Stereological Estimation of Dopaminergic Neuron Number in the Mouse Substantia Nigra Using the Optical Fractionator and Standard Microscopy Equipment
Summary
This work presents a step-by-step protocol for the unbiased stereological estimation of dopaminergic neuronal cell numbers in the mouse substantia nigra using standard microscopy equipment (i.e., a light microscope, a motorized object table (x, y, z plane), and public domain software for digital image analysis.
Abstract
In pre-clinical Parkinson's disease research, analysis of the nigrostriatal tract, including quantification of dopaminergic neuron loss within the substantia nigra, is essential. To estimate the total dopaminergic neuron number, unbiased stereology using the optical fractionator method is currently considered the gold standard. Because the theory behind the optical fractionator method is complex and because stereology is difficult to achieve without specialized equipment, several commercially available complete stereology systems that include the necessary software do exist, purely for cell counting reasons. Since purchasing a specialized stereology setup is not always feasible, for many reasons, this report describes a method for the stereological estimation of dopaminergic neuronal cell counts using standard microscopy equipment, including a light microscope, a motorized object table (x, y, z plane) with imaging software, and a computer for analysis. A step-by-step explanation is given on how to perform stereological quantification using the optical fractionator method, and pre-programmed files for the calculation of estimated cell counts are provided. To assess the accuracy of this method, a comparison to data obtained from a commercially available stereology apparatus was performed. Comparable cell numbers were found using this protocol and the stereology device, thus demonstrating the precision of this protocol for unbiased stereology.
Introduction
The quantification of neuronal cell number is pivotal in pre-clinical Parkinson's disease research to determine the level of neurodegeneration within the substantia nigra (SN)1,2. The unbiased stereological estimation of cell number in a region of interest is considered the gold standard3,4,5.
Before the advent of unbiased stereology, the number of neurons in sections was assessed by manipulating counted cell profiles to correct for the variable probabilities that neurons come into sight in a section. One of the most commonly used methods was the correction of quantified cell counts described by Abercrombie6. This method attempted to take into account that cells can be quantified more than once if fragments of the same cell are found in adjacent thin sections. Therefore, Abercrombie and other authors generated equations that required assumptions about the shape, size, and orientation of the counted cells7,8. However, these assumptions were usually not realized and therefore led to systematic errors and divergence from the actual cell number (i.e., bias). Moreover, the bias could not be reduced by additional sampling3.
For the stereological estimation of cell numbers using the optical fractionator, mathematical principles are applied to directly estimate the cell numbers directly in a defined, 3-dimensional volume. The advantage of this method is that it does not involve assumptions about the shape, size, and orientation of the cells being counted. Thus, the estimated cell numbers are closer to the true values and get closer as the sample size increases (i.e., unbiased)3. Because many rules must be followed when using stereology to keep the method unbiased, ready-to-use commercial stereology systems have been developed (for review, see Schmitz and Hof, 20054). Specialized stereology systems implement design-based stereological methods with a priori defined probes and sampling schemes for stereological assessments that lead to independence from shape, size, spatial distribution, and orientation of the cells to be analyzed4,9. However, commercially available stereology systems are expensive; this may limit implementation in new research.
The aim of this study was to develop a usable technique for the design-based stereological estimation of dopaminergic cell counts in the mouse SN, employing the optical fractionator method and using standard microscopy equipment (i.e., light microscope, standard microscope software, and a motorized x, y, z stage). For this, a step-by-step guide on how to process mouse brain tissue and how to estimate SN cell numbers using design-based unbiased stereology is presented. Moreover, templates for the calculation of the estimated cell numbers and coefficients of error (CE) are provided.
The method described here is not limited to the analysis of the SN, but can be adapted for use in other anatomically defined regions of the mouse or rat brain. For instance, unbiased stereology has been used to estimate neuronal cell numbers in the hippocampus10 and the locus coeruleus11. Additionally, cell types other than neurons, such as astrocytes12 and microglia13, can be assessed as well. Therefore, this method can be useful to scientists who intend to implement unbiased stereology in their research but are not willing to spend a lot of money to purchase a stereology system.
Protocol
All applicable international, national, and/or institutional guidelines for the care and use of animals were followed. The protocol was approved by local authorities at the Regierung von Unterfranken, Wuerzburg, Germany.
1. Tissue Processing and Immunohistochemistry
- Euthanize mice with CO2 or any other approved method.
- Perfuse six 12-week-old C56Bl/6N male mice transcardially with 10 mL of 0.1 M phosphate-buffered saline (PBS) using a 25-G needle and an infusion pump, followed by 70 mL of 4% paraformaldehyde (PFA) in 0.1 M PBS.
CAUTION: PFA is toxic, allergenic, and carcinogenic. - Dissect the mouse brain in the coronal plane with a brain matrix slicer at the region of +0.74 mm from Bregma (Figure 25, Paxinos and Franklin mouse brain atlas14).
- Put the dorsal part of the brain, including the SN, in 4% PFA in 0.1 M PBS for 1 d at 4 °C for immersion-fixation.
- Exchange the PFA for 30% sucrose/0.1 mol PBS solution for cryo-protection and incubate for another 2 d at 4 °C.
- Put the dorsal part of the brain into a cryomold filled with optimal cutting temperature (OCT) compound and slowly freeze the tissue in liquid dry ice-cooled isopentane.
CAUTION: Dry ice is extremely cold (-78 °C); use personal protective equipment (PPE), including gloves, while working with dry ice. Dry ice evaporates in CO2; therefore, work under a fume hood. - Serially cut 30-µm cryo-sections in the coronal plane and collect the sections, starting at 2.46 mm from Bregma (Figure 51, Paxinos and Franklin mouse brain atlas14) and ending at 4.04 mm from Bregma (Figure 64, Paxinos and Franklin mouse brain atlas14).
- Store four series in tubes that are filled with cryoprotectant (30% glycerol, 30% ethoxyethanol, and 40% PBS) at -20 °C. During the sectioning procedure, mark the right hemisphere by puncturing a small hole into the upper region of the brainstem.
- For immunohistochemical staining, select one series of sections per mouse. Preincubate free-floating sections for 1 h in 10% normal goat serum (NGS)/2% BSA/0.5% detergent in 0.1 mol PBS on a shaker. Incubate the sections with rabbit anti-mouse tyrosine hydroxylase (TH; 1:1,000 dilution) antibody diluted in 2% NGS/2% BSA/0.5% detergent in 0.1 mol PBS overnight at 4 °C.
- Apply secondary antibodies against rabbit Igs (1:100 dilution) for 2 h at room temperature, followed by avidin/biotin reagent (1:100 dilution). Incubate and stain with 3,3-diaminobenzidine-tetrahydrochloride (DAB) and H2O2. Mount the sections after staining them on coated object slides.
NOTE: TH+ dopaminergic SN neurons are shown in Figure 1a. By staining one series, an average of 13 SN sections, extending from the rostral to caudal portions of the SN pars compacta (SNpc) and reticulata, are stained per animal (Figure 1b). The sections are separated by 120 µm (1/4 series) (Figure 1c).
CAUTION: DAB is a suspected carcinogen. It is toxic by contact and inhalation. Use PPE when working with DAB.
2. Acquisition of Images
- Capture TH immunohistochemically stained SN sections digitally using imaging software that is coupled to a microscope. Separately analyze each section of one series.
- Use the scan slide option of the imaging software. Save in TIFF format for high-quality images (Figure 2).
NOTE: Image resolution is 312 pixels per cm. - Press "Acquire" to open the acquisition window (Figure 2a).
- Set the binning to "2" for grayscale images.
NOTE: The acquisition of grayscale images instead of color pictures reduces the size of the final stack image file (Figure 2a). - Click on "Show Live" in the acquisition window to open a new window showing the live image of the section (Figure 2a and b).
- Click on "Stage" in the acquisition window. Choose "Scan Slide" in the dropdown menu and check the box (Figure 2c).
NOTE: This enables the acquisition and alignment of highly magnified SN images (630X magnification) in the x,y-plane, as well as the acquisition of stack images with a distance between consecutive images of 1 µm in the z-plane, covering a total range of 13 µm. - Select the 2.5X objective to search for the SN.
- Change the objective to 63x.
- Define the upper left corner (TopLeft) (Figure 2c) and lower right corner (LowRight) (Figure 2d) of the section by clicking on "Set."
- Click on "Z-Series" and check the "Z-Plane series" box (Figure 2e).
- Determine the actual mounted thickness of the sections at three randomly selected counting sites per section using the imaging software. For this, click on "View Top Offset" to define the top of the section (Figure 2e) and then click on "Stop View Top" (Figure 2f). Thereafter, click on "View Bottom Offset" (Figure 2f) and then click on "Stop View Bottom" (Figure 2g). Write down the thickness of the section.
- Subtract 3 µm (guard zone) from the top offset and type the result into the top offset slot (Figure 2h). Subtract 13 µm (height of the optical disector) from the top offset number and type the result into the bottom offset slot (Figure 2i). Select the following parameters: steps, 14; size, 1.00 (Figure 2i).
NOTE: This corresponds to a thickness in the z-plane of 13 µm, with a 1-µm distance between consecutive images. - Select the directory to save the file (Figure 2j).
- Click on "Sequence" to start the acquisition (Figure 2j).
- Click on "Processing" and select the following parameters: "All Planes," from "Sequence;" "Montage;" and the "Fast" and "Stitching" boxes, with an image overlap of 10% (Figure 2k). Start to stitch the images by clicking on "start".
NOTE: This will generate stack images for analysis (Figure 2l, Figure 3a-e).
3. Sequence of Stereological Assessment
- Perform the analysis of SN stack images using the NIH ImageJ software version 4.7.
- Download the two plugins that are needed from the ImageJ.nih.gov website: the "Grid Overlay" plugin15 and the "Cell Counter" plugin16. Install both plugins as described.
- For unbiased stereological assessment, define the counting parameters as follows: grid size, 130 x 130 µm; counting frame, 50 x 50 µm; and optical disector height, 13 µm.
- Open the image by clicking on "File" → "Open" (Figure 4a) Set the image scale by using the "Analyze" → "Set Scale" command (Figure 4b). For this step, change the defined size from pixels to µm (Figure 4c).
NOTE: For the analyzed images, 425 pixels corresponds to 100 µm. - Select the "Plugins" → "Grid" → "Grid Type: Lines" command to randomly insert a grid. Check the "Random Offset" box. Define the size of one square within the grid (grid-square) as 130 µm x 130 µm = 16,900 µm2 (Figure 3b and Figure 4d and e).
- Change image type from 16-bit to RGB Color (Click on "Image" → "Type" → "RGB Color") (Figure 4f). Locate the SNpc, as described by Baquet et al.17. Encircle the SNpc with the "Paintbrush Tool" (brush width: 11) (Figure 3c and Figure 4g-i).
- Undo the "Set Scale" command by clicking "Analyze" → "Set Scale" → "Click to Remove Scale" (Figure 4j-l) to enable the performance of the calculations in pixels rather than µm.
- Make a screenshot (Figure 4m), save the image file, and open the new file with ImageJ (Figure 4n). Select "Point" (Figure 4o) and a brush width of 25 (Figure 4p). Mark every grid-square that contacts the SN for placement of an optical disector (Figure 4q). Perform analysis of the cell number in all optical disectors that are localized fully or partially within the outlined SN.
- Select one grid-square that includes parts of the SN. Measure the upper left corner of this square with the cursor in ImageJ to define the x and y coordinates to start with (Figure 4r).
NOTE: Coordinates will be inserted for optical disector positioning using the "optical disector position" (Figure 4s), as described in step 4. - Calculate the optical disector position by using the spreadsheet template entitled, "Optical disector position," as described in step 4.
- Calibrate the optical disector (macro written by Christopher S. Ward, Supplemental File) by clicking on "Plugins" → "Macros" → "Edit" (Figure 4t), open the "opt_dis_grid.txt" file (Figure 4u), and define the size of the "usergrid" in pixels that corresponds with the x,y dimension of the optical disector (Figure 4v). Select a size of 50 µm x 50 µm, so that the size of one side of the usergrid is defined as 212.5 pixels (50 x 4.25 pixels).
- Determine the position of the optical disector in the stack image by inserting the ImageX and ImageY coordinates that define the center of the optical disector (Figure 4v). Run the macro ("Macros" → "Run Macro") (Figure 4w).
NOTE: The grid will vanish from the stack image (Figure 3d and Figure 4x). The optical disector will persist when moving down in the z-plane (Figure 3e and Figure 4x). - Select "Plugins" → "Cell Counter" to start the Cell Counter plugin (Figure 4y). Initialize the Cell Counter and select a marker type (Figure 4z). Zoom in on the image (click on "+") and perform the stereological assessment of the cell numbers at a magnification of 200%.
- Identify dopaminergic neuronal perikarya by their rounded or ovoid shape and cell size (Figure 1a). Quantify the number of cells using stereological rules.
NOTE: Since the optical disector is a 3-dimensional cube (Figure 5a), define the exclusion sides as the front, left, and top sides of the cube (red). Therefore, don't count TH+ cells that come into contact with either the red line (left and front) or the top side. Add the results of the cell counts to the prepared spreadsheet file entitled, "Calculation of cell count" (step 5). - According to the SN image, calculate the next grid-square in which to place the optical disector for the next cell count, using x,y steps the size of the overlying grid (Figure 5b) and the "Optical disector position" spreadsheet template, as described in step 4.
- Perform an estimation of the cell number using the "Calculation of cell count" spreadsheet (step 5).
4. Calculation of Optical Disector Position Using the "Optical Disector Position" Spreadsheet Template
- Calculate the size and position of each optical disector to be placed into the overlying grid within the outline of the SN (Figure 5b) using the "Optical disector position.xlsx" spreadsheet file (Supplemental File).
- Insert the conversion of pixel per µm, as defined by the "Set Scale" command, into the yellow marked positions of the "Optical disector position.xlsx" file (Figure 6a and Supplemental File).
- Insert the planned size of the optical disector and the size of the grid-square of the overlying grid, defined by the length of one side (in µm), into the orange marked positions.
NOTE: Results are depicted in the red boxes next to the orange boxes. - Start with the grid-square within the SN outline that was determined as the starting point (one grid-square was selected to begin with, as in step 3.9, above).
- Insert the x and y coordinates, as measured in step 3.9, into the green boxes of the file (by using these values, the x,y coordinates for the center of the optical disector within this first square are calculated, and the results are depicted in the blue boxes). Run the opt_dis_grid.txt macro (Supplemental File).
- Calculate the x,y coordinates of the consecutive optical disectors by inserting the grid-square distance (measured in number of squares) relative to the first grid-square (brown boxes).
NOTE: +x: grid-square is located to the right; -x: grid-square is located to the left; +y: grid-square is located to the bottom; -y: grid-square is located to the top. Results are shown in the gray boxes.
5. Estimation of Cell Number Using the "Calculation of Cell Count" Spreadsheet
- Calculate the estimated number of TH+ SN cells per animal (N) using the formula published by West et al., 19913, where ∑Q– is defined as the total number of TH+ neurons counted in all optical disectors of one brain section.
NOTE: The height of the optical disector (h) in relation to the measured thickness of the section (t) is included in the equation. The area sampling fraction (asf) is defined as the proportion of the area (A) of the optical disector (A optical disector) frame size within the square size of the grid (A x,y step) (A optical disector/A x,y step) (Figure 3b), and the section sampling fraction (ssf) is defined as the proportion of sections of the whole serially cut brain:
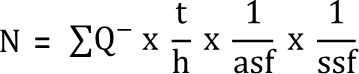
To ensure high-quality quantitative estimates, the coefficient of error (CE) should be lower than 0.10. Determine the CE using the formula by Keuker et al., 200110:
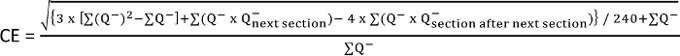
- For the stereological estimation of the cell number within the SN of each mouse, insert the indicated information regarding the total number of generated series per mouse, the height of the optical disector, the x,y area of the optical disector (A optical disector, in µm2), the x,y area of the grid-square (A x,y step, in µm2), and the mean measured thickness of the section (in µm) in the yellow boxes using the "Calculation of cell count.xlsx" spreadsheet file (Figure 6b and Supplemental File).
- Analyze each SN section from the same animal, as described in step 5.
- Insert cell counts for each optical disector in the gray boxes, each line referring to one SN section, using the "Calculation of cell count.xlsx" spreadsheet file (Supplemental File).
NOTE: The estimated cell number and the calculated CE are depicted in the blue boxes (Figure 6b).
6. Estimation of Cell Number Using a Commercially Available Stereology System
- Start the estimation of dopaminergic SNpc cell number by clicking on "Probes" → "Optical Fractionator Workflow" to open the Optical Fractionator Workflow window.
NOTE: A new window will pop-up to determine if a new series of SN is going to be counted. - Start a new series of TH+ SN sections by clicking "Start a new subject" in the pop-up window.
- Go through the different steps of the Optical Fractionator Workflow. Set up the subject in the first step. For this, insert the investigator's name, the subject's name (SN group), and other helpful information. Insert the numbers of sections to count for this SN. Insert the thickness of the cut tissue sections. Insert the section interval, which is four in this case. Insert the randomly determined section number to start with.
- In the second step, set the microscope to low magnification (2X objective) and select "2X Mag Lens" from the menu.
- In step 3, trace the region of interest with the left mouse button, which is the SNpc, as described by Baquet et al., 200917.
- Set the microscope to high magnification in step 4. For this, change the objective to 100X and select "100X Mag lens" from the dropdown menu.
- Measure the mounted thickness in step 5 by focusing to the top and to the bottom of the section.
- In step 6, define the counting frame size as 50 x 50 µm; this is the size of the optical disector (x,y).
- In step 7, set the size of the systematic random sampling (SRS) grid as 130 x 130 µm.
- Enter a 3-µm guard zone in step 8.
- Save the settings in step 9.
- Finally, count the TH+ dopaminergic cells in each of the optical disectors within the SRS grid in step 10 of the Optical Fractionator Workflow. For this, start by clicking on the "Counting" button.
- After finishing one section, click on the "Begin Next Section" button to start the Optical Fractionator Workflow of the next SN section of the same SN series.
- When the last SN section of one SN series has been quantified, click on "I've Finished Counting."
- Click on the "View Results" button to display the results of the stereology sampling.
Representative Results
Using the presented method, the estimated number of TH+ dopaminergic neurons in the right SN ranged between 7,363 and 7,987 cells and, in the left SN, between 7,446 and 7,904 cells. Thus, the mean number of dopaminergic neurons (± SEM) was 7,647 ± 83 cells for the right SN and 7,675 ± 66 for the left SN. The calculated CE for each animal was lower than 0.08 (range: 0.073-0.079) (Figure 7). To ascertain the comparability of this method with commercially available and specialized stereology systems, SN cell numbers were also quantified using the stereology setup (Figure 8). The counting parameters were: grid size, 130 x 130 µm; counting frame, 50 x 50 µm; guard zone, 3 µm. Sections were analyzed with a 100x/1.25 numerical aperture objective on a BX53 microscope. The estimated population of TH+ SN neurons using the mean section thickness ranged from 7,067 to 8,105 cells/SN and 7,164 to 8,015 cells/SN on the right and left sides, respectively. The mean neuronal number was calculated as 7,535 ± 155 cells for the right SN and 7,699 ± 128 cells for the left SN. The Gundersen CE (m=1) was ≤0.08 in each SN quantified (Figure 7). Statistical analysis using paired Student's t-test did not reveal any significant differences between the TH+ cell counts of the two methods in the right or in the left SN (mean ± SEM; right: t(5)=0.9524, P >0.05; left: t(5)=0.2928, P>0.05) (Figure 7).
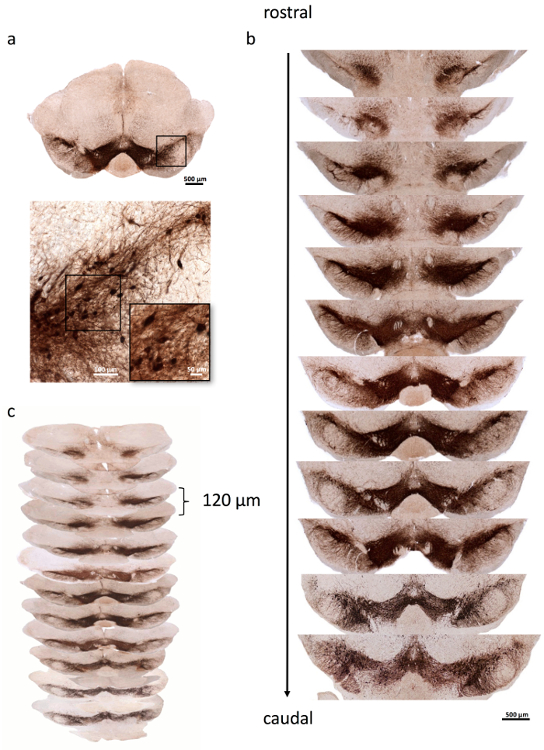
Figure 1. Representative images of mouse SN stained for dopaminergic neurons.
(a) An overview of a representative mouse SN is shown in the upper panel. Note the tiny hole in the upper part of the right brainstem that marks the right side. Higher magnification of the inset (black box) depicts TH+ dopaminergic neurons. (b) One series of TH-stained SN sections is shown, covering the whole mouse SN from rostral to caudal. (c) Each section is separated from the consecutive section by 120 µm. Scale bars: a, upper panel, 500 µm; lower panel, large image, 100 µm; lower panel, inset, 50 µm; b, 500 µm. Please click here to view a larger version of this figure.
Figure 2. Acquisition of images for stereological analysis using the imaging software.
(a) Red arrow: open the acquisition window; green arrow: set the binning to "2;" blue arrow: show the live image. (b) A new window showing the live image (red arrow) will open. (c) Red arrow: select the "Stage" menu; green arrow: select the "Scan Slide" option; blue arrow: check the "Scan Slide" option; black arrow: define the upper left corner. (d) Black arrow: define the lower right corner. (e) Red arrow: select the "Z-series" menu; green arrow: check the "Z-Plane series;" blue arrow: start the "View Top Offset" to search for the top of the section. (f) Red arrow: click on "Stop View Top" to define the top of the section; green arrow: start "View Bottom Offset" to search for the bottom of the section. (g) Red arrow: click on "Stop View Bottom" to define the bottom of the section. (h) Red arrow: substract the 3-µm guard zone from the "Top Offset" and insert the result into the "Top Offset" slot. (i) Red arrow: subtract 13 µm from the "Top Offset" number and insert the result into the "Bottom Offset" slot; green arrow: define 14 steps that corresponds to a z-plane thickness of 13 µm; blue arrow: define a size of 1.00 µm that corresponds to a 1-µm distance between consecutive images. (j) Red arrow: select the directory to save the file to; blue arrow: click on "Sequence" to start the acquisition of images. (k) Red arrow: click on "Processing;" check the following parameters: green arrow: "All planes;" blue arrow: "Sequence;" black arrow: "Montage;" gray arrow: "Stitching;" purple arrow: fast. (l) Click on the yellow arrow to start stitching the images. Please click here to download this file.
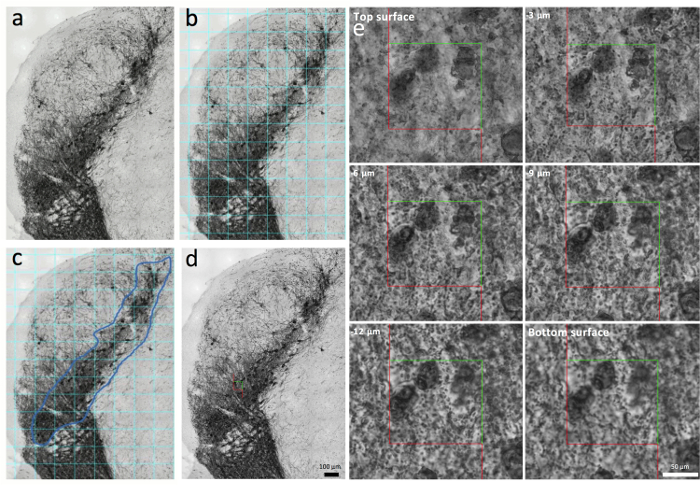
Figure 3. Processing the images for stereological analysis.
(a) An example stack image taken from a mouse SN. (b and c) After the randomized insertion of a grid overlying the SN (turquoise lines, b), the SN is outlined in a second step (blue line, c). (d) An optical disector is placed into the SN stack image. (e) Six consecutive focal planes within the SN stack image cover 13 µm total in the z-plane. Scale bars: a-d, 100 µm; e, 50 µm. Please click here to view a larger version of this figure.
Figure 4. Sequence of stereological assessment using ImageJ.
(a) Red arrow: click on "File" → "Open" to open a stack image. (b) Red arrow: click on "Analyze" → "Set Scale". (c) Red arrows: define the parameters for the "Distance in pixels," "Known distance," and "Unit of length." (d) Red arrow: select "Plugins" → "Grid." (e) Red arrow: select "Lines;" green arrow: check "Random Offset;" blue arrow: insert the size of the grid in µm2. (f) Red arrow: click on "Image" → "Type" → "RGB Color." (g) Red arrow: select the "Paintbrush Tool;" green arrow: define the "brush width" as 11. (h) Red arrow: start to encircle the SNpc. (i) Red arrows depict the encircled SNpc. (j) Red arrow: click on "Analyze" → "Set Scale" and remove the scale (k) by clicking on "Click to Remove Scale," depicted by the red arrow. Note the change from µm (k, green arrow) to pixels (l, red arrow). (m) Take a screenshot (red arrow points towards the screenshot) and (n) open the screenshot image file with ImageJ. (o) Red arrow: click on "Point." (p) Red arrow: insert a brush width of 25. (q) The red arrow depicts the marked grid-squares that contact the SNpc. (r) The red arrow points at the upper left corner of a grid-square that includes part of the SN. By putting the cursor at this point, the x,y coordinates for the "optical disector position" (step 4) are measured and inserted into the "optical disector position.xlsx" template (s, red arrow). (t) Red arrow: select "Plugins" → "Macros" → "Edit." (u) Red arrow: select the "opt_dis_grid.txt" file. A new window will open (v). Insert the size of the "usergrid," in pixels (v, red arrow), and the x,y coordinates (v, green arrow). (w) Run the "opt_dis_grid.txt" macro (red arrow). (x) An optical disector will appear in the middle of the previously defined grid-square (red arrow). (y) Red arrow: click on "Plugins" → "Cell Counter." (z) Initialize the Cell Counter (red arrow) and select a marker type (green arrow). Please click here to download this file.
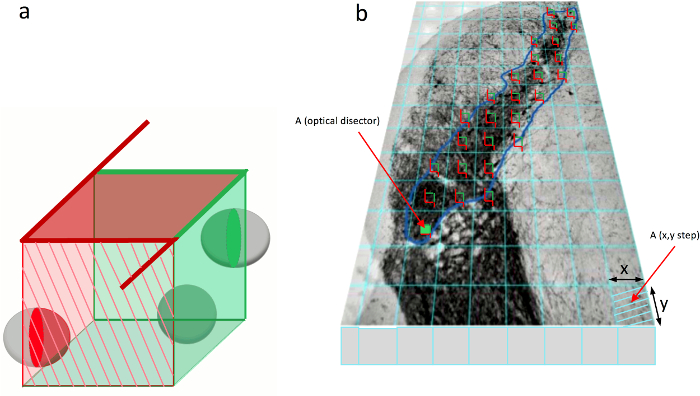
Figure 5. Placement of the optical disector.
(a) Illustration of the optical disector used for unbiased stereology. The optical disector is a 3-dimensional cube. Three schematic cells are visible within the optical disector. The stereological rules for the quantification of cells indicate that cells touching the red labeled sides of the optical disector are excluded (red cell), while cells that contact the green labeled sides are included in cell counting (green cells). (b) An optical disector is placed in every grid-square that comes in contact with the SN. Please click here to view a larger version of this figure.
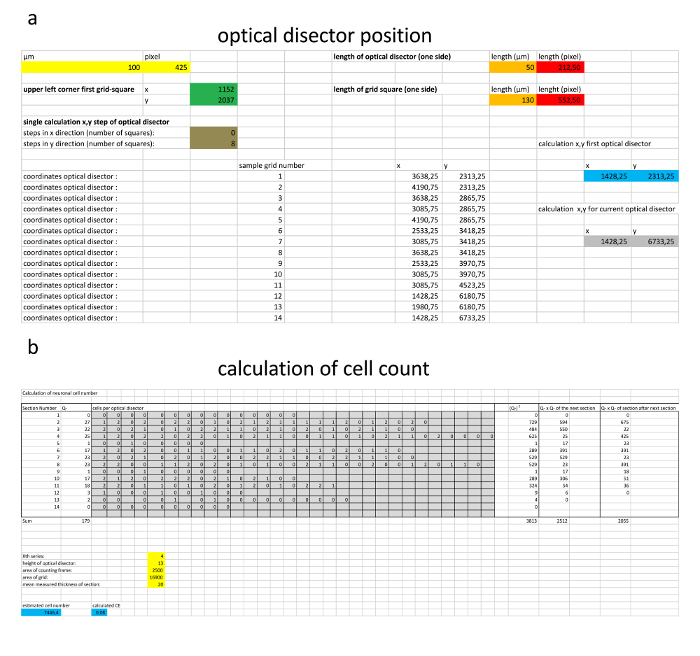
Figure 6. Calculation of the optical disector position and estimation of cell number.
(a) An example calculation performed for the positioning of the optical disector. (b) An example of the estimation of total cell number. The numbers of quantified cells are inserted in the gray boxes, while the parameters of stereological quantification are inserted in the yellow boxes. The estimated cell number and the CE are displayed in the blue boxes. Please click here to view a larger version of this figure.
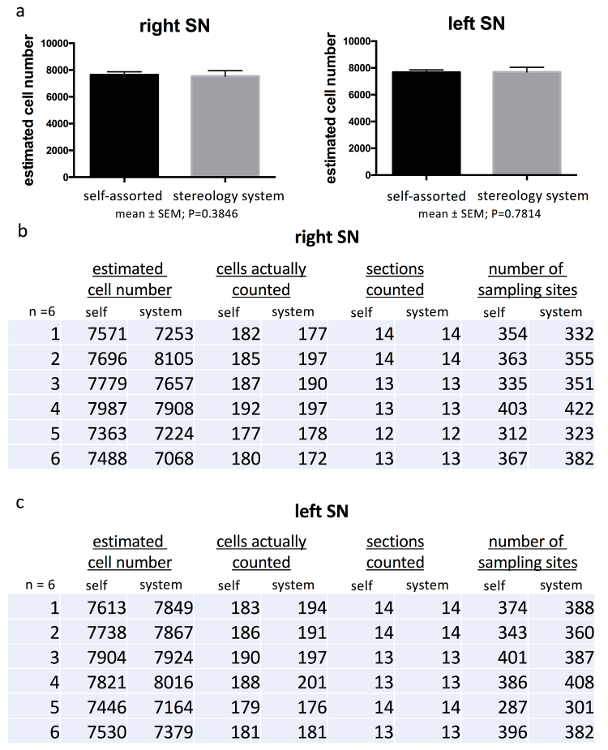
Figure 7. Comparison of cell counting parameter.
(a) Comparing the estimated TH+ cell number of the right and left mouse SN obtained from the described stereological method with the data acquired from analysis using the commercially available stereology system did not show any significant differences. A detailed list of the obtained data of the right (b) and left (c) SN is given with the estimated cell number per animal, the number of cells actually counted, the number of sections counted, and the number of sampling sites (i.e., the number of grid-squares counted) for both methods. The values in the bar graphs are the mean ± SEM of the data from 6 mice. Please click here to view a larger version of this figure.
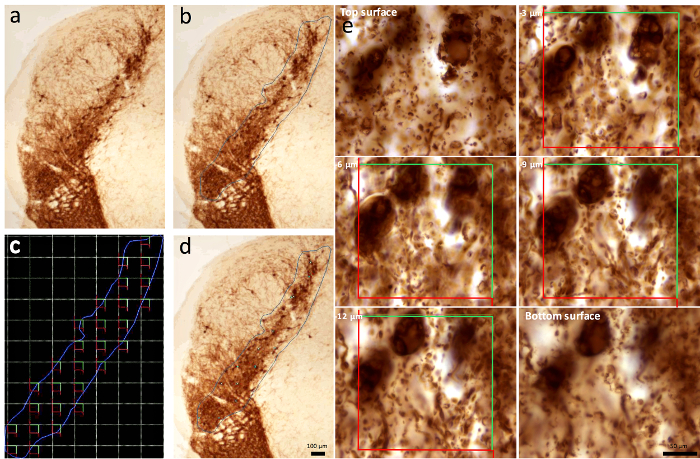
Figure 8. Quantification of cells with the commercially available stereology system.
(a-b) The outline of the SN is drawn and (c) the positions of the optical disectors are automatically determined. (d-e) Each optical disector is analyzed for cell number by focusing along the z-plane. Six consecutive focal planes within the SN are shown. The red and green lines correspond to the optical disector. Scale bars: a-d, 100 µm; e, 50 µm. Please click here to view a larger version of this figure.
Supplemental File 1. opt_dis_grid.txt Please click here to download this file.
Supplemental File 2. Optical disector position.xlsx Please click here to download this file.
Supplemental File 3. Calculation of cell count.xlsx Please click here to download this file.
Discussion
Stereology starts with tissue processing. The serial cutting of SN tissue must be performed carefully to prevent the loss of sections during stereological analysis. Additionally, one essential step is to mark one hemisphere in order to distinguish the right from the left SN when performing stereology. Placing a tiny hole at the upper part of the brainstem generated the best results in the presented study. Moreover, since working with the optical fractionator method demands that the tissue is cut in thick sections of about 30-40 µm, rather than the 5-10 µm more commonly used in immunohistochemistry, antibody and other incubation fluid tissue penetration during immunohistochemical staining must be ensured, such as by using a detergent.
Immunohistochemical staining for dopaminergic neurons using the TH antibody labels SN neurons, as well as neurons within the ventral tegmental area (VTA), which is located medially from the SN, and neurons in the retrorubral field. Thus, another critical point when performing stereology of the SN is knowledge of the mouse SN anatomy, since SN neurons cannot be visualized exclusively by standard immunohistochemical or histological staining. Instead, the recognition of anatomical landmarks, like the subthalamic nucleus, the retrorubral field, and the VTA, are important to determine the whole extent of the SN17,18,19.
Assessment of the neuronal number needs to be reliable and reproducible. Although various groups use commercially available stereology setups, numbers of TH+ dopaminergic SN neurons in wt mice, taken from different publications, vary in number17,20. In Baquet et al., design-based stereological analysis in 3- to 4-month-old male and female C57BL/6J wt mice using a commercially available stereology system revealed an estimated 8,716 ± 338 TH+ dopaminergic SN neurons (range: 7,546-9,869)17. In contrast, Smeyne et al. found fewer TH+ SN cells in 9- to 16-week-old male C57BL/6J mice that received 0.9% saline i.p., ranging from 6,302 ± 262 cells to 6,861 ± 338 cells, using a newer version of the same stereology system20. However, the first author was also the last author in the Baquet et al. paper, therefore excluding varying degrees of experience as the reason for the different results. These contradicting data reflect the need for a stereological quantification method that shows less variability. To verify the technique of quantification and the formula used, a comparison of the results of SN cell number count using the described method with a quantification using the commercially available stereology setup was performed. Statistical analysis did not show any significant differences when comparing the numbers of TH+ dopaminergic neurons in the right and left SN of wt mice determined using both techniques of cell quantification, thus demonstrating that this method is feasible for the stereological estimation of SN neurons in mice.
The main advantage of the described method is that it reduces the costs that are usually associated with the implementation of stereology techniques for cell counting. A second advantage is that, for the described method, stack images of the SN are saved and can be analyzed any time (even at the same time) by another investigator. Moreover, the acquisition and analyses of fluorescence images are possible with this method, because grayscale images can be colored with the "Lookup Tables" in ImageJ. However, there are limitations to this study. First, the quantification of cell number is more time consuming (around 50%) than quantification with the commercially available stereology system. This is mainly because of two reasons: 1. stack images of the SN must be taken and 2. calculation of the optical disector placement within each SN and calculation of the estimated cell number and CE must be performed manually using the calculation templates. Second, at least with the system available in our laboratory, the analysis can only be performed on grayscale images because of the large file size of color images, which could not be processed by ImageJ. This might pose a problem if there is a need to quantify two different cell populations at the same time in a double stain. It could be more difficult to distinguish two differently stained cell populations in grayscale images. Therefore, although a motorized stage (x, y, z plane) and the corresponding software are available in many laboratories working with histological specimens, the management and quality of stack images will depend on the microscope software and the performance of the attached computer.
There are possible ways to overcome the limitations. By using the calculation templates we developed, the programming of macros to automate some of the currently manually performed processes could reduce errors or inaccuracies during stereological assessment and accelerate the speed of work. Additionally, with the ongoing development of ImageJ and other imaging software, as well as with increasing computer processing power, larger image file sizes should not pose a problem in the near future. Therefore, analyses of bright-field color images will be manageable with the described technique.
The technique described is not restricted to analyses of TH+ SN cells, but can be expanded to analyze other cell types and brain regions of rodents. For this, the anatomical borders of the region of interest must be determined, and the respective staining must be performed. Additionally, this technique can be adapted to thicker sections. For this, the acquisition of stack images must be adapted to provide more stack images. For example, if 40 µm-thick section are required, the actual mounted thickness of the section after tissue staining must be assessed to determine the necessary height of the optical disector. Subtract 6 µm (3-µm guard zone at the top and the bottom) from the mean tissue thickness and take the result as the height of the optical disector.
Disclosures
The authors have nothing to disclose.
Acknowledgements
The authors are grateful to Keali Röhm, Louisa Frieß, and Heike Menzel for their expert technical assistance; to Helga Brünner for the animal care; and to Chistopher S. Ward for the generation and distribution of the optical disector grid plugin for the ImageJ software.
Materials
| Paxinos mouse atlas | The Mouse Brain George Paxinos Keith B.J.FranklinCopyright @2001 by Academic Press CD Rom designet & created by Paul Halasz | ||
| brain matrix slicer mouse | Zivic Instruments | BSMAS 001-1 | |
| paraformaldehyde | Merck | 1040051000 | |
| sucrose /D(+) Saccharose | Roth | 4621.1 | |
| isopentane | Roth | 3927.1 | |
| glycerol | Merck | 1040931000 | |
| Ethanol | Sigma Aldrich | 32205-1L | |
| Name | Company | Catalog Number | Comments |
| phosphate buffered saline ingredients: | |||
| sodium chloride | Sigma Aldrich | 31434-1KG-R | |
| potassium dihydrogen phosphate | Merck | 1048731000 | |
| di-sodium hydrogen phosphate dihydrate | Merck | 1065801000 | |
| potassium chloride | Merck | 1049360500 | |
| normal goat serum | Dako | X0907 | |
| bovine serum albumin | Sigma | A4503-100G | |
| Triton X-100 | Sigma Aldrich | X100-100ml | detergent |
| 3,3-Diaminobenzidine-tetrahydrochlorid/DAB tablets 10mg pH 7.0 | Kem En Tec | 4170 | |
| H2O2/ Hydrogen peroxide 30% | Merck | 1072090250 | |
| avidin/biotin reagent | Thermo Scientific | 32050 | Standard Ultra Sensitive ABC Staining Kit, 1:100 |
| rabbit anti mouse tyrosine hydroxylase antibody | abcam | Ab112 | 1:1000 |
| biotinylated goat-anti-rabbit IgG H+L | vector laboratories | BA-1000 | 1:100 |
| StereoInvestigator version 11.07 | MBF | ||
| BX53 microscope | Olympus | ||
| Visiview | Visitron Systems GmbH | 3.3.0.2 | |
| Axiophot2 | Zeiss | ||
| ImageJ software | NIH | Version 4.7 | |
| Tissue-TEK OCT | Sakura | 4583 | |
| dry ice | |||
| grid overlay plugin | Wayne Rasband | https://imagej.nih.gov/ij/plugins/graphic-overlay.html | |
| cell counter plugin | Kurt de Vos | https://imagej.nih.gov/ij/plugins/cell-counter.html). | |
| optical disector macro | Christopher Ward | ||
| C57Bl/6N male mice | Charles River, Germany | ||
| SuperFrost Plus coated object slides | Langenbrinck, Germany | ||
| 25G needle Microlance 3 | BD | 300400 | |
| REGLO Analog Infusion pump | Ismatec | ISM 829 | |
| StereoInvestigator system | StereoInvestigator version 11.07 | ||
| BX53 microscope | BX53 microscope | ||
| self-assorted stereology | Visiview | ||
| Axiophot2 | Axiophot2 |
References
- Ip, C. W., et al. AAV1/2-induced overexpression of A53T-alpha-synuclein in the substantia nigra results in degeneration of the nigrostriatal system with Lewy-like pathology and motor impairment: a new mouse model for Parkinson’s disease. Acta Neuropathol Commun. 5 (1), 11 (2017).
- Ip, C. W., Beck, S. K., Volkmann, J. Lymphocytes reduce nigrostriatal deficits in the 6-hydroxydopamine mouse model of Parkinson’s disease. J Neural Transm (Vienna). 122 (12), 1633-1643 (2015).
- West, M. J., Slomianka, L., Gundersen, H. J. Unbiased stereological estimation of the total number of neurons in thesubdivisions of the rat hippocampus using the optical fractionator. Anat Rec. 231 (4), 482-497 (1991).
- Schmitz, C., Hof, P. R. Design-based stereology in neuroscience. 신경과학. 130 (4), 813-831 (2005).
- Tieu, K. A guide to neurotoxic animal models of Parkinson’s disease. Cold Spring Harb Perspect Med. 1 (1), e009316 (2011).
- Abercrombie, M. Estimation of nuclear population from microtome sections. Anat Rec. 94, 239-247 (1946).
- Rose, R. D., Rohrlich, D. Counting sectioned cells via mathematical reconstruction. J Comp Neurol. 263 (3), 365-386 (1987).
- Weibel, E. R., Gomez, D. M. A principle for counting tissue structures on random sections. J Appl Physiol. 17, 343-348 (1962).
- West, M. J. Design-based stereological methods for counting neurons. Prog Brain Res. 135, 43-51 (2002).
- Keuker, J. I., Vollmann-Honsdorf, G. K., Fuchs, E. How to use the optical fractionator: an example based on the estimation of neurons in the hippocampal CA1 and CA3 regions of tree shrews. Brain Res Brain Res Protoc. 7 (3), 211-221 (2001).
- Mouton, P. R., et al. The effects of age and lipopolysaccharide (LPS)-mediated peripheral inflammation on numbers of central catecholaminergic neurons. Neurobiol Aging. 33 (2), e427-e436 (2012).
- Barreto, G. E., Sun, X., Xu, L., Giffard, R. G. Astrocyte proliferation following stroke in the mouse depends on distance from the infarct. PLoS One. 6 (11), e27881 (2011).
- Robinson, S., et al. Microstructural and microglial changes after repetitive mild traumatic brain injury in mice. J Neurosci Res. 95 (4), 1025-1035 (2017).
- Paxinos, G., Franklin, K. . The mouse brain in stereotaxic coordinates. , (2001).
- . Grid_Overlay.java Available from: https://imagej.nih.gov/ij/plugins/graphic-overlay.html (2010)
- . cell_counter.jar Available from: https://imagej.nih.gov/ij/plugins/cell-counter.html (2010)
- Baquet, Z. C., Williams, D., Brody, J., Smeyne, R. J. A comparison of model-based (2D) and design-based (3D) stereological methods for estimating cell number in the substantia nigra pars compacta (SNpc) of the C57BL/6J mouse. 신경과학. 161 (4), 1082-1090 (2009).
- Fu, Y., et al. A cytoarchitectonic and chemoarchitectonic analysis of the dopamine cell groups in the substantia nigra, ventral tegmental area, and retrorubral field in the mouse. Brain Struct Funct. 217 (2), 591-612 (2012).
- German, D. C., Manaye, K. F. Midbrain dopaminergic neurons (nuclei A8, A9, and A10): three-dimensional reconstruction in the rat. J Comp Neurol. 331 (3), 297-309 (1993).
- Smeyne, R. J., et al. Assessment of the Effects of MPTP and Paraquat on Dopaminergic Neurons and Microglia in the Substantia Nigra Pars Compacta of C57BL/6 Mice. PLoS One. 11 (10), e0164094 (2016).

