular mortality in general population cohorts: a collaborative meta-analysis. Lancet. 375 (9731), 2073-2081 (2010).
- Faul, C., Asanuma, K., Yanagida-Asanuma, E., Kim, K., Mundel, P. Actin up: regulation of podocyte structure and function by components of the actin cytoskeleton. Trends in Cell Biology. 17 (9), 428-437 (2007).
- Saleem, M. A., et al. Co-localization of nephrin, podocin, and the actin cytoskeleton – Evidence for a role in podocyte foot process formation. American Journal of Pathology. 161 (4), 1459-1466 (2002).
- Kestila, M., et al. Positionally cloned gene for a novel glomerular protein – nephrin – is mutated in congenital nephrotic syndrome. Molecular Cell. 1 (4), 575-582 (1998).
- Huber, T. B., et al. Podocin-mediated recruitment of nephrin into lipid rafts is required for efficient nephrin signaling. Journal of the American Society of Nephrology. 14, 8 (2003).
- Boute, N., et al. NPHS2, encoding the glomerular protein podocin, is mutated in autosomal recessive steroid-resistant nephrotic syndrome. Nature Genetics. 24 (4), 349-354 (2000).
- Galbraith, C. G., Galbraith, J. A. Super-resolution microscopy at a glance. Journal of Cell Science. 124 (10), 1607-1611 (2011).
- Willig, K. I., Rizzoli, S. O., Westphal, V., Jahn, R., Hell, S. W. STED microscopy reveals that synaptotagmin remains clustered after synaptic vesicle exocytosis. Nature. 440 (7086), 935-939 (2006).
- van de Linde, S., et al. Direct stochastic optical reconstruction microscopy with standard fluorescent probes. Nature Protocols. 6 (7), 991-1009 (2011).
- Rust, M. J., Bates, M., Zhuang, X. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM). Nature Methods. 3 (10), 793-795 (2006).
- Testa, I., et al. Multicolor fluorescence nanoscopy in fixed and living cells by exciting conventional fluorophores with a single wavelength. Biophysical Journal. 99 (8), 2686-2694 (2010).
- Wassie, A. T., Zhao, Y., Boyden, E. S. Expansion microscopy: principles and uses in biological research. Nature Methods. 16 (1), 33-41 (2019).
- Tillberg, P. W., et al. Protein-retention expansion microscopy of cells and tissues labeled using standard fluorescent proteins and antibodies. Nature Biotechnology. 34 (9), 987-992 (2016).
- Truckenbrodt, S., Sommer, C., Rizzoli, S. O., Danzl, J. G. A practical guide to optimization in X10 expansion microscopy. Nature Protocols. 14 (3), 832-863 (2019).
- Truckenbrodt, S., et al. X10 expansion microscopy enables 25-nm resolution on conventional microscopes. Embo Reports. 19 (9), (2018).
- Chang, J. B., et al. Iterative expansion microscopy. Nature Methods. 14 (6), 593-599 (2017).
- Chozinski, T. J., et al. nanoscale optical imaging of mouse and human kidney via expansion microscopy. Scientific Reports. 8 (1), 10396 (2018).
- Richter, K. N., et al. Glyoxal as an alternative fixative to formaldehyde in immunostaining and super-resolution microscopy. The EMBO Journal. 37 (1), 139-159 (2018).
- Rinschen, M. M., et al. Quantitative deep mapping of the cultured podocyte proteome uncovers shifts in proteostatic mechanisms during differentiation. American Journal of Physiology-Cell Physiology. 311 (3), 404-417 (2016).
- Asano, S. M., et al. Expansion microscopy: protocols for imaging proteins and RNA in cells and tissues. Current Protocols in Cell Biology. 80 (1), 56 (2018).
- Wen, G., et al. Evaluation of direct grafting strategies via trivalent anchoring for enabling lipid membrane and cytoskeleton staining in expansion microscopy. ACS Nano. 14 (7), 7860-7867 (2020).
- Chen, F., et al. Nanoscale imaging of RNA with expansion microscopy. Nature Methods. 13 (8), 679-684 (2016).
- Gambarotto, D., et al. Imaging cellular ultrastructures using expansion microscopy (U-ExM). Nature Methods. 16 (1), 71-74 (2019).
- Zhao, Y., et al. Nanoscale imaging of clinical specimens using pathology-optimized expansion microscopy. Nature Biotechnology. 35 (8), 757-764 (2017).

