PCR Mutagenesis, Cloning, Expression, Fast Protein Purification Protocols and Crystallization of the Wild Type and Mutant Forms of Tryptophan Synthase
Summary
This article presents a series of consecutive methods for the expression and purification of Salmonella typhimurium tryptophan synthase comp this protocol a rapid system to purify the protein complex in a day. Covered methods are site-directed mutagenesis, protein expression in Escherichia coli, affinity chromatography, gel filtration chromatography, and crystallization.
Abstract
Structural studies with tryptophan synthase (TS) bienzyme complex (α2β2 TS) from Salmonella typhimurium have been performed to better understand its catalytic mechanism, allosteric behavior, and details of the enzymatic transformation of substrate to product in PLP-dependent enzymes. In this work, a novel expression system to produce the isolated α- and isolated β-subunit allowed the purification of high amounts of pure subunits and α2β2 StTS complex from the isolated subunits within 2 days. Purification was carried out by affinity chromatography followed by cleavage of the affinity tag, ammonium sulfate precipitation, and size exclusion chromatography (SEC). To better understand the role of key residues at the enzyme β-site, site-direct mutagenesis was performed in prior structural studies. Another protocol was created to purify the wild type and mutant α2β2 StTS complexes. A simple, fast and efficient protocol using ammonium sulfate fractionation and SEC allowed purification of α2β2 StTS complex in a single day. Both purification protocols described in this work have considerable advantages when compared with previous protocols to purify the same complex using PEG 8000 and spermine to crystalize the α2β2 StTS complex along the purification protocol. Crystallization of wild type and some mutant forms occurs under slightly different conditions, impairing the purification of some mutants using PEG 8000 and spermine. To prepare crystals suitable for x-ray crystallographic studies several efforts were made to optimize crystallization, crystal quality and cryoprotection. The methods presented here should be generally applicable for purification of tryptophan synthase subunits and wild type and mutant α2β2 StTS complexes.
Introduction
The tryptophan synthase (TS) bienzyme complex (α2β2) is an allosteric enzyme, catalyzing the last two steps in the biosynthesis of the amino acid L-Tryptophan in bacteria, plants, and fungi1,2,3. Bacterium Salmonella enterica serovar typhimurium (St) causes a severe gastrointestinal infection in humans and other animals. Since humans and higher animals do not have TS (EC 4.2.1.20), the inhibition of S. typhimurium α2β2 TS complex (α2β2 StTS) has been explored as a potential drug target for the treatment of cryptosporidiosis and tuberculosis4, genital and ocular infections5, and for potential herbicide utilization in agriculture6. The α-subunit catalyzes the aldolytic cleavage of indole-3-glycerol-phosphate (IGP) to glyceraldehyde-3-phosphate (GAP) and indole, through the formation of an indolenine tautomer intermediate and subsequently carbon-carbon bond cleavage to produce GAP and indole3,6. The β-catalytic site contains a pyridoxal 5′-phosphate (PLP) cofactor molecule bound to β-Lys87 via a Schiff base, which functions as an electron sink in the course of the reactions at the enzyme β-subunit3,7. The β-site catalyzes the replacement of the L-Serine side-chain hydroxyl by indole to give L-Tryptophan and a water molecule in a PLP-dependent reaction. StTS serves as a longstanding model for the investigation of substrate channeling and allosteric communication within multi-enzyme complexes2,3. Bidirectional allosteric communication between the α- and β-subunits of TS is necessary to synchronize the catalytic steps and prevent indole release during L-Tryptophan synthesis3. To extend this effort, we have prepared several mutants (β-Gln114Ala, β-Lys167Thr, and β-Ser377Ala) by single point mutation to be used in further explorations of the relationship between enzyme structure, mechanism and function at the catalytic site of the StTS β-subunit.
Detailed research on the catalytic mechanism of α2β2StTS was initiated by the research group of Edith W. Miles. Early studies with native Escherichia coli α2β2 TS complex have focused on the purification and characterization of the isolated α-subunit8,9, isolated β-subunit10,11 and the reconstitution of the α2β2 TS complex from the isolated subunits12. Purification was carried out by ammonium sulfate precipitation, sample dialysis, DEAE-Sephadex chromatography, dialysis, and a second chromatographic round on a DEAE-Sephadex column12. In another protocol, the purification of the same complex was improved by loading the clarified cell lysate on a DEAE-Sephadex column followed by a chromatographic step on a Sepharose 4B column, ammonium sulfate precipitation and dialysis13. Both purification protocols last for 4-5 days. Escherichia coli α2β2 TS complex crystallized but crystals were not suitable for X-ray diffraction at that time.
In a novel study, recombinant and wild type forms of S. typhimurium α2β2 TS complex were purified and crystalized14,15. The recombinant α2β2 StTS complex was overexpressed in E. coli strain CB149 carrying the pEBA-10 expression vector. Initial crystallization and X-ray diffraction data collection and analysis of the α2β2 StTS complex were reported14. However, long and thin needle like α2β2 StTS crystals impaired structural studies. In an attempt to collect better X-ray diffraction data, another purification protocol was described to purify the wild type and mutant forms of the α2β2 StTS complex15. Purification was carried out with an initial precipitation using spermine and PEG 8,000 into the clarified cell lysate and a large bulky precipitate was removed by centrifugation. The supernatant fraction containing high amounts of α2β2 StTS complex was stored for 16-48 h at 4 °C until yellow crystals precipitated. Crystals were washed and extensively dialyzed against different buffers. Protein complex was recrystallized in buffer containing ammonium sulfate and dialyzed15. Although, protein crystallization depends on protein and precipitant concentrations in solution, it is difficult to monitor, predict, and reproduce purification for other mutant forms of α2β2 StTS complex in solution. This protocol has the advantage that it does not use any chromatographic methods; however, the disadvantages are the long purification time necessary to crystallize, dialyze, and recrystallize, typically requiring 5-7 days. To obtain crystals suitable for X-ray data collection, more than 600 crystallization conditions were evaluated using a combination and variation of protein concentration, temperature, precipitants (PEG 4,000, 6,000, and 8,000), and additives (CaCl2, MnCl2, ZnCl2, cadaverine, putrescine, spermine, or spermidine)15. Crystals had a better crystalline form and grew faster in conditions containing 12% PEG 8,000 and 2 mM spermine. Crystallization was more favorable at 25 °C rather than at 4, 30, or 42 °C and grew to maximum dimensions within 3 days15. Several α2β2 StTS crystal structures were reported at that time (1996-1999)16,17,18,19,20,21 and many other structures have been published to date.
Here, the main purpose is to present alternative protocols to purify tryptophan synthase and optimize protein crystallization. The present work shows significant improvements to purify the wild type isolated α-subunit (αStTS), isolated β-subunit (βStTS), reconstituted α2β2 StTS complex from the isolated subunits, and wild type and mutant forms of the α2β2 StTS complex. The advantages over past protocols are considerable since purification time was reduced significantly and crystallization and cryoprotection were optimized. Mutant forms of α2β2 StTS complex engineered in this work have crystallized near the same condition used for the wild type form. However, fine crystallization optimization was necessary to obtain large single crystals of sufficient quality for structure determination at near atomic resolution. To date, there are 134 tryptophan synthase crystal structures deposited in the Protein Data Bank (PDB), accounting 101, 31 and 2 crystal structures, respectively, for bacteria, archaea and eukaryote. Nicely, 73 structures belong to S. enterica serovar typhimurium and 5 crystal structures of the α2β2 StTS complex have resolution limits higher than 1.50 Angstroms. Not surprisingly, 4 out 5 were prepared in our research group (PDB IDs:5CGQ at 1.18 Å, 4HT3 at 1.30 Å, 4HPJ at 1.45 Å, 6DZ4 at 1.45 Å resolution). The refined crystal structures of mutant form of α2β2 StTS complex are anticipated to provide new insights into the mechanism and roles played by essential amino acid residues involved in L-Tryptophan synthesis.
Protocol
1. Fast protocol to purify the α- and β-subunit and the recombined α2β2 StTS complex
- DNA subcloning into pETSUMO expression vector
- Obtain the translationally coupling gene (trpA and trpB) encoding the α– and β-subunits of the tryptophan synthase from bacterium Salmonella enterica serovar typhimurium cloned in the pEBA-10 expression vector22. Use pEBA-10 vector as a DNA template.
NOTE: Alternatively, the listed primers below can be used to amplify both genes from the Salmonella enterica serovar typhimurium genome. All molecular biology steps were followed as described in Molecular Cloning: A Laboratory Manual23. - Use polymerase chain reaction (PCR) to amplify individually the full-length polynucleotide sequence of the α-subunit (αStTS) with primers αStTS-FW-Bam and αStTS-Rev-Eco and the full-length sequence of β-subunit (βStTS) with primers βStTS-FW-Bam and βStTS-Rev-Hind. Use a melting temperature of approximately 55 °C and a polymerase extension time of 2 min.
- Use high-fidelity DNA polymerase (e.g., Phusion) and the manufacturer’s protocol to amplify the DNA sequences. For a 50 µL PCR reaction add 34 µL of nuclease-free water, 10 µL of 5x reaction buffer, 1 µL of 10 mM dNTPs, 1 µL of 10 µM forward primer, 1 µL of 10 µM reverse primer, 1 µL of Template DNA (200 ng), 1.5 µL of DMSO, 0.5 µL of Phusion DNA polymerase.
- For the PCR program, use a hot start (180 seconds at 98 °C) followed by 30 amplification cycles (30 seconds at 98 °C, 30 seconds at 55 °C and 120 seconds at 72 °C), and a final extension (300 seconds at 72 °C).
NOTE: The italicized sequences correspond to the BamHI, EcoRI, BamHI and HindIII restriction sites, respectively. Enzyme cleavage efficiency close to the termini of PCR fragments were enhanced by adding extra bases (lowercase sequences).
αStTS-FW-Bam: 5′-cgcGGATCCATGGAACGCTACGAAAA-3′
αStTS-Rev-Eco: 5′-ccgGAATTCTTATGCGCGGCTGGC-3′
βStTS-FW-Bam: 5′-cgcGGATCCATGACAACACTTCTCAAC-3′
βStTS-Rev-Hind: 5′-cccAAGCTTTCAGATTTCCCCTC-3′
- Load PCR product on 0.8% agarose gel in 1x TAE buffer (40 mM Tris, 20 mM acetic acid and 1 mM EDTA) at 6 V/cm, gel extract the DNA band of interest, and gel purify the PCR fragment using a silica bead kit following the instructions from the manufacturer.
- Digest at least 200 ng of each DNA fragment with appropriate restriction enzymes and conditions recommended by the manufacturer for 2 hours at 37 °C.
- To set up a 50 μL restriction digestion reaction add 34 µL of nuclease-free water, 10 µL of DNA (200 ng), 5 μL of 10x reaction buffer, 0.5 µL of restriction enzyme 1, and 0.5 µL of restriction enzyme 2.
- Load the digestion product on 0.8% agarose gel on 1x TAE at 6 V/cm, gel extract, and gel purify the digested fragment using a commercial kit.
- Subclone individually each fragment into the E. coli expression modified vector pET SUMO, previously digested with appropriate enzymes and gel purified.
NOTE: This vector is a modified version of the commercial pET SUMO. This vector has been optimized for restriction enzyme cloning. The multi cloning site (MCS) of pET28b vector (BamHI, EcoRI, SacI, SalI, HindIII, NotI, and XhoI) was inserted in the pET SUMO cloning site. This vector contains an N-terminal 6x-Histidine tag in frame with the Small Ubiquitin-like Modifier protein (SUMO) and multiple cloning sites. - Ligate 100 ng of modified pET SUMO and 50 ng of PCR fragment with T4 DNA ligase for 2 hours at 25 °C.
- Transform the constructed plasmid into competent cells of E. coli strain DH10Bα cells. Plate cells on LB agar plates containing 35 μg/mL kanamycin. Incubate the plate inverted overnight at 37 °C.
- Select a single colony from each transformation, prepare ultra-pure plasmid DNA, and perform DNA sequencing to verify that αStTS or βStTS were cloned in frame with the N-terminal His6-SUMO tag.
NOTE: Prepare glycerol stocks of cell culture (turn cell suspension in 16% final concentration of sterile glycerol) and store them long-term at -80 °C. - Transform the expression plasmid SUMO-αStTS or SUMO-βStTS individually into competent cells of E. coli expression strain Rosetta (DE3) pLysS with a T7 promoter-based system. Plate the recombinant cells on Luria Bertani (LB) agar plates containing 35 μg/mL kanamycin and chloramphenicol. Incubate the plate inverted overnight at 37 °C.
- After successful colony formation, pick one single colony (without any satellite colonies) and disperse it in 5 mL of LB medium with both antibiotics. Culture cells overnight with shaking at 200 rpm at 37 °C.
NOTE: Prepare glycerol stocks of cell culture, store long-term at -80 °C or use immediately for recombinant protein expression.
- Obtain the translationally coupling gene (trpA and trpB) encoding the α– and β-subunits of the tryptophan synthase from bacterium Salmonella enterica serovar typhimurium cloned in the pEBA-10 expression vector22. Use pEBA-10 vector as a DNA template.
- Expression of the SUMO-αStTS and SUMO-βStTS subunits
- Inoculate fresh E. coli strain Rosetta (DE3) pLysS cells harboring SUMO-αStTS or SUMO-βStTS constructs or scrape some of the frozen glycerol stock into a 50 mL culture of LB containing 35 μg/mL kanamycin and chloramphenicol. Grow cells overnight with shaking at 200 rpm at 37 °C.
- Next morning, inoculate 5 mL of the overnight cell culture in a fresh and sterile 2x 1000 mL of LB containing 2% glycerol plus kanamycin and chloramphenicol (2.8 L Fernbach flask). Grow cell culture with shaking at 200 rpm at 37 °C.
NOTE: The expression of SUMO-αStTS or SUMO-βStTS in LB broth yields 125-150 mg of tagged protein per liter. Consider scaling protocol up or down to fulfil specific demands. - Induce recombinant protein expression when the OD600 reaches 0.6-0.8 by addition of isopropyl β-D-1 thiogalactopyranoside (IPTG) at a final concentration of 0.4 mM followed by incubation at 30 °C overnight with shaking at 200 rpm.
- Harvest the cells by centrifuging at 4,000 x g at 4 °C for 20 min. Remove the supernatant and re-suspend the cell pellets with cold lysis buffer 1 (50 mM Tris-Cl, pH 8.0, containing 500 mM NaCl, 5% glycerol, 10 mM 2-mercaptoethanol, and 40 mM imidazole-Cl) to a final volume of 60 mL.
NOTE: Cells can be stored long-term at -80 °C or used immediately for protein purification. To store cells, split the cell suspension into 2 x 50 mL disposable centrifuge conical tubes and keep cell pellets at -80 °C until the protein purification step.
- Purification of the α- and β-subunits of tryptophan synthase
NOTE: All procedures are to be conducted at 4 °C unless otherwise stated. To reduce purification time, equilibrate Ni-NTA agarose nickel-charged affinity columns and size exclusion column in buffer prior to protein purification or during recombinant protein expression.- Disrupt cell pellet by sonication using a digital sonifier with 1/2" Horn probe (or a similar equipment). Perform 20 cycles at 80% amplitude duty cycle on ice water bath using 10 s pulse and 20 s rest or until complete cell disruption.
- Centrifuge the cell lysate at 30,000 x g for 30 min. Aspirate supernatant, ensuring the pellet does not dislodge from tube. Filter the supernatant with a 0.45 μm filter unit on ice and flow it through a 15 mL Ni-NTA agarose nickel-charged affinity column pre-equilibrated in lysis buffer 1.
NOTE: Each Ni-NTA agarose column will be used to purify either SUMO-αStTS or SUMO-βStTS recombinant protein. Purification can be performed in a 2x 5 mL Ni-NiTA column attached to a fast protein liquid chromatography system. Purify one protein at a time. - Wash Ni-NTA agarose column in 100 mL of lysis buffer 1.
- Proceed with an 80 mL one-step elution with buffer E1 (25 mM Tris-Cl buffer, pH 7.8, containing 200 mM NaCl, 5% glycerol, and 400 mM imidazole-Cl).
NOTE: SUMO-protease tolerates up to 300 mM imidazole and the membrane of centrifugal filter devices tolerates up to 100 mM imidazole. We recommend performing an ammonium sulfate precipitation to remove high amounts of imidazole and decrease the purification time instead dilute the protein sample and waste time with protein concentration. - Assess the initial volume of the supernatant fraction. Slowly add small amounts of ammonium sulfate at a time, until a 60% saturation (39.48 g/ 100 mL) at 25 °C is reached. Gently stir the solution for 30 min and avoid bubbles. Centrifuge at 10,000 x g for 15 min.
- Aspirate and discard the supernatant fraction carefully. Resuspend the pellet fraction in 20 mL of sample buffer (20 mM Tris-Cl, pH 8.0, 300 mM NaCl and 5% glycerol).
- For His-SUMO-tag cleavage with SUMO-protease, add the recombinant fragment of Ubl-specific protease 1 from Saccharomyces cerevisiae in a 1:1000 ratio and incubate the mixture for 2 hours at 4 °C.
- Centrifuge the digestion product at 10,000 x g in 25 °C for 20 min to remove protein aggregates prior to load the sample through an affinity chromatography column.
- Remove traces of His6-SUMO tag passing the 20 mL resuspended sample on a 15 mL Ni-NTA agarose column, previously equilibrated in lysis buffer 1.
- Collect the pass-through sample containing the non-tagged αStTS or βStTS.
- Wash the Ni-NTA column in 20 mL of buffer 1 to collect leftovers of non-tagged αStTS or βStTS.
- Concentrate each subunit separately with a 15 mL 10 kDa cutoff centrifugal filter unit by spinning at 3,000 x g at 4 °C. Transfer the concentrated protein to a fresh 2.0 mL tube and microcentrifuge (10,000 x g, 10 min, 4 °C) to remove aggregates. Determine protein concentration24, prepare 1 mL aliquots at 20-25 mg mL-1, label, flash-freeze in liquid nitrogen, and store them at -80 °C.
NOTE: Pre-purified protein samples can be stored long-term at -80 °C or used immediately for protein purification on a size exclusion chromatography column (SEC). - Take a protein aliquot (20 mg) and microcentrifuge at 10,000 x g for 10 min to remove aggregates prior SEC.
- Load sample on a size exclusion chromatography column (e.g., HiPrep 16/60 Sephacryl S-200 HR) attached to a fast protein liquid chromatography at a flow rate of 0.5 mL min-1, previously equilibrated in SEC buffer (10 mM Tris-Cl, pH 7.8, 100 mM NaCl, 5% glycerol, 0.1 mM pyridoxal phosphate).
NOTE: To purify higher amounts of isolated αStTS or βStTS subunit and decrease the number of SEC rounds, load a 5 mL sample at 20-25 mg mL-1 on a size exclusion chromatography column at a flow rate of 1.5 mL min-1. - Assess the quality of αStTS or βStTS in the peak fraction using a 12% or a 15% sodium dodecyl sulfate-gel electrophoresis (SDS-PAGE), respectively, stained with Coomassie brilliant blue stain25.
- Concentrate protein with fresh 15 mL 10 kDa cutoff centrifugal filter units, determine protein concentration24, prepare 0.5 mL aliquots at 20-25 mg mL-1, label, flash-freeze in liquid nitrogen, and store them at -80 °C.
- Purification of the α2β2 StTS complex from the α- and β-subunits
NOTE: Equilibrate the size exclusion chromatography column (Sephadex S-200 HR or Superdex 200 pg) in SEC buffer (10 mM Tris-Cl, pH 7.8, 100 mM NaCl, 5% glycerol, 0.1 mM pyridoxal phosphate).- To purify the wild-type α2β2 StTS complex from the α- and β-subunits, thaw concentrate samples of αStTS and βStTS subunits at 4 °C.
- Combine aliquots (1.2 αStTS: 1.0 βStTS molar ratio) and incubated at 4 °C for 1 h.
NOTE: Excess of αStTS is necessary to ensure that most of βStTS will be incorporated into the α2β2 StTS complex. - Remove protein aggregates by microcentrifugation (10,000 x g, 10 min, 4 °C).
- Load the clarified supernatant onto the size exclusion column.
- Assess the quality of αStTS or βStTS in the peak fraction using a 12% or a 15% sodium dodecyl sulfate-gel electrophoresis (SDS-PAGE), respectively, stained with Coomassie brilliant blue stain25.
- Determine protein concentration24, prepare 250-1000 μL aliquots at 15-20 mg mL-1, flash-freeze in liquid nitrogen, and store at -80 °C.
2. Purification of the wild type or mutant form of the α2β2 StTS complex
- Site-directed mutagenesis to prepare mutant βStTS
- Use construct pEBA-10 expression vector22 as the DNA template during two-steps of polymerase chain reaction to introduce specific point-mutations in the β-chain TS polynucleotide sequence.
NOTE: This protocol can be used to introduce a single point mutation in either α-or β-subunit from Salmonella typhimurium tryptophan synthase. For additional, new oligonucleotide primers containing the desirable mutation must be appropriately designed. In this work, mutations βQ114A, βK167T, βS377A are listed. - Perform PCR reactions using pairs of nucleotide primers TS-FW-NcoI/Q114A-Rev, TS-FW-NcoI/K167T-Rev, and TS-FW-NcoI/S377A-Rev to generate fragments A1, B1, and C1, respectively.
NOTE: Oligonucleotide TS-NcoI-FW and TS-SacI-Rev, respectively, anneals upstream and downstream of the αβ StTS polynucleotide sequence cloned in the pEBA-10 vector.- Use high-fidelity DNA polymerase (e.g., Phusion) and the manufacturer’s protocol to amplify the DNA sequences. For a 50 µL PCR reaction, add 34 µL of nuclease-free water, 10 µL of 5x reaction buffer, 1 µL of 10 mM dNTPs, 1 µL of 10 µM forward primer, 1 µL of 10 µM reverse primer, 1 µL of Template DNA (200 ng), 1.5 µL of DMSO, 0.5 µL of DNA polymerase.
- For the PCR program, use a hot start (180 seconds at 98 °C) followed by 30 amplification cycles (30 seconds at 98 °C, 30 seconds at 55 °C and 120 seconds at 72 °C), and a final extension (300 seconds at 72°C).
NOTE: All molecular biology steps were followed as described in Molecular Cloning: A Laboratory Manual23. While a lowercase sequence corresponds to a restriction site, a bold and italic sequence corresponds to a mutation.
TS-FW-NcoI: 5’-CAA TTT CAC ACA GGA AAC AGA cca tgg-3’
Q114A-Rev: 5’-C AGA GGC GAC GCC GTG CGC ACC GGC GCC GGT TTC-3’
K167T-Rev: 5’-CTC GTT ACA GGC ATC TGT TAG CGT AGC GGA GCC-3’
S377A-Rev: 5’-C TTT ATC TCC GCG GCC AGC GAG ATT GAC CAC CAG-3’
- Perform PCR reactions using pairs of nucleotide primers TS-Rev-SacI/Q114A-FF, TS-Rev-SacI/K167T-FF, and TS-Rev-SacI/S377A-FF to generate fragments A2, B2, and C2, respectively.
TS-Rev-SacI (5’-TTA tgc gcg GCT GGC GGC TTT CAT GGC TGA G-3’
Q114A-FF 5’-GAA ACC GGC GCC GGT GCG CAC GGC GTC GCC TCT G-3’
K167T-FF 5’-GGC TCC GCT ACG CTA ACA GAT GCC TGT AAC GAG-3’
S377A-FF 5’-CTG GTG GTC AAT CTC GCT GGC CGC GGA GAT AAA G-3’ - Use polymerase chain reaction to individually amplify the partial fragments. Use a melting temperature of 55 °C and a polymerase extension time of 2 min.
- To perform the second round of PCR reactions, load the PCR product above on 0.8% agarose gel on 1x TAE at 6 V/cm and gel extract and gel purify the fragments of interest.
- Mix fragments A1/A2, B1/B2, and C1/C2 separately in equimolar amounts, heat denature the mixture for 10 min at 96 °C, and anneal at 25 °C. Extend the recombinant strands with a high-fidelity polymerase and deoxyribonucleotides according to the manufacturer’s protocol.
- To generate a high copy number of the full-length mutant DNA fragment, add oligo primers TS-FW-NcoI and TS-Rev-SacI to perform a second PCR.
- Load PCR product on 0.8% agarose in 1x TAE buffer at 6 V/cm, gel extract the DNA band of interest, gel purify the PCR fragment, and proceed with appropriate restriction digestion for 2 hours at 37 °C following the appropriate buffer and conditions recommended by the manufacturer.
- Digest at least 200 ng of each DNA fragment with appropriate restriction enzymes and conditions recommended by the manufacturer for 2 hours at 37 °C. To set up a 50 μL restriction digestion reaction add 34 µL of nuclease-free water, 10 µL of DNA (200 ng), 5 μL of 10x reaction buffer, 0.5 µL of restriction enzyme 1, and 0.5 µL of restriction enzyme 2. Load digestion product on 0.8% agarose in 1x TAE buffer at 6 V/cm and purify the digested PCR fragment.
- Digest 200 ng of pEBA-10 construct with restriction enzymes NcoI and SacI following the manufacturer’s recommendation. Load the digestion product on 0.8% agarose gel in 1x TAE buffer at 6 V/cm, excise the band correspondent to the vector, and gel purify the digested vector.
- Ligate 100 ng of vector with 50 ng of PCR fragment, previously digested and purified, with T4 DNA ligase for 2 hours at 25 °C. Transform the constructed plasmid into competent cells E. coli strain DH10B cells. Plate cells on LB agar plates containing 50 μg/mL ampicillin. Incubate the plate inverted overnight at 37 °C.
- Pick a single colony (without any satellite colonies) from each cell transformation and disperse it in 5 mL of LB medium containing ampicillin. Grow cells overnight with shaking at 200 rpm at 37 °C. Prepare 10 μg of ultra-pure plasmid DNA from each engineered construct. Prepare glycerol stocks of cell culture (turn cell suspension in 16% final concentration of sterile glycerol) and store long-term at -80 °C.
- Perform DNA sequencing to confirm the full-length sequence encoding the α- and β-subunits of tryptophan synthase. Confirm each individual single mutation and discard any plasmid construct containing undesirable random mutation. The oligonucleotide primers used in this study are listed below.
TS-1F: 5’-ATGACAACACTTCTCAAC-3’
TS-1R: 5’-GAAATGCCAGAACATTAC-3’
TS-2F: 5’-CAGTCGCCGAACGTC-3’
TS-3F: 5’-GATGATGCAAACAGC-3’
TS-4F: 5’-CTGGCATTGAACAGTC-3’
TS-5F: 5’-CGTTGCATCATCTCATTG-3’
NOTE: Oligonucleotide primers TS-1F, TS-1R, TS-2F, and TS-3F anneal on the polynucleotide sequence of the β-subunit (GenBank accession code: CP051286.1). Primers TS-4F and TS-5F anneal on the polynucleotide sequence of the α-subunit (GenBank accession code: CP053865.1). - Use 200 ng of each plasmid construct (pEBA-10-βQ114A, pEBA-10-βK167T, and pEBA-10-βS377A) to transform competent cells of E. coli expression strain CB149, lacking the trp operon26. After successful colony formation, pick one single colony and grow the cells in 5 mL of LB medium containing 50 μg/mL ampicillin. Culture in the bacterial incubator at 37 °C overnight. Prepare glycerol stocks of cell culture, store long-term at -80 °C or use immediately for recombinant protein expression.
NOTE: A single a single point mutation in either α-or β-subunit from Salmonella typhimurium tryptophan synthase may impair enzyme function or lower synthesis of L-Tryptophan in E. coli strain CB149. Please consider using either E. coli strain BL21(DE)pLys-S or Rosetta (DE3)pLys-S if no expression or lower amounts of recombinant α2β2 StTS complex is detected in a SDS-PAGE gel.
- Use construct pEBA-10 expression vector22 as the DNA template during two-steps of polymerase chain reaction to introduce specific point-mutations in the β-chain TS polynucleotide sequence.
- Expression of wild type and mutant form of α2β2 StTS complex in E. coli
- Grow E. coli expression strain harboring the desirable construct in a fresh and sterile 50 mL of LB medium containing 50 μg/mL ampicillin. Grow cells overnight with shaking at 200 rpm at 37 °C.
- Add 5 mL of the overnight cell culture in a fresh and sterile 2x 1000 mL of LB containing 2% glycerol plus ampicillin (2.8 L Fernbach flask). Grow cell culture with shaking at 200 rpm at 37 °C.
- Induce recombinant protein expression when the OD600 reaches 0.6-0.8 by addition of IPTG at a final concentration of 0.4 mM followed by incubation at 30 °C overnight with shaking at 200 rpm.
- Harvest the cells by centrifuging at 4,000 x g in 4 °C for 20 min. Remove the supernatant and re-suspend the cell pellets with cold lysis buffer 2 (50 mM Tris-Cl, pH 7.80, containing 100 mM NaCl, 5 mM dithiothreitol, 1 mM EDTA, and 1 mM PMSF) to a final volume of 50 mL buffer.
NOTE: Cells can be stored long-term at -80 °C or used immediately for protein purification. To store cells, split the cell suspension into 2 x 50 mL disposable centrifuge conical tubes and keep cell pellets at -80 °C until the protein purification step. The expression of mutant and wild type form of α2β2 StTS complex in LB broth yields 125-150 mg of protein per liter. Consider scaling protocol up or down to fulfill specific demands.
- Purification of wild type or mutant form of α2β2 StTS complex
NOTE: The following protocol is intended to purify the non-tagged recombinant wild type or mutant form of the recombinant α2β2 StTS complex within 1 day, depending on skill set and efforts. To reduce purification time, equilibrate size exclusion column in buffer prior protein purification/expression. Purification of wild type or mutant α2β2 StTS complex is carried out by a two-step purification comprising ammonium sulfate fractionation and a size exclusion chromatography. This protocol yields 60-100 mg of pure complex from 1 L of LB medium.- Disrupt cell pellet by sonication using a digital sonifier with 1/2" Horn probe (or a similar equipment). Perform 20 cycles at 80% amplitude duty cycle on ice water bath using 10 s pulse and 20 s rest or until complete cell disruption.
- Centrifuge the cell lysate at 30,000 x g for 30 min at 25 °C. Aspirate supernatant, ensuring the pellet does not dislodge from tube. Filter the clarified supernatant fraction with a 0.45 μm filter at room temperature.
- Assess the initial volume of the clarified supernatant fraction. Slowly add small amounts of ammonium sulfate at a time, until a 20% saturation is reached (11.51 g / 100 mL). Carry out ammonium sulfate fractionation out at 25 °C. Gently stir the solution for 10 min and avoid bubbles.
- Centrifuge at 30,000 x g for 10 min at 25 °C. Transfer the 20% supernatant fraction into a clean flask. Gently resuspend 20% pellet fraction in 20 mL of sample buffer 2 (50 mM Tris-Cl, pH 7.80, containing 100 mM NaCl, 1 mM EDTA, and 2 mM dithiothreitol) and prepare a sample to run SDS-PAGE gel. Discard the 20% pellet fraction.
- Assess the initial volume of the 20% supernatant fraction. Add ammonium sulfate to 30% saturation is reached (5.94 g / 100 mL), stir solution, avoid bubbles, and centrifuge as before. Transfer the 30% supernatant fraction into a clean flask. Gently resuspend 30% pellet fraction in 20 mL of sample buffer 2 and prepare a sample to run SDS-PAGE gel. Discard the 30% pellet fraction.
- Assess the initial volume of the 30% supernatant fraction. Add ammonium sulfate at a 40% saturation is reached (6.14 g / 100 mL), stir solution, avoid bubbles, and centrifuge as before. Transfer the 40% supernatant fraction into a clean flask. Gently resuspend 40% pellet fraction in 10 mL of sample buffer 2 and prepare a sample to run SDS-PAGE gel. Discard the 40% supernatant fraction.
NOTE: Assess the quality of αβStTS complex using samples of the 30% and 40% supernatant and pellet fractions in a 12% SDS-PAGE gel25. If you verify that any other mutant αβStTS complex is in the 40% supernatant fraction, proceed with a 60% saturation of ammonium sulfate (13.0 g / 100 mL). Pre-purified protein aliquots of the pre-purified α2β2 StTS complex can be stored long-term at -80 °C or used immediately for protein purification on a size exclusion chromatography column (SEC). - Microcentrifuge the protein sample at 10,000 x g, 20 min, 4 °C and load the supernatant fraction on a HiPrep 16/60 Sephacryl S-200 HR column attached to a fast protein liquid chromatography at a flow rate of 0.5 mL min-1, previously equilibrated in SEC buffer (10 mM Tris-Cl, pH 7.8, 100 mM NaCl, 5% glycerol, 0.1 mM pyridoxal phosphate).
NOTE: To purify large amounts of complex, load a 5 mL sample at 20-25 mg mL-1 on a HiLoad 26/600 Superdex 200 pg column at a flow rate of 1.5 mL min-1. - Assess samples collected along the ammonium sulfate fractionation and peak fractions from the SEC column on a 12% SDS-PAGE gel stained with Coomassie brilliant blue stain25.
- Concentrate the wild type or mutant α2β2 StTS complex with a 15 mL 100 kDa cutoff centrifugal filter unit by spinning at 3,000 x g at 4 °C. Transfer the concentrated protein to a fresh 2.0 mL tube and microcentrifuge (10,000 x g, 10 min, 4 °C) to remove aggregates.
- Determine protein concentration24, prepare 0.5 mL aliquots at 20-25 mg mL-1, label, flash-freeze in liquid nitrogen, and store them at -80 °C.
3. Optimized crystallization for wild type and mutant form of the α2β2 StTS complex
NOTE: The initial crystallization condition for the α2β2 StTS complex was previously reported in conditions containing 12% PEG 8,000 and 2 mM spermine22.
- Prepare stock solutions prior to the crystallization assays to achieve a better crystallization reproducibility. To perform structural studies with TS, crystallize protein with Na+ or Cs+ ion at the metal coordination site of the bienzyme complex.
- Prepare 5 mL stock solution of 200 mM spermine in water and keep 500 µL aliquots at -20 °C.
- Prepare 50 mL stock solution of 30% (w/v) PEG 8000 in water and keep 25 mL aliquots in a 50 mL disposable centrifuge conical flasks at 25 °C.
- Prepare a 25 mL stock solution of 1 M CsCl in water and keep at 25 °C.
- Prepare 25 mL stock solution of 1 M NaCl in water and keep at 25 °C.
- Prepare 3x 50 mL stock solution of 1 M bicine and titrate with CsOH or NaOH to obtain buffered solutions at pH 7.6, 7.8, and 8.0. Keep 25 mL aliquots at 4 °C.
- Thaw a sample of α2β2 StTS complex (20-25 mg mL-1) on an ice bath.
- Microcentrifuge sample at 10,000 x g for 10 min at 25 °C to remove protein aggregates.
- Transfer the cleared supernatant fraction into a clean microcentrifuge tube.
- Estimated protein concentration24 and dilute protein aliquots (150-200 µL) at 15 mg mL-1 with 50 mM bicine-CsOH or -NaOH, pH 7.8 containing 50 mM CsCl or NaCl. Keep protein sample at 25 °C.
- Prepare the 500 µL reservoir solutions for 3x 24-well sitting drop plates in sterile 1.5 mL labeled microcentrifuge tubes. The reservoir solution contains 50 mM bicine-CsOH or -NaOH, 50 mM CsCl or NaCl, and PEG 8000. Vary the concentration of PEG 8000 (6-11%) on the plate columns and the concentration of spermine (2-8 mM) on the plate rows. Change buffer pH (pH 7.6, 7.8 and 8.0) for each set.
- Cap the tubes and vortex vigorously at least 10 sec. Centrifuge tubes at 10,000 x g for 10 min at 25 °C to remove bubbles. Dispense 500 µL buffered solution in each labeled reservoir.
- Use a P-10 micropipette and dispense 5 µL of protein solution at 15 mg mL-1 per each sitting drop well. Avoid bubbles. Add 5 µL of each correspondent reservoir solution to the protein drop. Avoid bubbles during mixing and do not pipette up and down to homogenize the mixture.
- Tape the plate with a transparent adhesive tape and store the plate at 25 °C. Crystals appear in 2-5 days and grow to their full dimensions within two weeks.
4. X-ray diffraction data collection and α2β2 StTS complex structure solution
NOTE: Prior to X-ray diffraction data collection, prepare cryoprotectant solution for each crystal in advance. Use the specific reservoir solution to prepare 3 aliquots containing increasing concentrations of dimethyl sulfoxide in solution (10, 20, and 30% v/v) and specific ligand (s). Dimethyl sulfoxide was found to be a better cryoprotectant than glycerol, ethylene glycol, and PEG 200-300.
- Harvest a large single crystal using a cryoloop under the stereoscopic microscope.
- Pipette 2 μL of each cryoprotectant solution containing the precipitation solution plus higher concentrations of dimethyl sulfoxide and ligand (s) onto a new cover slide.
- Sequentially, soak crystal in each drop and let the crystal equilibrate for 30 s in the cryoprotectant solution.
- Flash-cooling the cryoprotected crystal using a gaseous nitrogen stream at -173 °C (100 K) or immerse in liquid nitrogen for long-term storage in pucks.
NOTE: Fill a foam Dewar with liquid nitrogen, pre-cool a crystal puck, store crystals in the cryogenic storage Dewar, and ship crystals to the synchrotron using a dry shipper. - Proceed with X-ray diffraction data collection at -173 °C. Record X-ray diffraction data using a 0.5-4.0 s exposure time and 0.5° oscillations. Rotate crystal 180-360°.
- Process the X-ray diffraction images with iMosflm27 to generate reflection file in the appropriate space group. Generally, α2β2 StTS crystals belong to the space group C 121 (C2).
- Scale together multiple observations of reflections with Scala28, implemented in the CCP4 package29.
- Solve the structure of the high-resolution α2β2 StTS complex by molecular replacement using MolRep30 and an appropriate search model. Inspect the model and the electron density map in Coot31 after a successful structure solution.
NOTE: There are many TS crystals structures deposited in the Protein Data Bank with different ligand (s). For a better molecular replacement step and decreased time fitting the newer crystal structure, use the best search model containing ligand (s) of interest. Proceed with crystal structure refinement in CCP429,30 or Phenix31. - Make manual adjustments to the model using Coot32, followed by automatically refinement with Refmac29,33 or phenix.refine31,34. Build and refine the final model by running iterative rounds of model building in Coot32 and automated refinement.
- Proceed with coordinate and structure factors deposition in the Protein Data Bank web site.
Representative Results
Purification of the α– and β-subunits of the tryptophan synthase
The α-subunit (αStTS) and the β-subunit (βStTS) of the Salmonella typhimurium tryptophan synthase were subcloned in the modified pET SUMO vector. Figure 1A shows representative SDS-PAGE results of two strong bands corresponding to the His6-SUMO-αStTS (lane αON) and His6-SUMO-βStTS (lane βON) fusion protein. The purification protocol described in this work allowed purification of both subunits individually within 2 days. The first day was used to purify each protein by Ni-NTA affinity chromatography, ammonium sulfate precipitation followed by His-SUMO-tag cleavage, removal of His-SUMO-tag traces, and protein concentration. Figure 1B and 1C show representative SDS-PAGE results of the α-subunit and the β-subunit purification, respectively. On the second day, the concentrate α-subunit, β-subunit, and the α2β2 StTS complex from the α- and β-subunits were loaded on a size exclusion chromatography column. Figure 1D shows a typical elution profile of αStTS, βStTS, and α2β2StTS complexes on a S-200 HR size exclusion column. Figure 1E shows a representative SDS-PAGE result of the collected peak fractions. The purest peak fractions were pooled, concentrated, and the α2β2 StTS complex was used for protein crystallization studies.
Purification of the wild type and mutant α2β2 StTS complex
Another rapid and efficient protocol to purify the wild type and mutant form of the α2β2 S. typhimurium tryptophan synthase complex is described in this work. Figure 2 shows a representation of the pEBA-10 construct containing the wild type translationally coupling gene (trpA and trpB) encoding for the α– and β-subunits22. The two-step PCR mutagenesis protocol to generate mutant forms of the α2β2 StTS complex is depicted in Figure 3.
The coding regions of mutant α2β2 StTS complex in pEBA10 were confirmed by DNA sequencing and used to transform E. coli strain CB149 cells26. The wild type and mutant form of the α2β2 StTS complex were overexpressed and the recombinant proteins were purified successfully within 1-2 days. Ammonium sulfate fractionation at room temperature readily removed most of the contaminant proteins from the heterologous expression system (Figure 4A, lanes 20P, 30P and 40S). A representative elution profile with relative elution position of α2β2 StTS (143.06 kDa) complex on a HiPrep 16/60 Sephacryl S-200 HR size exclusion chromatography column is shown in Figure 4B. The purity of the excluded peak fractions was SDS-PAGE analyzed before pooling (Figure 4C).
Optimization of wild type and mutant α2β2 tryptophan synthase complex crystallization
Aliquots of wild type and mutant α2β2 StTScomplex at 15 mg ml-1 were used to set up 24-well sitting drop plates. Typically, droplets consisting of 5 µL protein solution and the equivalent volume of reservoir solution were equilibrated against 500 µL of reservoir solution (Figure 5). Spermine is required to crystallize the wild type and mutant α2β2 StTS complex22. While the final concentration of spermine to crystallize the wild type is 2 mM, the concentration of spermine to crystalize the mutant complex in this work showed to be slightly higher (4-8 mM).
Large single crystals were obtained through a fine crystallization optimization, varying PEG 8000 (6-11%) and bicine buffer pH. Crystals with different morphologies appeared in 2-5 days and crystals grew to full size within two weeks (Figure 6). Prior to X-ray diffraction data collection, crystals were soaked in cryoprotectant solution (reservoir buffer containing up 30% dimethyl sulfoxide). The optimized process resulted in quality crystals suitable for X-ray diffraction measurements at near atomic resolution.
X-ray diffraction data analysis
A crystal structure of the wild type α2β2 StTS complex was prepared with methods described in this article and X-ray diffraction data was collected at near atomic resolution. The crystal was soaked in cryoprotective solution containing F9 inhibitor (2- ({[4- (Trifluoromethoxy)Phenyl]Sulfonyl}Amino)Ethyl Dihydrogen Phosphate) and L-Tryptophan.
A complete X-ray diffraction data set was collected on the SIBYLS synchrotron beamline 12.3.1 at the Advanced Light Source (Berkeley-CA) by rotating the crystal 360° in increments of 0.5°. X-ray diffraction intensities were processed, and data-collection statistics are summarized in Table 1. Symmetry analysis indicates that the crystal belonged to the monoclinic space group C2. The unit-cell parameters are a = 182.55, b = 59.30, c = 67.37Å, α = 90.00, β = 94.82, γ = 90.00°. The calculated value of the Matthews coefficient (Vm = 2.57 Å3 Da-1) suggests the presence of one TS heterodimer molecule (αβ StTS) in the asymmetric unit of the crystal with a solvent content of 52.08%35,36. All X-ray data were collected at low temperatures (100 K) to improve the diffraction quality and decrease the radiation decay. The α2β2 StTS crystal structure in complex was solved by the molecular replacement method using the wild type αβ StTS model in complex with the inhibitor F9 at the α-site and cesium ion at the metal coordination site (PDB ID code: 4HT3). The final coordinate file and the structure factors were deposited in the PDB with accession code 5CGQ (Figure 7A). The crystal structure of the wild type α2β2 StTS complex with inhibitor F9 at the enzyme α-site (Figure 7B), cesium ion at the metal coordination site (Figure 7C), the cofactor pyridoxal 5'-phosphate covalently bonded to βLys87 (Figure 7D), and the product L-tryptophan at the enzyme β-site (Figure 7E) was solved at 1.18 Angstrom resolution. Model 5CGQ is the highest resolution α2β2 StTS crystal structure deposited in the PDB to date.
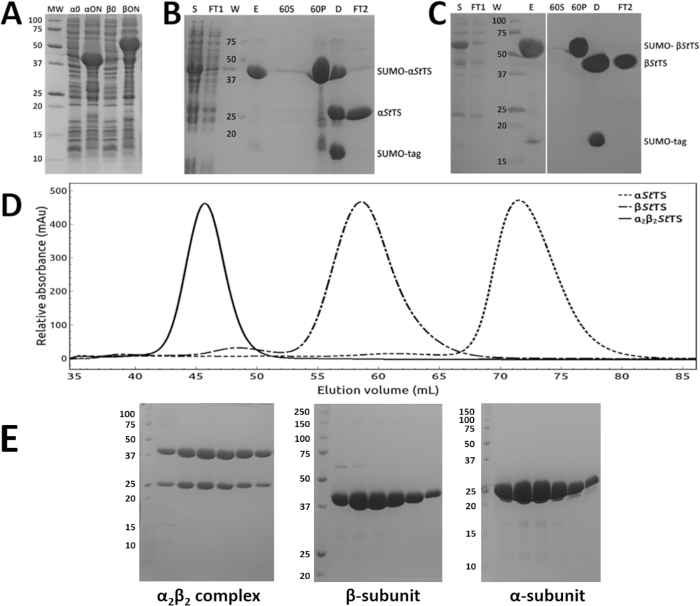
Figure 1: Purification of the α- and β-subunits and the α2β2StTS complex. (A) Recombinant protein expression. 12% SDS-PAGE gel of the overnight expression profile of SUMO-αStTS (αON) and SUMO-βStTS (βON) after IPTG induction at 30 °C (α/β0 prior IPTG induction). (B, C) Ni-NTA affinity chromatography followed by ammonium sulfate precipitation (60% saturation), (S) clarified crude extract (FT1) Ni-NTA column pass through sample (W) column wash sample (E) eluate sample (60S) and (60P) supernatant and precipitate fractions after high-speed centrifugation, respectively (D) SUMO-protease digestion product (FT2) Ni-NTA column pass through sample containing the tag-less αStTS or βStTS subunit. (D) elution profile of αStTS subunit (28.67 kDa), βStTS subunit (42.86 kDa), and α2β2 StTS complex (143.06 kDa) with a HiPrep 16/60 Sephacryl S-200 HR size exclusion chromatography column. Each run was performed separately. (E) SDS-PAGE gels of the excluded peak fractions from each individual chromatography. While 15% SDS-PAGE gels were prepared to analyze α2β2 StTS complex and αStTS subunit, a 12% SDS-PAGE gel was prepared to analyze the βStTS subunit. Lane MW, molecular-weight markers in kDa. Please click here to view a larger version of this figure.
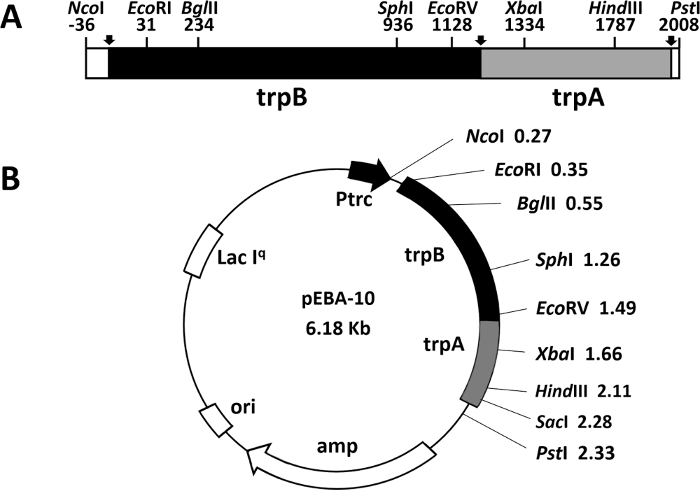
Figure 2: Representation of the construct pEBA-10. (A) Representation of the wild type translationally coupling gene (trpA and trpB) encoding the α– and β-subunits of the tryptophan synthase from bacterium Salmonella enterica serovar typhimurium (Yang, Ahmed et al. 1996). (B) The vector contains an ampicillin resistance (amp) gene, a replication origin (ori), a lacIq gene to better shutdown a lac promoter in absence of IPTG inducer, and the LacI-repressed promoter. Please click here to view a larger version of this figure.
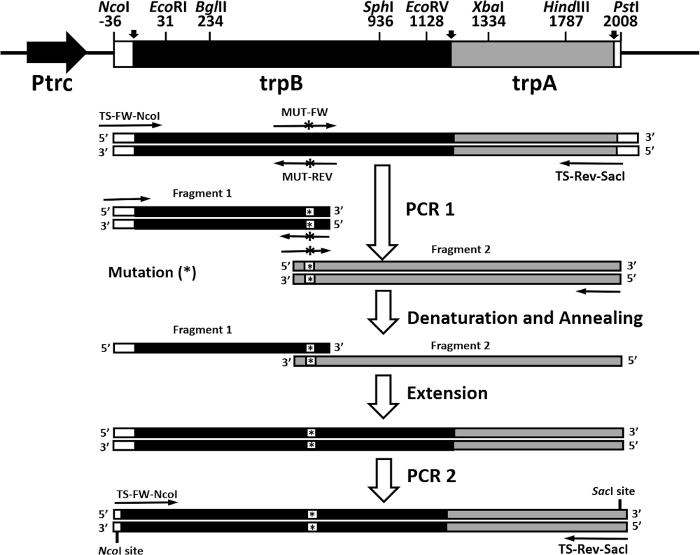
Figure 3: Overall representation of the two-step PCR mutagenesis protocol. The pEBA-10 vector was used as a DNA template. The first round of PCR was prepared with primers TS-FW-NcoI and MUT-REV (a reverse primer containing a mutation) to generate the first fragment and primers TS-Rev-SacI and MUT-FW (a forward primer containing a mutation) to generate the second fragment). Fragments were gel purified and equimolarly combined, heat denatured, and annealed. The recombinant strands were extended with polymerase and deoxyribonucleotides. The second round of PCR was prepared with primers TS-FW-NcoI and TS-Rev-SacI. Please click here to view a larger version of this figure.
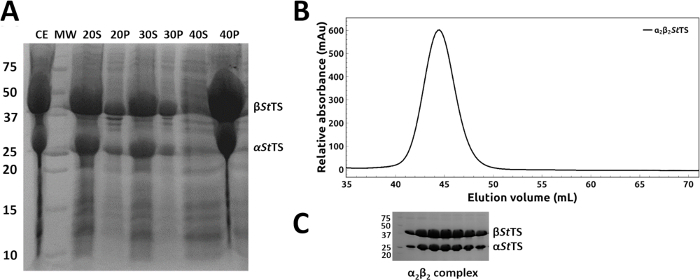
Figure 4: Purification of wild type and mutant form of α2β2 StTS complex. (A) 12% SDS-PAGE gel of samples collected along the ammonium sulfate precipitation using 20, 30, 40 and 50% ammonium sulfate saturation at room temperature: (CE) crude extract (S) and (P) supernatant and precipitate fractions after high-speed centrifugation. (B) Elution profile of α2β2 StTS (143.06 kDa) complex with a HiPrep 16/60 Sephacryl S-200 HR size exclusion chromatography column. (C) 12% SDS-PAGE gel picture of the excluded peak fractions. Lane MW, molecular-weight markers in kDa (Precision Plus Protein Unstained Standards, Bio-Rad). Please click here to view a larger version of this figure.
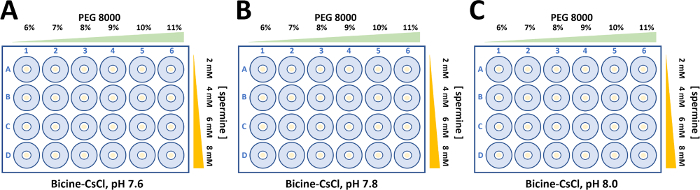
Figure 5: Crystallization optimization for wild type and mutant form of α2β2 StTS complex. Crystals were grown in 50 mM Bicine-CsOH buffer containing 50 mM CsCl2. The concentration of polyethylene glycol 8000 (6-11%) and spermine (2-8 mM) were varied to obtain single large crystals forms to perform structural studies by X-ray protein crystallography. (A) Bicine-CsOH, pH 7.6. (B) Bicine-CsOH, pH 7.8. (C) Bicine-CsOH, pH 8.0. Please click here to view a larger version of this figure.
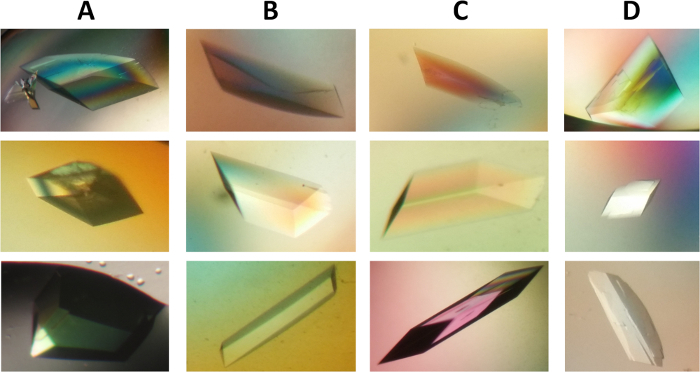
Figure 6: Photomicrograph of crystals of wild type and mutant form of α2β2 StTS complex. Crystals differ in morphology, but they belong to the space group C2. The crystals grew to their full dimensions in the final conditions after two weeks. Crystals of approximately 0.20 x 0.15 x 0.10 mm in size. (A-D) PLP holo-crystals in complex with cesium ion at the metal coordination site of the wild type form (column A), mutant form α2β2 βQ114A (column B), α2β2 βK167T (column C), and α2β2 βS377A (column D). Please click here to view a larger version of this figure.
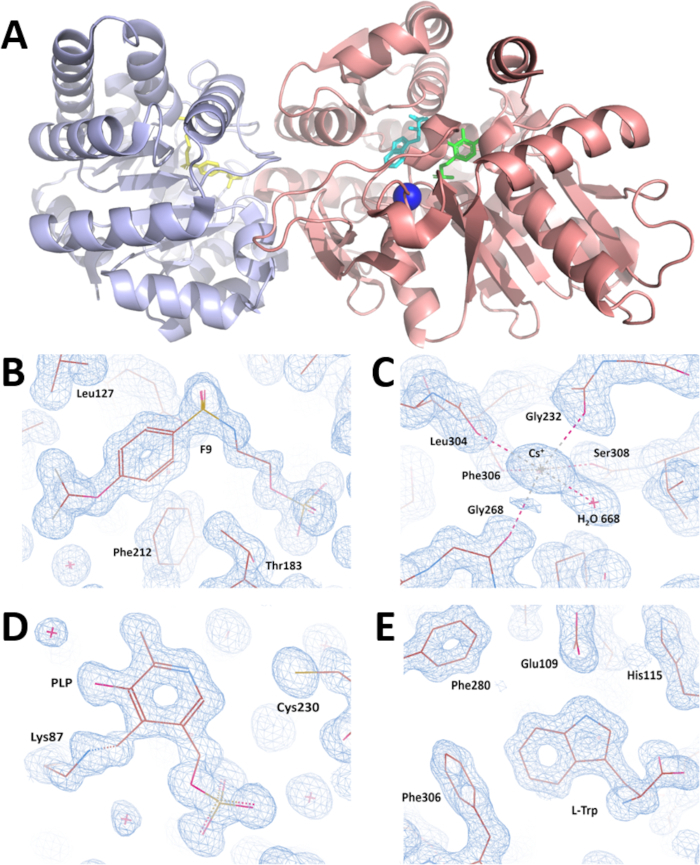
Figure 7: Overall visualization of crystal structure and validation of electron density maps obtained after crystal structure refinement. (A) crystal structure of the wild type α2β2 StTS complex with inhibitor F9 at the enzyme α-site (yellow colored), cesium ion at the metal coordination site (blue colored), the cofactor pyridoxal 5'-phosphate covalent bonded to βLys87 (green colored), and the product L-tryptophan at the enzyme β-site (cyan colored) at 1.18 Angstrom resolution. While the α-subunit is colored in light blue, the β-subunit is colored in salmon. (B-E) Electron density maps contoured at 1.0 r.m.s. level around (B) inhibitor F9 (C) cesium ion (D) pyridoxal-5′-phosphate, and (E) L-Tryptophan. Please click here to view a larger version of this figure.
| Data collection and processing | |
| X-ray source / Beam line | ALS Beamline 12.3.1 |
| Wavelength (Å) | 10,000 |
| Resolution (Å) | 40.00 – 1.18 (1.24 – 1.18) |
| Total number of reflections | 2151280 (252941) |
| Total number unique reflections | 231646 (32187) |
| Space group for indexing, scaling and merging | C 1 2 1 |
| Cell dimensions | |
| a, b, c (Å) | 182.55, 59.30, 67.37 |
| α, β, γ (°) | 90.00, 94.82, 90.00 |
| Mosaicity | 0.61 |
| Matthews volume VM (Å3 Da-1) | 2.57 |
| Rmeas (%) | 8.6 (93.0) |
| <I/σ(I)> | 14.7 (3.0) |
| CC1/2 (%) | 0.999 (0.778) |
| Completeness (%) | 98.6 (94.2) |
| Multiplicity | 9.3 (7.9) |
| Refinement statistics | |
| Rwork/Rfree (%) | 14.04 / 16.05 |
| RMSD bond length (Å) | 0.0120 |
| RMSD bond angle (°) | 14,059 |
| Ramachandran favored | 515 (96.44%) |
| Ramachandran allowed | 16 (3.00%) |
| Ramachandran disallowed | 3 (0.56%) |
Table 1: Data collection and processing. Values in parentheses are for the outer shell.
Discussion
We have successfully engineered mutant form α2β2 βQ114A, α2β2 βK167T, and α2β2 βS377A StTS complexes for structure-function correlation studies. Initially, we have tried to purify the mutants using a previous purification protocol22, which requires α2β2 StTS complex crystallization with PEG 8000 and spermine during purification. Although crystallization rate depends on the mutant form and on the concentration of the complex in solution, being difficult to predict when crystals appear in a large solution volume. Crystallization could be achieved either after long periods (48-96 h) or being necessary the addition of extra amounts of PEG 8000 after crystallization initiation15.
Unfortunately, mutant forms of α2β2 StTS complex presented in this work were not successfully purified using this protocol since they failed to crystallize during the initial steps of the protocol, impairing crystallographic studies. Therefore, we have created a simple and efficient purification protocol comprising ammonium sulfate fractionation and size exclusion chromatography, which give high yields of wild type and mutant form of α2β2 StTS complex. This protocol is faster (1-2 days) and reproducible when compared with the previous protocol (5-7 days)15,22, since there is no crystallization requirements and protocol troubleshooting along purification. In addition, we have created new expression constructs and protocol to purify high amounts of the α-subunit, β-subunit and the reconstitute α2β2 StTS complex from the isolates.
Future application includes the recombination of wild type and mutant sub-units to perform functional and structural studies. Mutant α2β2 βQ114A, α2β2 βK167T, and α2β2 βS377A complex crystallized in conditions containing higher concentrations of spermine (4-8 mM) when compared with the wild type form (2 mM). Therefore, it is worthwhile spending time on improving the quality of protein crystals by varying the concentration of precipitants and buffer pH. Single large crystal forms randomly grew in bicine buffered solution (pH 7.6-8.0) containing 6-11% PEG 8,000. The methods described in this work will be used to prepare crystal structures of the wild type and mutant forms of the α2β2 complex with different ligands within the α- and β-active sites, mimicking different intermediates and transition states involved in the conversion of indole and serine to tryptophan. The crystal structures of these mutants are anticipated to provide new insights into the mechanism and roles played by key residues in L-Tryptophan synthesis.
Disclosures
The authors have nothing to disclose.
Acknowledgements
This work was supported by the US National Institute of Health (GM097569).
Materials
| 15 mL 10 kDa filter | MilliporeSigma | UFC901024 | centrifugal filter unit |
| 15 mL 100 kDa filter | MilliporeSigma | UFC910024 | centrifugal filter unit |
| 2 mL cryogenic vials | Corning | CLS430489 | Cryogenic vials |
| 2 mL microcentrifuge tubes | Fisher Scientific | 05-408-141 | microcentrifuge tubes |
| 24-well Cryschem Plate | Hampton Research | HR3-158 | 24-well sitting drop plates |
| 2-mercaptoethanol | Fisher Scientific | O3446I-100 | Chemical |
| 50 mL centrifuge conical tubes | Thermo Scientific | 12-565-270 | centrifuge conical tubes |
| AB15 ACCUMET Basic | Fisher Scientific | 13-636-AB15 | pH meter |
| Agarose | Fisher Scientific | BP1356-100 | Agarose gel |
| ammonium sulfate | Fisher Scientific | A702-500 | Chemical |
| Ampicillin | Fisher Scientific | BP1760-5 | Antibiotic |
| Bacterial incubator | Fisher Scientific | S35836 | incubator. |
| BamHI | New England Biolabs | R0136S | Restriction enzyme |
| bicine | Fisher Scientific | BP2646100 | Chemical |
| Branson 450 Digital Sonifier | Brason | B450 | Cell disruptor |
| Cesium chloride | Fisher Scientific | BP210-100 | Chemical |
| Cesium hydroxide | Acros Organics | AC213601000 | Chemical |
| Chloramphenicol | Fisher Scientific | BP904-100 | Antibiotic |
| dimethyl sulfoxide | Fisher Scientific | D1391 | Chemical |
| dithiothreitol | Fisher Scientific | BP172-5 | Chemical |
| DNA Polymerase | Thermo Scientific | F530S | HF polymerase |
| dNTP Set | Invitrogen | 10-297-018 | dNTPs set |
| EcoRI | New England Biolabs | R0101S | Restriction enzyme |
| Ethylenediaminetetraacetic acid | Fisher Scientific | S311-100 | Chemical |
| Excella E25R Orbital Shaker | Eppendorf New Brunswick | M1353-0004 | Orbital incubator |
| GE AKTA Prime Plus | GE Healthcare | 8149-30-0004 | FPLC |
| Gel Extraction Kit | Invitrogen | K210012 | DNA purification kit |
| Glycerol | Fisher Scientific | G33-500 | Chemical |
| HindIII | New England Biolabs | R0104S | Restriction enzyme |
| His-Trap columns | GE Healthcare | GE17-5255-01 | 5 mL Histrap column |
| imidazole | Fisher Scientific | O3196-500 | Chemical |
| IPTG | Thermo Fisher Scientific | R0392 | Inducer |
| Kanamycin | Fisher Scientific | BP906-5 | Antibiotic |
| Kelvinator Series-100 | Kelvinator | discontinued | Ultra low freezer |
| LB broth | Fisher Scientific | BP1426-500 | Liquid broth |
| Luria Bertani agar | Fisher Scientific | BP1425-2 | Solid broth |
| NaCl | Fisher Scientific | S271-500 | Chemical |
| NcoI | New England Biolabs | R0193S | Restriction enzyme |
| Ni-NTA affinity beads | Thermo Fisher Scientific | R90115 | Ni-NTA agarose beads |
| PEG 8000 | Fisher Scientific | BP233-100 | Chemical |
| phenylmethylsulfonyl fluoride | Fisher Scientific | 44-865-0 | Chemical |
| pyridoxal phosphate | Acros Organics | AC228170010 | Chemical |
| S-200 HR | Cytiva | 45-000-196 | Size exclusion column |
| SacI | New England Biolabs | R0156S | Restriction enzyme |
| Sodium hydroxide | Fisher Scientific | S318-100 | Chemical |
| Sorvall RC-5B centrifuge | Sorvall | 8327-30-1004 | Floor cetrifuge |
| Spermine | Acros Organics | AC132750010 | Chemical |
| Superdex 200 prep grade | Cytiva | 45-002-491 | Size exclusion column |
| T4 DNA ligase | New England Biolabs | M0202S | DNA liagse |
| Tris | Fisher Scientific | BP152-500 | Chemical |
| Ubl-specific protease 1 | Thermo Scientific | 12588018 | SUMO Protease |
References
- Miles, E. W. Tryptophan synthase: a multienzyme complex with an intramolecular tunnel. Chemical Record. 1 (2), 140-151 (2001).
- Raboni, S., Bettati, S., Mozzarelli, A. Tryptophan synthase: a mine for enzymologists. Cellular and Molecular Life. 66 (14), 2391-2403 (2009).
- Dunn, M. F. Allosteric regulation of substrate channeling and catalysis in the tryptophan synthase bienzyme complex. Archives of Biochemistry and Biophysics. 519 (2), 154-166 (2012).
- Chaudhary, K., Roos, D. S. Protozoan genomics for drug discovery. Nature Biotechnology. 23 (9), 1089-1091 (2005).
- Caldwell, H. D., et al. Polymorphisms in Chlamydia trachomatis tryptophan synthase genes differentiate between genital and ocular isolates. Journal of Clinical Investigation. 111 (11), 1757-1769 (2003).
- Kulik, V., et al. On the structural basis of the catalytic mechanism and the regulation of the alpha subunit of tryptophan synthase from Salmonella typhimurium and BX1 from maize, two evolutionarily related enzymes. Journal of Molecular Biology. 352 (3), 608-620 (2005).
- Barends, T. R., et al. Structure and mechanistic implications of a tryptophan synthase quinonoid intermediate. Chembiochem. 9 (7), 1024-1028 (2008).
- Hatanaka, M., White, E. A., Horibata, K., Crawford, I. P. A study of the catalytic properties of Escherichia coli tryptophan synthetase, a two-component enzyme. Archives of Biochemistry and Biophysics. 97, 596-606 (1962).
- Henning, U., Helinski, D. R., Chao, F. C., Yanofsky, C. The A protein of the tryptophan synthetase of Escherichia coli. Purification, crystallization, and composition studies. Journal of Biological Chemistry. 237, 1523-1530 (1962).
- Wilson, D. A., Crawford, I. P. Purification and properties of the B component of Escherichia coli tryptophan synthetase. Journal of Biological Chemistry. 240 (12), 4801-4808 (1965).
- Adachi, O., Miles, E. W. A rapid method for preparing crystalline beta 2 subunit of tryptophan synthetase of Escherichia coli in high yield. Journal of Biological Chemistry. 249 (17), 5430-5434 (1974).
- Adachi, O., Kohn, L. D., Miles, E. W. Crystalline alpha2 beta2 complexes of tryptophan synthetase of Escherichia coli. A comparison between the native complex and the reconstituted complex. Journal of Biological Chemistry. 249 (24), 7756-7763 (1974).
- Higgins, W., Fairwell, T., Miles, E. W. An active proteolytic derivative of the alpha subunit of tryptophan synthase. Identification of the site of cleavage and characterization of the fragments. 생화학. 18 (22), 4827-4835 (1979).
- Ahmed, S. A., Miles, E. W., Davies, D. R. Crystallization and preliminary X-ray crystallographic data of the tryptophan synthase alpha 2 beta 2 complex from Salmonella typhimurium. Journal of Biological Chemistry. 260 (6), 3716-3718 (1985).
- Miles, E. W., et al. The beta subunit of tryptophan synthase. Clarification of the roles of histidine 86, lysine 87, arginine 148, cysteine 170, and cysteine 230. Journal of Biological Chemistry. 264 (11), 6280-6287 (1989).
- Rhee, S., Parris, K. D., Ahmed, S. A., Miles, E. W., Davies, D. R. Exchange of K+ or Cs+ for Na+ induces local and long-range changes in the three-dimensional structure of the tryptophan synthase alpha2beta2 complex. 생화학. 35 (13), 4211-4221 (1996).
- Rhee, S., et al. Crystal structures of a mutant (betaK87T) tryptophan synthase alpha2beta2 complex with ligands bound to the active sites of the alpha- and beta-subunits reveal ligand-induced conformational changes. 생화학. 36 (25), 7664-7680 (1997).
- Schneider, T. R., et al. Loop closure and intersubunit communication in tryptophan synthase. 생화학. 37 (16), 5394-5406 (1998).
- Rhee, S., Miles, E. W., Davies, D. R. Cryo-crystallography of a true substrate, indole-3-glycerol phosphate, bound to a mutant (alphaD60N) tryptophan synthase alpha2beta2 complex reveals the correct orientation of active site alphaGlu49. Journal of Biological Chemistry. 273 (15), 8553-8555 (1998).
- Weyand, M., Schlichting, I. Crystal structure of wild-type tryptophan synthase complexed with the natural substrate indole-3-glycerol phosphate. 생화학. 38 (50), 16469-16480 (1999).
- Sachpatzidis, A., et al. Crystallographic studies of phosphonate-based alpha-reaction transition-state analogues complexed to tryptophan synthase. 생화학. 38 (39), 12665-12674 (1999).
- Yang, L. H., Ahmed, S. A., Miles, E. W. PCR mutagenesis and overexpression of tryptophan synthase from Salmonella typhimurium: On the roles of beta (2) subunit Lys-382. Protein Expression and Purification. 8 (2), 126-136 (1996).
- Green, M. R., Sambrook, J. . Molecular cloning: a laboratory manual. , (2012).
- Lowry, O. H., et al. Protein Measurement with the Folin Phenol Reagent. Journal of Biological Chemistry. 193 (1), 265-275 (1951).
- Laemmli, U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 227 (5259), 680-685 (1970).
- Kawasaki, H., Bauerle, R., Zon, G., Ahmed, S. A., Miles, E. W. Site-specific mutagenesis of the alpha subunit of tryptophan synthase from Salmonella typhimurium. Changing arginine 179 to leucine alters the reciprocal transmission of substrate-induced conformational changes between the alpha and beta 2 subunits. Journal of Biological Chemistry. 262 (22), 10678-10683 (1987).
- Battye, T. G., Kontogiannis, L., Johnson, O., Powell, H. R., Leslie, A. G. iMOSFLM: a new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallographica Section D Structural Biology. 67, 271-281 (2011).
- Evans, P. Scaling and Assessment of Data Quality. Acta Crystallographica Section D-Biological Crystallography. 62, 72-82 (2006).
- Winn, M. D., et al. Overview of the CCP4 suite and current developments. Acta Crystallographica Section D-Biological Crystallography. 67, 235-242 (2011).
- Vagin, A., Teplyakov, A. Molecular replacement with MOLREP. Acta Crystallographica Section D Structural Biology. 66, 22-25 (2010).
- Adams, P. D., et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallographica Section D Structural Biology. 66, 213-221 (2010).
- Emsley, P., Lohkamp, B., Scott, W. G., Cowtan, K. Features and development of Coot. Acta Crystallographica Section D Structural Biology. 66, 486-501 (2010).
- Murshudov, G. N., Vagin, A. A., Dodson, E. J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallographica Section D Structural Biology. 53, 240-255 (1997).
- Afonine, P. V., et al. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallographica Section D Structural Biology. 68, 352-367 (2012).
- Matthews, B. W. Solvent content of protein crystals. Journal of Molecular Biology. 33 (2), 491-497 (1968).
- Kantardjieff, K. A., Matthews Rupp, B. coefficient probabilities: Improved estimates for unit cell contents of proteins, DNA, and protein-nucleic acid complex crystals. Protein Science. 12 (9), 1865-1871 (2003).

