Generalized Psychophysiological Interaction (PPI) Analysis of Memory Related Connectivity in Individuals at Genetic Risk for Alzheimer’s Disease
Özet
This manuscript describes how to implement a psychophysiological interaction analysis to reveal task-dependent changes in functional connectivity between a selected seed region and voxels in other regions of the brain. Psychophysiological interaction analysis is a popular method to examine task effects on brain connectivity, distinct from traditional univariate activation effects.
Abstract
In neuroimaging, functional magnetic resonance imaging (fMRI) measures the blood-oxygenation-level dependent (BOLD) signal in the brain. The degree of correlation of the BOLD signal in spatially independent regions of the brain defines the functional connectivity of those regions. During a cognitive fMRI task, a psychophysiological interaction (PPI) analysis can be used to examine changes in the functional connectivity during specific contexts defined by the cognitive task. An example of such a task is one that engages the memory system, asking participants to learn pairs of unrelated words (encoding) and recall the second word in a pair when presented with the first word (retrieval). In the present study, we used this type of associative memory task and a generalized PPI (gPPI) analysis to compare changes in hippocampal connectivity in older adults who are carriers of the Alzheimer’s disease (AD) genetic risk factor apolipoprotein-E epsilon-4 (APOEε4). Specifically, we show that the functional connectivity of subregions of the hippocampus changes during encoding and retrieval, the two active phases of the associative memory task. Context-dependent changes in functional connectivity of the hippocampus were significantly different in carriers of APOEε4 compared to non-carriers. PPI analyses make it possible to examine changes in functional connectivity, distinct from univariate main effects, and to compare these changes across groups. Thus, a PPI analysis may reveal complex task effects in specific cohorts that traditional univariate methods do not capture. PPI analyses cannot, however, determine directionality or causality between functionally connected regions. Nevertheless, PPI analyses provide powerful means for generating specific hypotheses regarding functional relationships, which can be tested using causal models. As the brain is increasingly described in terms of connectivity and networks, PPI is an important method for analyzing fMRI task data that is in line with the current conception of the human brain.
Introduction
The term "connectome" was coined in 2005 marking a paradigm shift in neuroscience that continues to this day1. The brain is increasingly described in terms of functional networks, connectivity and interactions between and among regions on a large scale. Nevertheless, the delineation of regional functional specialization and associations between fMRI-measured activity and task demands are still valid and useful approaches. In light of the growing interest in connectomics, functional connectivity approaches to task fMRI analysis are growing in popularity. One approach to measuring functional connectivity changes dependent on task demands makes use of the concept of PPI. A PPI is the interaction of an active task phase or particular task demand ("psycho") with the functional connectivity ("physio") of a region of interest or "seed" in the brain. PPI differs from bivariate, correlation-based analysis of functional connectivity, which generally measures the degree of correlation between the activity in two regions without any constraints related to task demands.
The concept and framework of a PPI analysis was originally described by Friston and colleagues in 19972. The authors asserted that their approach was important because it would allow the investigation of connectivity to be more functionally specific and allow for inferences that activity in a distal seed could be modulating activity resulting from a task demand. In 2012, McLaren and colleagues added to this original framework and described a gPPI approach in which all task phases and their interactions are included in a single model3. This approach leads to results that are more sensitive and specific to the task phase and interaction being investigated. It is this updated gPPI approach that we employ in the present study (see step 6.2.2 in Protocol). The gPPI approach has now been cited in over 200 studies. For clarity, hereafter we use 'PPI' to describe common features of both the standard and generalized version. 'gPPI' will be used to discuss specific advances associated with the newer framework.
The overall goal of a PPI analysis is to understand how the demands of a cognitive task influence or modulate the functional connectivity of a seed region. A PPI analysis requires a strong a priori hypothesis. Activity in the seed region must be modulated by the task in order for the PPI approach to work effectively4. For example, in the present study, we based our seed selection on the strong evidence that hippocampal activity is modulated by the cognitive demands of a memory task. Using PPI, regions that are significantly more or less functionally connected to the hippocampus during specific task phases can be identified. In short, we ask the question, "in which regions is activity more correlated with the seed during context A as compared with baseline?" We can also ask the logical opposite (as it is important to understand the difference): "in which regions is activity less correlated with the seed during context A as compared to baseline?" When interpreting group differences in PPI effects, it is important to examine the data and whether positive or negative change in functional connectivity, or both, is driving group differences.
The PPI approach has been used to study dynamic task control hubs in healthy controls, how modulation of functional connectivity is related to cognitive performance in Alzheimer's disease (AD), intelligence in individuals with autism, motor network connectivity in individuals with Parkinson's disease, face processing in individuals with body dysmorphic disorder and anorexia, emotion regulation, memory, and many other specific questions related to connectivity5,6,7,8,9,10,11. In the present study, we compare changes in functional connectivity of subregions of the hippocampus during memory encoding and retrieval between a group of individuals at increased genetic risk for AD to a group without the risk factor12. The following describes the protocol that we used, applying the gPPI approach, to allow us to test if task-elicited changes in functional connectivity differ in association with the presence of APOEε4, a genetic risk factor for AD.
Protocol
The present study was performed in compliance with the UCLA Institutional Review Board (IRB) protocols and approved by the UCLA Human Subjects Protection Committee. All participants gave written informed consent in order to enroll in this study.
1. Participant Selection
- Obtain IRB approval to perform the study.
- Screen individuals aged 55 and older for cognitive decline using a standardized neuropsychological battery. Include tests of General Intelligence (Subtests of the WAIS-III)13, Fluency (Fruits and Vegetables)14, Attention (Digits Forward and Backward)13, Language (Boston Naming Test)15, Verbal Memory (Buschke-Fuld Selective Reminding Task)16, WMS-III Logical Memory and Verbal Paired Associates learning13, and Visual Memory (Rey-Osterrieth Figure test)17.
- Have the participants complete mood questionnaires such as the Hamilton Depression and Anxiety Inventories18,19 as well as the Mini Mental State Exam (MMSE)20.
- Include participants that score 26 or above on the MMSE and perform better than two standard deviations below normal for their age on cognitive tests. Exclude participants with clinical anxiety, depression or any other neuropsychiatric or neurological illness. Exclude participants who do not meet MRI safety criteria or who do not consent to a blood draw.
NOTE: In the present study, 93 participants met these criteria (mean age = 67.4 years, 31M/49F).
2. Genotyping
- Have a trained phlebotomist or other medical professional draw blood from each participant.
- Isolate 200 µg genomic DNA from 10 mL of the sample as described21.
- Carry out single nucleotide polymorphism (SNP) genotyping using real-time PCR at two loci, rs429358 and rs7412 to discriminate APOE alleles22.
- Incorporate reporter dyes for rs429358 and rs7412 into a SNP genotyping assay. After each PCR amplification cycle is completed, plot fluorescent signals on a graph showing distribution of reporter and quencher dyes. Perform the experiment in duplicate to confirm results.
- Analyze the SNP genotyping data using a software package developed for the real-time PCR procedure output23.
NOTE: The program used in the present study calculates the affinity of the sample to one of the two reporter dyes that, in turn, represents one APOE SNP over the other. In the present study, 34 carriers of the AD risk allele, APOEε4 (heterozygous ε3/ε4) and 46 non-carriers (homozygous ε3/ε3) were enrolled for a total of 80 study participants. Exclude carriers of the APOEε2 allele because there is evidence that this allele may have a protective effect related to AD.
3. Functional and Structural Imaging Data Collection
- Use a 3 Tesla (3T) MRI system to acquire whole-brain imaging data.
- For functional imaging, collect axial slices using an echo planar imaging (EPI) sequence. To facilitate registration of the functional images, acquire axial slices of T2-weighted, co-planar structural images. For high-resolution structural imaging, collect axial slices using a 3D T1-weighted sequence.
NOTE: In the present study, a 3T magnet was used with a 12-channel head coil. The parameters below were designed for a specific scanner and coil. See Table of Materials for more information.- Acquire functional imaging data using the following sequence parameters: repetition time (TR) = 2,500 ms, echo time (TE) = 21 ms, field of view (FOV) = 200 mm x 200 mm, flip angle = 75°, matrix = 64 x 64, 33 slices, slice thickness = 3 mm, interslice gap = 0.75 mm, voxel size = 3.125 x 3.125 x 3.75 mm.
- Trigger the unrelated words associative memory task to begin with the third volume of the functional imaging sequence. To account for steady-state equilibrium, exclude the first two volumes of each functional scan from analyses.
NOTE: The unrelated words associative memory task has been described elsewhere12,24. Briefly, it is a block-design functional task with encoding and retrieval blocks. Participants are instructed to learn pairs of unrelated words. - Acquire T2-weighted, co-planar structural imaging data using the following sequence parameters: TR = 5,000 ms, TE = 34 ms, FOV = 200 mm x 200 mm, flip angle = 90°, matrix = 128 x 128, 28 slices, slice thickness = 3 mm, interslice gap = 1 mm and voxel size = 1.56 x 1.56 x 4 mm.
- Acquire high-resolution structural (anatomical) imaging using the following Magnetization Prepared Rapid Gradient Echo (MPRAGE) sequence parameters: TR = 1,900 ms, TE = 2.26 ms, TI = 900 ms, FOV = 250 mm x 218 mm, flip angle = 9°, matrix = 256 x 215, 176 slices, slice thickness = 1 mm, zero-filled to a matrix of 256 x 224 resulting in a voxel size = 1 x 0.976 x 0.976 mm.
- For functional imaging, collect axial slices using an echo planar imaging (EPI) sequence. To facilitate registration of the functional images, acquire axial slices of T2-weighted, co-planar structural images. For high-resolution structural imaging, collect axial slices using a 3D T1-weighted sequence.
4. fMRI BOLD Data Preprocessing
- Preprocess the functional data using Functional MRI of the Brain (FMRIB) Software Library (FSL) version 6.0 (http://fsl.fmrib.ox.ac.ul) as follows:
- For each participant's dataset, remove head motion artifact from the data using the Motion Correction FMRIB's Linear Image Registration Tool (MCFLIRT)25.
- Remove non-brain tissue from the images using brain extraction tool (BET) with the optional -F flag26.
- Use the FSL Motion Outliers tool to identify any volumes in the functional data where there is excessive motion based on frame displacement between volumes. Flag volumes where motion is measured as an outlier (above the 75th percentile + 1.5 times the inter-quartile range) compared to the rest of the scan and use the output of this program to downweight those volumes in analyses.
NOTE: Before running group comparisons, check that average motion, as measured by FSL Motion Outliers, does not differ across the two groups. This will help ensure that findings are not driven by group-related differences in motion.
- Set up the preprocessing and first-level general linear model (GLM) using the graphical user interface (GUI) for FSL fMRI Expert Analysis Tool (FEAT) for the first participant.
NOTE: Repeat this step for each study participant. To save time, after setting up one run for one participant, write a script to run preprocessing for the remaining study participants' data by altering the "design.fsf" file (FSL FEAT output) for each participant to reference that participant's specific data.- In the data tab, click on "add 4D data" and navigate to the motion-corrected and brain-extracted file. Set the TR to 2.5 s (corresponding to the TR of the functional sequence acquired). Use the default high pass filter (set to 100 s).
NOTE: High pass filtering will remove low frequency signals of no interest. - In the pre-stats tab, click "none" under "motion correction" (as it was already performed in step 4.1). Uncheck "BET brain extraction" (as it was already completed in step 4.1). Type "5" in the box to set 5 mm full-width half-maximum (FWHM) Gaussian kernel for spatial smoothing.
NOTE: The FWHM for the smoothing kernel should generally be set at about twice the size of the functional scan voxel size. - Use the output (6 columns, rows = # of TRs in the scan) of MCFLIRT to create 6 single-column text files that describe the motion correction performed at each volume within the dataset. These will be added to the model as regressors in the next step.
- In the stats tab under "full model setup", add the 6 motion parameters and their temporal derivatives as regressors or explanatory variables (EVs) in the GLM. For each motion EV choose "custom" (1 entry per volume) for basic shape, "none" for convolution and check "apply temporal filtering."
NOTE: Motion parameters do not need to be convolved by any function because they reference the realignment performed at each functional volume during motion correction and thus do not need to be adjusted.
- In the stats tab under "full model setup", add the 6 motion parameters and their temporal derivatives as regressors or explanatory variables (EVs) in the GLM. For each motion EV choose "custom" (1 entry per volume) for basic shape, "none" for convolution and check "apply temporal filtering."
- In the stats tab, select the output of FSL Motion Outliers from step 4.1 under the "add additional confound EVs".
NOTE: This output is a matrix denoting each volume that was flagged for excessive motion and, by adding the confound file, will be deweighted in the GLM. - In the stats tab, click "full model set-up". Create the task timing text files denoting the onset and offset of different task phases and add these as EVs in the GLM by choosing 1 column format and navigating to the relevant text file (include one for the encoding phase of the task and one for the retrieval phase). For "convolution" choose the "double-gamma HRF" option from the drop down list for both of them. Do not model the baseline or non-active portions of the task in the GLM.
NOTE: HRF stands for hemodynamic response function. Convolving the task EV by the HRF shifts the timing of the task EV to be more consistent with expected task-induced BOLD signal changes in the brain. - In the registration tab, check "expanded functional image" and "main structural image" for a two-step registration.
- Select the participant's co-planar T2-weighted structural scan for the first step, in which functional data is registered to the co-planar structural data. Choose 6 degrees of freedom (DOF) for this step by clicking on the second drop down box under this step and choosing "6 DOF".
- For the next step, in which the T2-weighted image is registered to the high resolution T1-weighted MPRAGE, select boundary based registration (BBR) from the drop down box27.
NOTE: BBR uses intensity differences between white matter and gray matter to register structural and functional scans and has been shown to perform better than FLIRT and other alternative methods. - For the final step, in which the high-resolution structural data is registered to the standard MNI152 template, choose 12 degrees of freedom and a linear transformation by choosing "12 DOF".
NOTE: When all the steps in section 4 are complete the functional data are preprocessed and ready for further analysis.
- In the data tab, click on "add 4D data" and navigate to the motion-corrected and brain-extracted file. Set the TR to 2.5 s (corresponding to the TR of the functional sequence acquired). Use the default high pass filter (set to 100 s).
5. Hippocampal Seeds
- Generate a mask of the left hippocampus in each participant's high resolution structural space using FSL's FMRIB Integrated Registration and Segmentation Tool (FIRST) segmentation algorithm28.
NOTE: Other regions, including right hippocampus, would be interesting and valid seeds for further analyses. - Using a statistical software platform, write code in to calculate the length of the anterior and posterior thirds of the structure29. Specifically, use the length of the volumetric hippocampal mask in the anterior-posterior plane to find the coordinates demarking the anterior and posterior thirds of this plane.
NOTE: A recently published method of segmenting the hippocampus along the longitudinal axis might be an alternative seed creation approach30. - Based on these coordinates, create anterior and posterior hippocampal mask images. Register the anterior and posterior hippocampal masks into native functional space using the "example_func2highres" matrix in the registration directory of the FEAT output.
NOTE: Using the anterior and posterior thirds prevented signal blurring across the two hippocampal seeds after registration to functional space. There is evidence of functional specialization along the longitudinal axis of the hippocampus31,32,33,34. Anterior regions are input regions and associated with encoding, while the posterior hippocampus is an output region associated with memory retrieval and consolidation35,36,37. Thus, using these regions allows assessment of functional involvement of anterior versus posterior hippocampus in encoding versus retrieval phases of the memory task. - Use FSL mean timeseries (fslmeants) to extract the denoised average timeseries from the anterior and posterior hippocampal seeds (Figure 1). Follow the program instructions and use either the anterior or posterior hippocampal seed as the mask and the denoised, preprocessed functional data as the main image.
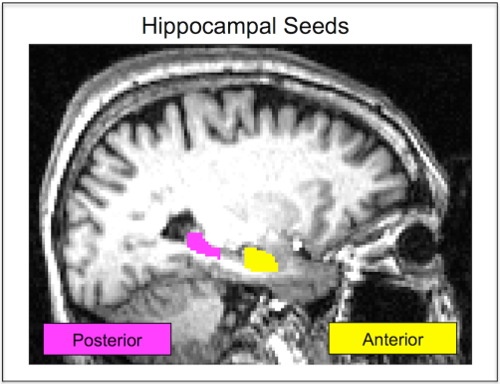
Figure 1: Hippocampal Seeds. In native space, a single participant's anterior hippocampus seed is shown in yellow. The posterior hippocampus seed for the same participant is shown in pink. Seeds are defined in each participant's unique structural image and then registered to their functional scan. Seeds are never in a standardized space, which improves the accuracy of the hippocampal segmentation. This figure has been reprinted with permission12. Please click here to view a larger version of this figure.
6. PPI Model
- Use the GUI for FSL FEAT to load the preprocessed functional data.
- In the data tab, choose the "filtered_func_data" denoised image (output from the steps completed in section 4) as the input file. In the pre-stats tab, set motion correction and brain extraction to "none." Unclick boxes to perform temporal filtering and spatial smoothing.
- PPI Model Set-Up (Table 1).
- In the stats tab, select "full model set-up". In the EVs tab, add all the EVs from the first level model: 6 motion correction EVs, confound EV matrix from FSL Motion Outliers and task timing EVs. Click the up arrow to add EVs. Include in this model an EV for the physiological timecourse from the seed (the text file output of fslmeants in step 5.4) as a covariate of no interest by clicking on the up arrow.
- Create the PPI terms.
- Choose "interaction" in the basic shape menu and select the seed timecourse EV and one task EV. For the "make zero" option, choose "mean" for the seed timecourse EV and "centre" for the task EV. Repeat this procedure for the other task phase(s). Run a separate model for each seed region.
NOTE: These new EVs are the PPI terms for the phase of the task selected (psycho) and the seed (physio). In the present study, a PPI term for the encoding phase and a second PPI term for the retrieval phase were included in each PPI model. The "centre" option ensures that the "on" and "off" phases of the block design task are treated equally. The "mean" option is always applied to the seed timecourse and results in the mean being subtracted from this regressor.
- Choose "interaction" in the basic shape menu and select the seed timecourse EV and one task EV. For the "make zero" option, choose "mean" for the seed timecourse EV and "centre" for the task EV. Repeat this procedure for the other task phase(s). Run a separate model for each seed region.
- In the contrasts and F-tests tab, model the following specific effects by entering "1" in the corresponding EV cell: psych_enc (encoding task phase), psych_ret (retrieval task phase), phys (seed timecourse), PPI_enc (PPI of seed and encoding), PPI_ret (PPI of seed and retrieval). Lastly, enter a "-1" to model negative PPIs for each task phase.
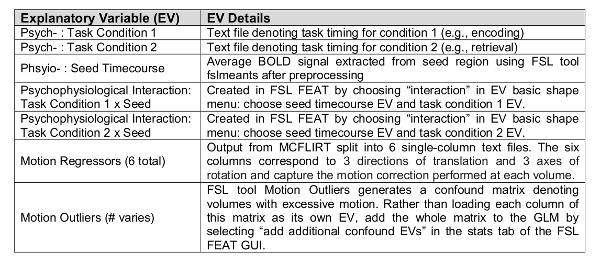
Table 1: gPPI model set-up.
7. Group Comparisons
- Select "higher-level analysis" in FSL FEAT to run a simple group model comparing APOEε4 carriers to non-carriers for each task-seed combination.
NOTE: These comparisons are run to generate the relevant group 4D residuals images ("res4d") needed to estimate the smoothness of the dataset. Statistically significant results from this group comparison are valid, but in the steps below another thresholding approach using AFNI and SPM8 to set a significant cluster minimum based on Monte Carlo simulations is described. - Use Analysis of Functional Neuroimaging (AFNI)
- Use AFNI's 3dFWHMx (any version after December 2015) at the command line to estimate the smoothness of the group 4D residuals images generated using FSL.
NOTE: A bug was discovered in AFNI's 3dClustSim and corrected in May 2015. In December 2015, AFNI's 3dFWHMx was updated to more accurately model auto-correlations. Thus, versions of these tools released in December 2015 or later should be used. - Use AFNI's 3dClustSim (any version after December 2015) to determine cluster extent minimums reaching significance at different voxel-level thresholds. Include the smoothness estimates from the previous step in the command line invocation of 3dClustSim. From the table generated by 3dClustSim, based on the study hypotheses regarding the expected effects' height and extent, choose a voxel-level threshold and corresponding cluster minimum size.
NOTE: In general, larger clusters minimize false positives.
- Use AFNI's 3dFWHMx (any version after December 2015) at the command line to estimate the smoothness of the group 4D residuals images generated using FSL.
- Use Statistical Parametric Mapping (SPM8)
- Using the SPM8 GUI, select "specify 2nd-level". The batch editor will open. Select "two sample t-test" under design. Navigate to the directory with the parameter estimate images for group 1 (APOEε4 carriers) and select by clicking on them. Next, add group 2 (APOEε4 non-carriers) images. Run this comparison by clicking on the green play button.
- Return to the SPM GUI, select "estimate", and navigate to the SPM.mat file created in the previous step to run the model estimation process.
- Select "results" and run group comparison contrasts: APOEε4 carriers > APOEε4 non-carriers, APOEε4 non-carriers > APOEε4 carriers.
- Click on "define a new contrast", choose "T-contrast" under "type" and enter "1 -1" in the "contrast" box for APOEε4 carriers > APOEε4 non-carriers. Click "done". Choose "none" for Apply Masking, and manually set the voxel-level threshold and the cluster size minimum according to the determination made in step 7.2.2.Enter "-1 1" for APOEε4 non-carriers > APOEε4 carriers.
Note: In the present study, a voxelwise threshold of p <0.005 was used and clusters thresholded at alpha <0.05.
- Click on "define a new contrast", choose "T-contrast" under "type" and enter "1 -1" in the "contrast" box for APOEε4 carriers > APOEε4 non-carriers. Click "done". Choose "none" for Apply Masking, and manually set the voxel-level threshold and the cluster size minimum according to the determination made in step 7.2.2.Enter "-1 1" for APOEε4 non-carriers > APOEε4 carriers.
Representative Results
With two different active task phases (encoding and retrieval) and two seed regions (anterior and posterior hippocampus) there are four conditions to report results for each group. The within-group task activation maps (not shown here, see Harrison et al., 201612) show that the occipital lobe, auditory cortex, large regions of parietal lobe, frontal language areas, superior temporal gyrus, and caudate (more pronounced during retrieval) have significant BOLD signal increases during encoding and retrieval in both experimental groups. Within-group PPI analyses revealed that there are no significant increases in functional connectivity with either anterior or posterior hippocampal seeds for either APOEε4 carriers or non-carriers. Within-group PPI analyses revealed significant decreases in functional connectivity in APOEε4 carriers for both task conditions and hippocampal subregions (Figure 2). In APOEε4 non-carriers, significant decreases in functional connectivity were only observed with posterior hippocampus during encoding (Figure 2). The positive and negative PPI maps show a divergence between APOEε4 carriers and non-carriers in how hippocampal functional connectivity changes during a memory task. To determine if the divergence is statistically significant, it is necessary to directly compare the groups for each of the four results38.
For the sake of brevity, group comparison results showing APOE-mediated differences only for one region and task phase, anterior hippocampus during retrieval, are presented here (non-carriers > carriers, Figure 3). During retrieval, the divergence of anterior hippocampus connectivity changes observed within group (Figure 2) results in significant between group differences in bilateral supramarginal gyrus, right angular gyrus and right precuneus.
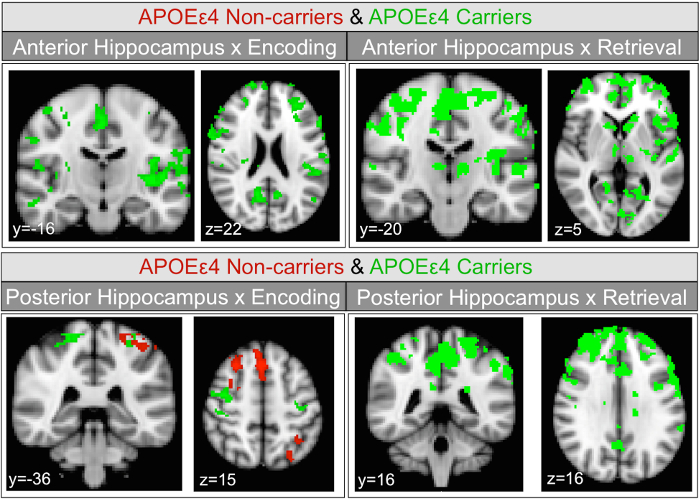
Figure 2: Hippocampal seeds task-dependent negative functional connectivity change maps. Coronal and axial views of group average task-dependent negative functional connectivity change of hippocampal subregions in APOEε4 non-carriers and carriers separately, within group. Task-dependent connectivity decreases with the anterior hippocampus seed are shown in the upper panels. The lower panels show task-dependent connectivity decreases with the posterior hippocampus. Maps were thresholded at z = 2.3, cluster corrected at p <0.05. Voxels meeting threshold in APOEε4 non-carriers (in red) and carriers (in green) are overlaid. This figure has been reprinted with permission12. Please click here to view a larger version of this figure.
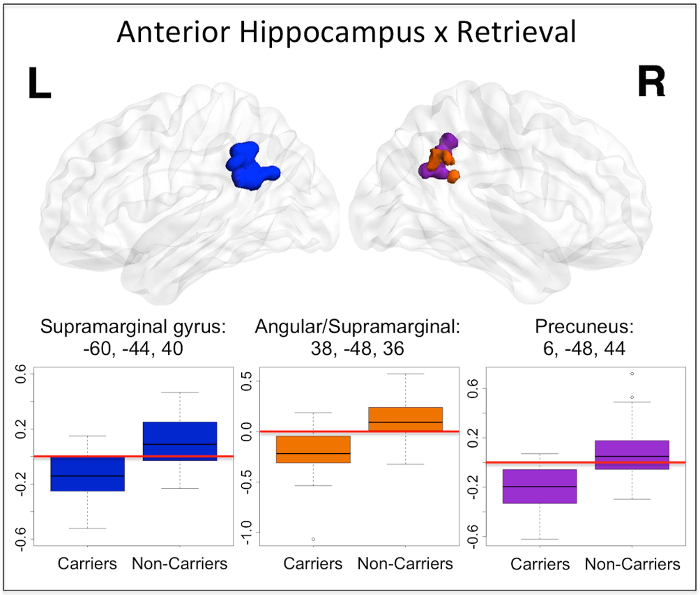
Figure 3: Anterior hippocampal seed connectivity change differences between APOEε4 carriers and non-carriers during retrieval. During retrieval, significant differences between APOEε4 carriers and non-carriers were found in left supramarginal gyrus (dark blue), right supramarginal/angular junction (orange) as well as right precuneus (purple). The results from this two-sample t-test were thresholded to reveal clusters significant at alpha <0.05 with a voxelwise threshold of p <0.005. The peak coordinate for each cluster is reported in MNI space, in x, y, z planes (mm). For illustration of the direction and magnitude of the difference between groups, contrasts of parameter estimates from each cluster are plotted by group. The red horizontal lines indicate zero and highlight that carriers have decreased (negative) functional connectivity to anterior hippocampus in these regions during retrieval. The band within the boxes represents the median while the upper and lower edges of the boxes represent the first and third quartiles, respectively. The whiskers extend up to 1.5 times the interquartile range. Data points outside this range are plotted as outliers. This figure has been reprinted with permission12. Please click here to view a larger version of this figure.
Discussion
Early task-based fMRI studies were designed to uncover statistical relationships between particular cognitive processes or demands and changes in the BOLD signal relative to a baseline measurement. This traditional approach is useful for identifying specific regions in the brain where activity is modulated by an experimental task. In contrast, a PPI analysis is chiefly concerned with the modulation of functional connectivity, or synchrony of activity, that results from a task-induced cognitive process. PPI measures context dependent functional connectivity between a defined region of interest (seed) and other regions of the brain, not just activity increases and decreases in localized areas. The selection of the seed region must be hypothesis-driven as PPI analyses will perform optimally when activity in the seed region is modulated, in a univariate framework, by the task-induced cognitive context. Then, the PPI framework can be used to explore how seed region activity becomes more or less synchronized with other regions in response to specific task contexts, such as memory encoding or retrieval. Differences between groups, therefore, are limited to the functional connectivity changes between the seed and other regions that are modulated by a particular task phase.
A thorough understanding of the GLM is essential for implementing a PPI analysis. A complete, group comparison PPI study has three levels of linear modeling: the first level (preprocessing, task, and motion modeling), the mid-level PPI model (add seed timecourse and task interaction EVs) and the higher level group comparison model (group contrasts of parameter estimates). At each step, an output image is used as the input for the following step. The gPPI approach proposed in 2012 and employed in the present study utilizes features of the GLM to ensure that contrasts are specific to interactions with the task phase of interest3. In the classic PPI, one models two conditions and an assumption is made that the two conditions are on the opposite side of baseline (if there is a baseline condition). gPPI allows one to accurately model all conditions and does not make any assumptions about how the conditions relate to the baseline condition. Another critical component of any PPI analysis is the appropriate selection of a seed region. Seed regions can be chosen based on prior evidence in the literature, such as in the present study in which the hippocampus was used as the seed region for a memory task. Another method of seed selection is to choose a region where activity significantly increases during a particular task phase. With this method, the seed region is defined not anatomically but using a group of suprathreshold voxels in univariate activation maps. With this approach to seed selection, PPI analyses avoid circularity because the main effect of the task is accounted for and the PPI only reveals effects that are distinct from (over and above) the main effect of the task.
Since PPI was first proposed, the concept of functionally connected, spatially distant brain regions has become broadly accepted. Through the use of resting state fMRI, it has been shown that the brain has intrinsic networks, or sets of regions that are functionally connected at rest. Thus, resting state fMRI studies often investigate functional connectivity, the same term used in PPI studies. The interpretation of functional connectivity, however, differs in resting state fMRI and PPI studies. PPI findings are, by definition, explanatory effects of an interaction between task and seed region that cannot be explained by the task design, the seed timecourse or any other confounding variable4. In resting state fMRI, differences in network activity might be caused by changes in connectivity between specific regions or by overall changes in network activity. Thus, if the goal of a study is to compare changes in functional connectivity between two groups, a PPI approach is better. In contrast, if the goal of a study is to describe differences in intrinsic connectivity between two groups, resting state fMRI analyses are better.
One major limitation of the original PPI framework is the lack of statistical power inherent in the approach4. Because the PPI term (EV) is created using two EVs also included in the model, it is likely to be correlated to both. In a GLM, the variance that can be explained by more than one predictor or EV is not assigned to a single EV. Thus, the PPI term only has power to detect effects that cannot be explained by the task or the seed timecourse, which are both correlated to the PPI term. Because of this, it is likely that false negatives occur in PPI analyses. gPPI, however, has been shown to minimize the number of false negatives and is more sensitive to small effect size findings3,39.
PPI can uncover task-dependent changes in functional connectivity between two regions, but it cannot determine whether activity in one region causes a change in activity in the other. In other words, a PPI analysis cannot be used to explore causality in functional connectivity changes. Other methods, such as dynamic causal modeling, are better suited for analyses of causality in functional data40. PPI analyses can inform the design of experiments using these techniques. In sum, PPI is a useful approach for examining task-specific changes in functional connectivity of a relevant seed region and comparing these changes between groups. Results from PPI studies can lead to a better understanding of the dynamic nature of functional connectivity in health, disease and risk for disease.
Açıklamalar
The authors have nothing to disclose.
Acknowledgements
This work was supported by the National Institute of Aging (grant number R01AG013308 to SYB, F31AG047041 to TMH). The authors used computational and storage services associated with the Hoffman2 Shared Cluster provided by UCLA Institute for Digital Research and Education’s Research Technology Group.
Materials
| 3T manetic resonance imaging scanner | Siemens Medical Solutions | MAGNETOM Trio, A Tim System | 3T MRI Scanner |
| FSL (FMRIB Software Library) | Oxford University | Version 6.0 | Functional Imaging Processing Software |
| AFNI (Analysis of Functional Neuroimaging) | National Institute of Mental Health, National Institutes of Health | Any version after May 2015 | Functional Imaging Processing Software |
| SPM8 (Statistical Parametric Mapping) | University College of London | SPM8 | Functional Imaging Processing Software |
| Matlab Software | The Mathworks, Inc | Version R2012a | Computing Software |
| SDS Software | Applied Biosystems, Inc | 7900HT Fast Real-Time PCR System | Real Time PCR |
| Taqman Assays | ThermoFisher Scientific | Specific to SNP | SNP Genotyping |
Referanslar
- Sporns, O., Tononi, G., Kötter, R. The Human Connectome: A Structural Description of the Human Brain. PLoS Comput Biol. 1 (4), 42 (2005).
- Friston, K. J., Buechel, C., Fink, G. R., Morris, J., Rolls, E., Dolan, R. J. Psychophysiological and modulatory interactions in neuroimaging. NeuroImage. 6 (3), 218-229 (1997).
- McLaren, D. G., Ries, M. L., Xu, G., Johnson, S. C. A generalized form of context-dependent psychophysiological interactions (gPPI): a comparison to standard approaches. NeuroImage. 61 (4), 1277-1286 (2012).
- O’Reilly, J. X., Woolrich, M. W., Behrens, T. E. J., Smith, S. M., Johansen-Berg, H. Tools of the trade: psychophysiological interactions and functional connectivity. Soc Cogn Affect Neurosci. 7 (5), 604-609 (2012).
- Moody, T. D., Sasaki, M. A., et al. Functional connectivity for face processing in individuals with body dysmorphic disorder and anorexia nervosa. Psychol Med. 45 (16), 3491-3503 (2015).
- Simard, I., Luck, D., Mottron, L., Zeffiro, T. A., Soulières, I. Autistic fluid intelligence: Increased reliance on visual functional connectivity with diminished modulation of coupling by task difficulty. NeuroImage Clin. 9, 467-478 (2015).
- Yan, L. -. R., Wu, Y. -. B., Zeng, X. -. H., Gao, L. -. C. Dysfunctional putamen modulation during bimanual finger-to-thumb movement in patients with Parkinson’s disease. Front Hum Neurosci. 9, 516 (2015).
- Cole, M. W., Reynolds, J. R., Power, J. D., Repovs, G., Anticevic, A., Braver, T. S. Multi-task connectivity reveals flexible hubs for adaptive task control. Nat Neurosci. 16 (9), 1348-1355 (2013).
- McLaren, D. G., Sperling, R. A., Atri, A. Flexible modulation of network connectivity related to cognition in Alzheimer’s disease. NeuroImage. 100, 544-557 (2014).
- Morawetz, C., Bode, S., Baudewig, J., Heekeren, H. R. Effective amygdala-prefrontal connectivity predicts individual differences in successful emotion regulation. Soc Cogn Affect Neurosci. , 169 (2016).
- Takashima, A., Bakker, I., van Hell, J. G., Janzen, G., McQueen, J. M. Richness of information about novel words influences how episodic and semantic memory networks interact during lexicalization. NeuroImage. 84, 265-278 (2014).
- Harrison, T. M., Burggren, A. C., Small, G. W., Bookheimer, S. Y. Altered memory-related functional connectivity of the anterior and posterior hippocampus in older adults at increased genetic risk for Alzheimer’s disease. Hum Brain Mapp. 37 (1), 366-380 (2016).
- Wechsler, D. . Wecshler Adult Intelligence Scale, 3rd Edition. , (1997).
- Cauthen, N. R. Verbal fluency: normative data. J Clin Psychol. 34 (1), 126-129 (1978).
- Goodglass, H. P., Kaplan, E. P. . Boston Naming Test, 3rd Edition. , (2001).
- Buschke, H., Fuld, P. A. Evaluating storage, retention, and retrieval in disordered memory and learning. Neurol. 24 (11), 1019-1025 (1974).
- Osterrieth, P. A. Le test de copie d’une figure complex: Contribution à l’étude de la perception et de la memoir. Archives de Psychologie. 30, 286-356 (1944).
- Hamilton, M. The assessment of anxiey states by rating. Br J Med Psychol. 32, 50-55 (1959).
- Hamilton, M. A rating scale for depression. J Neurol Neurosurg Psychiatry. 23, 56-62 (1960).
- Folstein, M. F., Robins, L. N., Helzer, J. E. The Mini-Mental State Examination. Arch Gen Psychiatry. 40 (7), 812 (1983).
- O’Brien, D., Campbell, K. A., Morken, N. W., Bair, R. J., Heath, E. M. Automated Nucleic Acid Purification for Large Samples. J Lab Autom. 6 (2), 67-70 (2001).
- Lehmann, M., Ghosh, P. M., et al. Greater medial temporal hypometabolism and lower cortical amyloid burden in ApoE4-positive AD patients. J Neurol Neurosurg Psychiatry. 85 (3), 266-273 (2014).
- . TaqMan® SNP Genotyping Assays User Guide Available from: https://tools.thermofisher.com/content/sfs/manuals/TaqMan_SNP_Genotyping_Assays_man.pdf (2014)
- Bookheimer, S. Y., Strojwas, M. H., et al. Patterns of brain activation in people at risk for Alzheimer’s disease. N Engl J Med. 343 (7), 450-456 (2000).
- Jenkinson, M., Bannister, P., Brady, M., Smith, S. Improved optimization for the robust and accurate linear registration and motion correction of brain images. NeuroImage. 17 (2), 825-841 (2002).
- Smith, S. M. Fast robust automated brain extraction. Hum Brain Mapp. 17 (3), 143-155 (2002).
- Greve, D. N., Fischl, B. Accurate and robust brain image alignment using boundary-based registration. NeuroImage. 48 (1), 63-72 (2009).
- Patenaude, B., Smith, S. M., Kennedy, D. N., Jenkinson, M. A Bayesian model of shape and appearance for subcortical brain segmentation. NeuroImage. 56 (3), 907-922 (2011).
- . Learn MATLAB Basics Available from: https://www.mathworks.com/support/learn-with-matlab-tutorials.html?s_tid=hp_ff_I_tutorials (2017)
- Lerma-Usabiaga, G., Iglesias, J. E., Insausti, R., Greve, D. N., Paz-Alonso, P. M. Automated segmentation of the human hippocampus along its longitudinal axis. Hum Brain Mapp. 37 (9), 3353-3367 (2016).
- Salami, A., Eriksson, J., Nyberg, L. Opposing effects of aging on large-scale brain systems for memory encoding and cognitive control. J Neurosci. 32 (31), 10749-10757 (2012).
- Schacter, D. L., Wagner, A. D. Medial temporal lobe activations in fMRI and PET studies of episodic encoding and retrieval. Hippocampus. 9 (1), 7-24 (1999).
- Strange, B., Dolan, R. Functional segregation within the human hippocampus. Mol Psychiatry. 4 (6), 508-511 (1999).
- Strange, B. A., Fletcher, P. C., Henson, R. N., Friston, K. J., Dolan, R. J. Segregating the functions of human hippocampus. Proc Natl Acad Sci U S A. 96 (7), 4034-4039 (1999).
- Eldridge, L. L., Engel, S. A., Zeineh, M. M., Bookheimer, S. Y., Knowlton, B. J. A dissociation of encoding and retrieval processes in the human hippocampus. J Neurosci. 25 (13), 3280-3286 (2005).
- Strange, B. A., Witter, M. P., Lein, E. S., Moser, E. I. Functional organization of the hippocampal longitudinal axis. Nat Rev Neurosci. 15 (10), 655-669 (2014).
- Zeineh, M. M., Engel, S. A., Thompson, P. M., Bookheimer, S. Y. Dynamics of the hippocampus during encoding and retrieval of face-name pairs. Science. 299 (5606), 577-580 (2003).
- Nieuwenhuis, S., Forstmann, B. U., Wagenmakers, E. -. J. Erroneous analyses of interactions in neuroscience: a problem of significance. Nat Neurosci. 14 (9), 1105-1107 (2011).
- Cisler, J. M., Bush, K., Steele, J. S. A comparison of statistical methods for detecting context-modulated functional connectivity in fMRI. NeuroImage. 84, 1042-1052 (2014).
- Friston, K. J., Harrison, L., Penny, W. Dynamic causal modelling. NeuroImage. 19 (4), 1273-1302 (2003).

