A Step Beyond BRET: Fluorescence by Unbound Excitation from Luminescence (FUEL)
Özet
Expanding the foundation and applicability of Fluorescence by Unbound Excitation from Luminescence (FUEL) by surveying the relevant principles and demonstrating its compatibility with a multitude of fluorophores and antibody-targeted conditions.
Abstract
Fluorescence by Unbound Excitation from Luminescence (FUEL) is a radiative excitation-emission process that produces increased signal and contrast enhancement in vitro and in vivo. FUEL shares many of the same underlying principles as Bioluminescence Resonance Energy Transfer (BRET), yet greatly differs in the acceptable working distances between the luminescent source and the fluorescent entity. While BRET is effectively limited to a maximum of 2 times the Förster radius, commonly less than 14 nm, FUEL can occur at distances up to µm or even cm in the absence of an optical absorber. Here we expand upon the foundation and applicability of FUEL by reviewing the relevant principles behind the phenomenon and demonstrate its compatibility with a wide variety of fluorophores and fluorescent nanoparticles. Further, the utility of antibody-targeted FUEL is explored. The examples shown here provide evidence that FUEL can be utilized for applications where BRET is not possible, filling the spatial void that exists between BRET and traditional whole animal imaging.
Introduction
The genetic modification of organisms, such as viruses1,2, bacteria3, or small mammals4 to either induce or constitutively express bioluminescence, has been highly successful and widely demonstrated5-7. Bioluminescence, an in vivo chemiluminescent reaction involving naturally occurring reagents, has the advantage of producing light without the need for an external light source. As such, bioluminescent imaging does not suffer from the common drawbacks of auto- and non-specific signal found from fluorescence imaging8. Consequently, bioluminescence has a significant signal-to-noise ratio since any detected signal originates solely from the intended source. While many models have exploited the lux operon from Photorhabdus luminescens (emission maximum centered between 480 and 490 nm) for in vitro and in vivo applications9, its use in small mammals has been problematic due to the very nature of the imaging conditions; the pervading existence of optical absorbers, such as hemoglobin, and scattering agents, such as tissue and bone, strongly affect blue to yellow wavelengths3. The expression of an engineered firefly luciferase (emission maximum at 617nm) has been recently developed and incorporated, providing a tool that greatly overcomes optical absorption10, but is still subject to scattering effects.
In response, there have been multiple attempts to red-shift the emitted signal into the desired optical window of 650-900 nm, a region of minimized absorption and scatter, using bioluminescence resonance energy transfer (BRET)11-13. As a tool to enhance signal detection, BRET, which uses a bioluminescent source as the donor and an added fluorophore as the acceptor, has found limited success. As a seminal example of this phenomenon, "self-illuminating quantum dots" (SIQDs)14 consist of modified Renilla reniformis luciferases bound to the external polymer-lysine layer of commercially available quantum dots (QDs). Upon substrate addition, the resulting bioluminescent reaction induces fluorescence emission from the QDs, generating a significant production of red photons. However, these SIQDs have limited applicability to in vivo visualization of physiologically relevant events. This limited applicability is likely due to the difficulty of linking the dual probe to the organ, cell or gene of interest, since the SIQDs cannot be genetically encoded and therefore would require a secondary modification of the polymer shell. To improve their applicability, alternative SIQDs, where the luciferases are bound directly to the luminescent core, have recently been employed15. Building off of the SIQD concept, a more applicable BRET system was achieved by attaching Cypridina luciferase to an indocyanine dye16, which was capable of specifically targeting tumors in mice while producing a substantial red shift from 460 nm to 675 nm. To undergo non-radiative energy transfer, BRET follows the same primary constraints as its fluorescent counterpart: there must be a strong spectral overlap between the donor emission and the acceptor excitation spectra and the working distance between the two moieties must be on the order of the Förster radius (5-14 nm depending on the donor-acceptor pair, with an effective maximum distance of twice the Förster radius17). This distance dependence greatly limits the types of events that can be observed using BRET as a means to enhance detection.
Recently a new approach was identified and demonstrated under both in vitro and in vivo conditions. Building off the foundation of BRET, Fluorescence by Unbound Excitation from Luminescence (FUEL)18,19 also requires a strong spectral overlap between the luminescent and fluorescent components. However, unlike BRET, FUEL is a completely radiative process whereby the emitted photon from the luminescent source is absorbed by an optically accessible fluorophore, which subsequently emits a red-shifted photon according to the fluorophore quantum yield. Akin to BRET, this approach can also be used to overcome the constraints of imaging in the presence of optical absorbers. The resulting red shift provides an overall increase and specificity in the detected signal due to a decrease in attenuation and a reduction of optical scattering effects. FUEL has been reported to occur between bioluminescent Escherichia coli expressing the lux operon and QDs18,19. While experimentally similar to the SIQDs, a fundamental difference exists: in FUEL, it is not necessary for the luminescent source to be physically bound to the fluorophore, which allows for genetic encoding of the luminescent probe. Due to the successful detection of FUEL between luminescent bacteria and QDs, it is possible that this technique could be applied to both superficial (skin) and deep tissue (lung, liver) infections such as Staphylococcus aureus and Klebsiellia pneumoniae.
Since the report of its experimental significance, FUEL has evolved to include a robust mathematical model20 that can be used to predict acceptable luminescent and fluorescent pairs, and its applications have expanded to include use in identification of photophysical characteristics such as quantum yield. We describe below some of the basic techniques of FUEL. First, we show evidence for this phenomenon over both short (µm) and long (cm) working distances, which fundamentally distinguishes FUEL from BRET. Second, we expand upon the possible FUEL pairs by examining a wide variety of fluorophores and fluorescent nanoparticles. Third, FUEL applications are investigated by comparing targeted and non-targeted FUEL pairs.
Protocol
1. Reagents
- Purchase or develop luminescent bacteria and appropriate culture media such as modified Escherichia coli expressing the luxABCDE21, Vibrio fischeri, Photobacterium sp. Klebsiella pneumoniae22.
- Prepare physiological saline (0.9% NaCl) and culture media solutions according to standard recipes. E. coli and K. pneumoniae were grown in Luria Bertani (LB) at 37 °C, and V. fischeri and Photobacterium sp in LB supplemented with 0.5 M NaCl at 22 °C.
- Purchase or acquire fluorescent probes such as Q-tracker 705 (40 nm diameter, referred to as QD705), Q-tracker 800 (40 nm diameter, referred to as QD800), fluorescent (40 nm diameter) and non-fluorescent polystyrene microspheres (48 nm diameter), and conventional fluorescent dyes.
- Prepare the necessary reagents for bacterial labeling.
- Ensure access to a whole animal bioluminescence imager, such as an IVIS Spectrum, that is capable of detecting signal under a wide range of emission wavelengths and exposure times. A plate reader capable of bioluminescence measurements will suffice for most of the experiments described here.
2. Basic Recapitulation of FUEL
- Starting from individual colonies on standard culture plates, start overnight cultures of luminescent bacteria. Here, V. fischeri were used.
- The day of experimentation, initiate fresh subcultures and allow them to progress until an OD600 of 1-1.5 is achieved. In order to produce comparable results it is best to use bacteria in similar growth states in which they produce intense signals. This can be assured by keeping the OD600 constant.
- Combine aliquots of 100 μl from each with either 5 μl of QD705 or physiological saline (PS), and then add to 895 μl of PS into standard spectroscopic cuvettes. Place the filled cuvettes into the IVIS Spectrum and take the measurements under the appropriate filter sets. Here, the 710-730 nm emission filter was used.
3. FUEL Over Varying Distances
- Fill two reduced volume (1 ml) plastic photometric cuvettes with either 50 μl of QD705 or 97.2 μl of non-fluorescent 48 nm polystyrene microspheres in a total volume of 1 ml PS. This ensures similar solid surface area per total volume between the two entities.
- Prepare a light source cuvette by encasing a third cuvette with standard black tape or some other opaque material capable of blocking light. Carefully prepare two identical optical windows on opposite sides of the cuvette.
- Place the previously filled cuvettes directly onto either side of the light source cuvette. Add a 1 ml aliquot of V. fischeri or other culture (i.e. luminescent E. coli or Photobacterium sp) from a fresh subculture into the light source cuvette and cover it with an opaque material, such as black paper to reduce any light contamination.
- Visualize the three cuvettes under the appropriate emission filters such as the Total Light and 710-730 nm emission filters, with exposure times of 10, 30, and 30 sec, respectively.
- Reposition both external cuvettes at equivalent further distances from the central cuvette, and then visualize again. Repeat until a final face-to-face (central to reduced volume cuvette) distance of 3cm is achieved. At each step, acquire a fluorescence image (450-480 nm excitation, 710-730 nm emission) to validate the QD705 location.
4. Investigating Potential FUEL Pairs
- Fill two reduced volume cuvettes with 1 ml solutions containing either a fluorophore or a fluorescent nanoparticle of interest in one, and PS or PS with non-fluorescent nanoparticles as the negative control in the other. In the work demonstrated here, a variety of potential commercially available FUEL fluorophores and fluorescent nanoparticles were investigated as outlined in Table 1.
- Add a 1 ml aliquot of fresh V. fischeri or other luminescent bacteria to a blacked-out third cuvette containing two physically equal optical windows situated on opposite sides. Cover the cuvette with an opaque substance such as a piece of black paper.
- At a short but equal distance place the two reduced volume cuvettes on either side of the blacked-out cuvette and visualize under the appropriate filters. Here, a distance of 0.7 cm was used. Repeat with the other fluorophores.
5. Targeted FUEL
- Prepare biotinylated antibodies specific to the luminescent bacteria. For example, here primary antibodies specific to K. pneumoniae (α-Kp) were biotinylated using a solution containing EZ-Link Sulfo-NHS-LC-Biotin.
- Remove excess reagent from the antibodies using desalting columns under centrifugation (1,500 x g) for 2 min.
- Use a standard Bradford assay to determine the protein concentration and determine the degree of antibody biotinylation by HABA assay.
- Obtain the α-Kp-biot biotinylated antibody (Ab) batches by mixing 100 μl (10 mM, dissolved in 1x PBS) with 40 μl α-Kp antibody, resulting in a final degree of substitution of 16 biotin residues per Ab and a final protein concentration of 10 mg ml-1.
- Using multiple overnight replicate cultures, target the washed bacteria with the aforementioned antibodies following the appropriate protocol.
- Specifically, wash the K. pneumoniae in 1x PBS, and resuspend in 1x PBS to an OD of 4 (ca. 4 x 108 CFU ml-1 OD-1).
- For each replicate, incubate 100 μl PBS, α-Kp antibodies (10 μl α-Kp-biotB or 10 μl PBS for the control) and 100 μl cells for 90 min at 30 °C. Wash the cells three times in 1x PBS, and resuspend in 196 μl PBS.
- Label the cells with the QD705 streptavidin conjugate and divide them into two equivalent volumes.
- Wash one solution three times and then resuspend it into the appropriate volume.
- Distribute 100 μl of the washed and unwashed solutions into individual wells of a black 96-well plate. Measure the resulting luminescence under the Total Light, 490-510 nm, and 710-730 nm filters. Fluorophore concentration can be determined using 450-480 nm excitation and 710-730 nm emission.
Representative Results
Resonance Energy Transfer (RET) is a non-radiative interaction between a luminescent donor and a fluorescent acceptor, whereby the energy from the exited donor is capable of inducing a fluorescence response from the acceptor through a strong dipole-dipole interaction13. RET, which has been described using fluorescent23, chemiluminescent24, and bioluminescent13 donors, principally requires: 1. Strong spectral overlap between the donor emission and accepter excitation spectra; 2. Appropriate rotational alignment between the two entities; and 3. A working distance no greater than 0.5- to 2-times the Förster radius, R0, between the donor and acceptor17. Contrasting with RET, FUEL occurs when a luminescent source, such as a bioluminescent bacteria, emits a photon that is absorbed and re-emitted by a second entity, such as a fluorophore or a fluorescent nanoparticle, red-shifting the emission spectrum of the original luminescent source. Thus, FUEL follows a standard excitation-emission process akin to standard epifluorescent conditions, yet without the use of a focused excitation. Evidence of this can be readily observed by the simple mixing of luminescent bacteria and highly-fluorescent quantum dots. In the presence of the Vibrio sp, a significant increase in red signal is observed when QD705 were also in solution compared to a non-fluorescent polystyrene microsphere control (Figure 1A). The components necessary to create bioluminescence, essentially the aldehyde substrate, luciferase, and ATP, are all produced and contained within the cytoplasm. Furthermore, the enzymatic luminophore production occurs within the bacterial cytoplasm (Figure 1B). As has been reported elsewhere, the distance between the inner and outer membrane of bacteria is typically greater than 30 nm25-27, a distance that does not allow for significant RET to occur. As well, the fluorescent nanoparticles do not cross into the bacterial cytoplasm since there is no endocytosis or other means of uptake. Together, these constraints indicate that FUEL is the dominant excitation-emission phenomenon that occurs when luminescent bacteria are mixed with fluorescent nanoparticles.
As was previously shown, FUEL exists beyond the range of RET. To investigate the FUEL dependence on distance, standard reduced volume spectrophotometric cuvettes can be used, with one containing a fluorophore solution, a second containing an appropriate control that can control for scatter, and the third containing an aliquot of fresh luminescent solution. It is essential that the central cuvette be enveloped using black tape or another opaque material, save for at least two identical optical windows located on opposite faces, to reduce any potential light contamination from the luminescent source. Further, the two remaining cuvettes need to be placed equidistantly onto either side of the central cuvette. Finally, a fresh luminescent solution, using bacteria or chemiluminescence, should be used with each experiment to ensure maximum light production. Upon appropriate placement, acquire the luminescence signal under the desired filters. The Total Light and 710-730 nm filters were used in this case, though only the data from the latter is shown. After each acquisition, increase incrementally the face-to-face distance from 1.0 cm to 3 cm (Figure 2). Finally, normalize all the data to the brightest point. In the examples used here, this occurred at the shortest distance (D = 1 cm) between the luminescent source and the cuvette containing the QD705. Using this approach, viable FUEL signal can be observed up to the final acquisition suggesting that, in the absence of any optical absorber, FUEL can occur at distances beyond any possible resonance energy transfer and can only be explained to be a purely radiative effect. Fitting the data reveals a decrease in signal as a function of distance D following a D-1 to D-2 dependence. Depending upon the geometric configuration, the former corresponds to the capacitor distance dependence and the latter to the inverse square law for point sources. This result is therefore consistent with our overall acquisition setup, given the size of the cuvette aperture and the distance between the luminescent source and the fluorophore.
FUEL not only applies to quantum dots, but also can be observed using a wide range of fluorophores ranging from the Alexa series to fluorescent microspheres (Table 2). In order to be comparable to the control, equal concentrations of the various fluorophores should be used and the total surface area of the nanoparticles held constant (Table 1). By comparing the fluorophores and nanoparticles to their appropriate controls (PS or PS with non-fluorescent polystyrene microspheres), a significant increase in signal is observed at the emission maximum of each fluorescent entity. The largest relative increase in signal was found to occur where the fluorescent emission maximum was farthest from the luminescent emission maximum. This is most likely due to the increased specificity of the fluorescent signal compared to the broad emission spectrum of the luminescent source. On the contrary, the least reliable FUEL signal was found when the two emission maxima were not well separated. Interestingly, a discernable FUEL signal was found with the Yellow microspheres even though the spectral difference is minimal, due most likely to the substantial quantity of the fluorophore present per bead (350 fluorescein equivalents). The results shown here indicate that under appropriate conditions FUEL can be achieved with a variety of fluorophores, which enables the tailoring of probes chosen for more relevant applications under both in vitro and in vivo conditions. The significance of the observed FUEL signal was determined using a standard two-tailed Student's t-test. As such, only the Alexa555 was found to be incompatible with the luminescent source used.
In order to explore the effects of targeting on FUEL, biotinylated antibodies were used to target luminescent bacteria and streptavidin-linked QDs. It is important that the bacteria or luminescence source provide a layer thick enough to minimize the possibility of RET between the luminophore and the corresponding fluorophore. After incubation and washing to remove unbound antibodies, the biotin-labeled bacteria are then exposed to either streptavidin-conjugated QD705 or non-functionalized QD705 as the control, the solutions divided and one set exposed to further washes in order to remove any non-adhered QD705. Here, for all four conditions, the resulting luminescence was observed under the Total Light, 490-510 nm, and 710-730 nm filters, though only the latter two filters are used for the data analysis. The presence of the QD705 was compared using 450-480 nm excitation and 710-730 nm emission filters. Upon investigation we have found little to no difference in red-shifted signal when the solutions were left in an unwashed state (Figure 3). The resulting fluorescence signal under this condition is also found to be quite similar, suggesting that an equal number of QD705 was present under both the targeted and non-targeted state. However, washing the samples to remove any unbound QD705 provides a nearly two-fold increase in relative red signal for the targeted bacteria compared to their non-targeted control. Investigation by fluorescence can be used to verify the presence or absence of the QD705. Comparatively, the targeted washed condition resulted in a fluorescence intensity that was almost three times less than the unwashed condition, yet the resulting red-shift decreased by only 30%, suggesting that under purely bound conditions the targeting of the bacteria will result in an increase in red-shifted emission. This strongly implicates the utility of targeted FUEL for future applications.
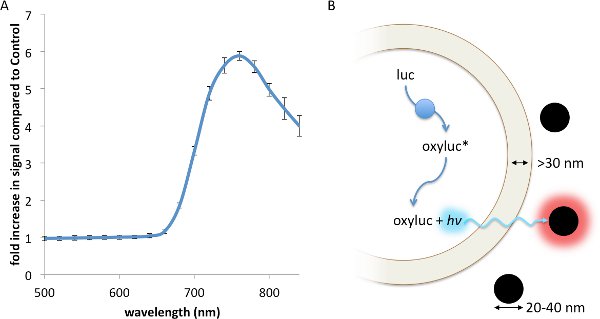
Figure 1. A FUEL interaction between luminescent bacteria and commercially available quantum dots leads to an increase in red photon production. Two spectrophotometric cuvettes were filled with solutions containing 100 μl aliquots of fresh V. fischeri culture with an OD600 of 1-1.5, 895 μl of PS, and either 5 μl of QD705 or physiological saline (PS), before being placed into an IVIS Spectrum and the emission spectrum acquired. A significant increase in red signal is achieved due to the presence of the QD705, as can be noted by the increase in detected photons•sec-1•cm-2 (p•sec-1•cm-2) compared to the control under the 710-730 nm emission filter (A). Here, the dual membrane of the bacteria excludes the interaction of the luminescent moieties (blue circles) and the QD705 (black circles). The former are found only in the bacterial cytoplasm while the latter are freely distributed in the bulk solution (B). N=3 for each bacterial solution. Please click here to view a larger version of this figure.
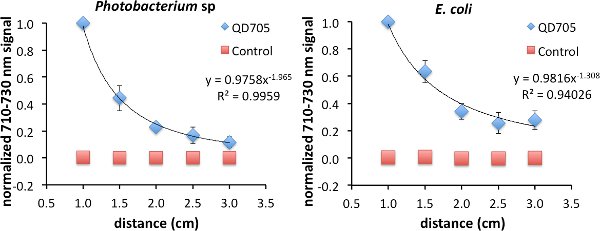
Figure 2. Evidence of FUEL occurring over distances completely excluding resonance energy transfer. Spectrophotometric cuvettes were filled with solutions containing either 50 μl of QD705 and 950 μl of physiological saline (PS) or 97.2 μl of non-fluorescent 48 nm polystyrene microspheres and 902.8 μl of PS, and placed equidistantly on opposite sides of a central black cuvette containing 1 ml of fresh luminescent Photobacterium sp at an OD600 of 1-1.5. The central cuvette had two identical optical windows allowing the emitted photons to freely disperse. Acquiring images at distances (D) ranging from 1.0 cm to 3 cm, the observed production of red light decreased as a function of distance displaying evidence that FUEL can occur at distances not achievable by resonance energy transfer. The intensity appeared to have D-2 dependence, similar to the inverse square law (left). The same protocol was followed for E. coli (right). The resulting trend lines hold form to the concept that the bacteria are acting as point sources of light. A total of three independent distance measurements were acquired with each using a unique subculture. Error bars are present at each point. In some cases the error bars were smaller than the symbols used that indicate the normalized intensity. Please click here to view a larger version of this figure.
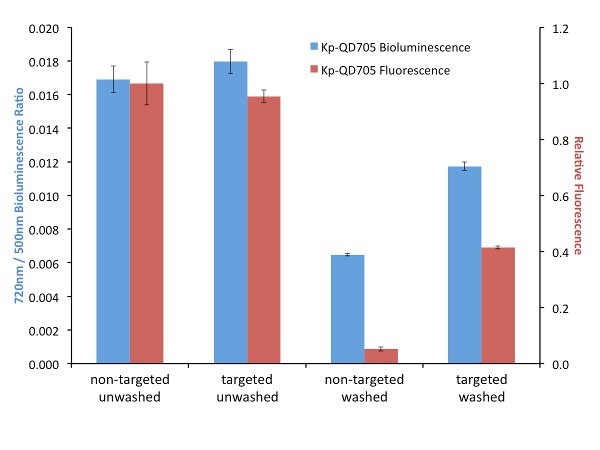
Figure 3. A comparison between targeted (specific) and non-targeted (bulk solution) FUEL. K. pneumoniae were functionalized with biotinylated antibodies and then exposed to either streptavidin-labeled (Targeted) or non-labeled (Non-Targeted) QD705. The resulting solutions were equally divided and one set washed three times with PBS before being resuspended into its initial volume in PBS. The solutions were then dispensed into individual wells of a black 96-well plate and the bioluminescence observed under the 490-510 nm (500 nm) and 710-730 nm (720 nm) emission filters (indicated as blue bars). The p•sec-1•cm-2 found from the 720 nm filter for each well was normalized by the respective p•sec-1•cm-2 bioluminescence intensity from the 500 nm of the same well. The relative fluorescence intensity resulting from an epifluorescent excitation was determined using 450-480 nm excitation and 710-730 nm emission filters (indicated as red bars). Little to no difference in bioluminescent or fluorescent signal was observed under the unwashed conditions (Non-Targeted Unwashed and Targeted Unwashed). This suggests that the bound and free-floating QD705 were equally excited by the bioluminescent photons. Upon washing, nearly a two-fold difference in relative red signal was observed for the QD705-labeled bacteria (Targeted Washed) compared to the control (Non-Targeted Washed). The absence of QD705 in the Non-Targeted Washed was confirmed by the lack of fluorescent signal and verified the QD705 labeling in the Targeted Washed state. The data is from four independent cultures of K. pneumoniae. For clarity, the legend indicates the source of the QD705 excitation. Please click here to view a larger version of this figure.
| Fluorophore | Concentration | μl | PS (μl) | λmax ex/em (nm) | Emission Filter (nm) |
| Alexa 555 | 37.2 µM | 4.77 | 995.23 | 555/565 | 570 – 590 |
| Alexa 568 | 37.2 µM | 4.77 | 995.23 | 578/603 | 590 – 610 |
| Alexa 633 | 37.2 µM | 4.77 | 995.23 | 632/637 | 650 – 670 |
| Alexa 700 | 37.2 µM | 4.77 | 995.23 | 702/723 | 710 – 730 |
| Nonfluorescent | 2.62% solids | 9.72 | 990.28 | ||
| µSph Pink | 5% solids | 4.77 | 995.23 | 580/605 | 610 – 630 |
| µSph Yellow | 5% solids | 4.77 | 995.23 | 505/515 | 510 – 530 |
| QD705 | 2 µM | 5 | 995 | 465/705 | 710 – 730 |
| QD800 | 2 µM | 5 | 995 | 465/705 | 790 – 810 |
Table 1. Properties of fluorophores used throughout the FUEL demonstrations.
| Control | filter | fluorophore | Control | p value | |||
| p·sec-1·cm-2 | SD | p·sec-1·cm-2 | SD | ||||
| A555 | PS | 580 | 1.08 x 107 | 3.31 x 106 | 9.09 x 106 | 1.75 x 106 | 0.233 |
| A568 | PS | 600 | 8.47 x 106 | 4.23 x 106 | 4.49 x 106 | 9.71 x 105 | 0.094 |
| A633 | PS | 660 | 2.40 x 106 | 1.25 x 106 | 7.85 x 105 | 2.39 x 105 | 0.046 |
| A700 | PS | 720 | 5.53 x 105 | 2.46 x 105 | 1.54 x 105 | 6.05 x 104 | 0.026 |
| MSph Yellow | non-fluorescent µspheres | 520 | 1.19 x 108 | 4.85 x 107 | 5.79 x 107 | 1.99 x 107 | 0.057 |
| MSph Pink | non-fluorescent µspheres | 620 | 2.37 x 107 | 1.36 x 107 | 2.16 x 106 | 8.00 x 105 | 0.026 |
| QD705 | non-fluorescent µspheres | 720 | 1.76 x 107 | 7.33 x 106 | 2.08 x 105 | 7.16 x 104 | 0.007 |
| QD800 | non-fluorescent µspheres | 800 | 7.79 x 106 | 4.72 x 106 | 3.60 x 104 | 1.52 x 104 | 0.023 |
Table 2. Identification of fluorophores and fluorescent nanoparticles compatible with FUEL.
Discussion
The fundamental demonstration of FUEL can be achieved simply by mixing luminescent bacteria with fluorescent nanoparticles or QDs. The two entities will be physically separated and remain beyond any efficient RET distance. More difficult is the FUEL signal optimization both in vitro and in vivo. Under in vitro conditions, both with and without an optical absorber present, usually the addition of excess fluorophore will be sufficient to maximize the FUEL response. However, at high concentrations phenomena such as static or collisional quenching can lead to a loss of fluorescent signal. Performing a dilution series by independently varying the concentration of the luminescent source and the fluorophore will help to optimize the desired concentrations. The establishment and optimization of FUEL under in vivo concentrations is much more difficult and needs to be addressed on a case-by-case basis. It can be difficult to create a condition where the fluorescent entity can be accessed optically by the luminescent source. As such, beginning with direct co-injections of the two moieties can provide information regarding the success of FUEL under optimal conditions.
Standard protocols exist for labeling bacteria and eukaryotic cells with fluorescent entities such as the Alexa series and QDs. Often this requires surface functionalization or activation with antibodies, which can lead to unwanted effects like reduced cell viability or altered metabolic activity. To overcome this, it is important to determine the optimal amount of antibody or activation agent needed that minimizes cellular perturbations while maximizing the fluorescent labeling. The use of QDs is advantageous because of their characteristically broad excitation spectra, narrow and tunable emission spectra, and the possibility of a large Stokes shift. However, QDs can be cytotoxic and may not be desirable in some cases.
FUEL is a phenomenon that is present in many BRET experiments13 and is applicable to a variety of luminescent and fluorescent sources. Until now, the photons resulting from FUEL were considered the product of non-specific interactions or an unfortunate background signal resulting from poorly designed BRET experiments. It is only with the type of experiments demonstrated here that we were able to identify the utility of this unwanted signal. In the shown examples, the luminescent bacteria act as a diffuse excitation source capable of eliciting a standard fluorescent response from a wide variety of fluorescent entities. Furthermore, due to the substantial working distance, it is safe to conclude that while FUEL can be constructed without the occurrence of BRET, in general BRET cannot be observed without a contribution from FUEL. Importantly, due to the lack of a targeting requirement, FUEL can be used to cover the spatial gap that exists between BRET and conventional whole-animal imaging techniques.
Açıklamalar
The authors have nothing to disclose.
Acknowledgements
The authors would like to extend their gratitude for financial support from the Pasteur Foundation of New York (to J.D., C.S., C.S.), the EU-FP7 Program “Automation” (to S.L.S.), the Institut Carnot Program 11 (to J.D., A.H., A.R., R.T., S.L.S.) and Project IMNOS (to R.T., S.L.S.), the Conny-Maeve Charitable Foundation (S.L.S.), the European Masters in Molecular Imaging (to I.T.), the Region Ile de France programs MODEXA (S.L.S.), SESAME (S.L.S.) and DimMalInf (S.L.S., R.T.), the ANR Program Grandes Investissement de l’avenir Infrastructures Nationales en Biologie-Santé: France LifebioImaging (FLI) France Life Imaging (R.T., S.L.S.), France Bioimaging (J.D., S.L.S) and the Institut Pasteur, Paris. Further, the authors would like to thank, José Bengoechea and Herbert Schweizer for reagents. Further, The authors would like to thank Cindy Fevre who generated the antibodies.
Materials
| Name of Material/ Equipment | Company | Catalog Number | Comments/Description |
| Escherichia coli expressing the luxABCDE operon | kindly provided by José A. Bengoechea with permission from Herbert P. Schweizer | ||
| Klebsiella pneumoniae 52145 | 52145 is a serotype K2 reference strain | ||
| Luria Bertani (LB) | standard growth media | ||
| Q-Tracker 705 | Life Sciences | Q21061MP | |
| Q-Tracker 800 | Life Sciences | Q21071MP | |
| Alexa 555 | Life Sciences | S21381 | |
| Alexa 568 | Life Sciences | S11226 | |
| Alexa 633 | Life Sciences | S21375 | |
| Alexa 700 | Life Sciences | S21383 | |
| Non-fluorescent microspheres | Polysciences, Inc | 15913 | |
| Pink microspheres | Life Sciences | F8887 | 40nm diameter |
| yellow microspheres | Life Sciences | F8888 | 40nm diameter |
| Ivis Spectrum | PerkinElmer | ||
| EZ-Link Sulfo-NHS-LC-Biotin | Thermo Scientific Pierce | 21425 | |
| Zeba Spin Desalting Columns 7K MWCO | Thermo Scientific Pierce | 21425 | |
| HABA assay kit | Thermo Scientific Pierce | 28005 | |
| Bradford assay | Bio-Rad | 500-0201 | |
Referanslar
- Hu, K., et al. A human immunodeficiency virus type 1 protease biosensor assay using bioluminescence resonance energy transfer. J Virol Methods. 128, 93-103 (2005).
- Contag, C. H., et al. Visualizing gene expression in living mammals using a bioluminescent reporter. Photochem Photobiol. 66, 523-531 (1997).
- Badr, C. E., Tannous, B. A. Bioluminescence imaging: progress and applications. Trends in Biotechnology. 29, 624-633 (2011).
- Maywood, E. S., et al. Analysis of core circadian feedback loop in suprachiasmatic nucleus of mCry1-luc transgenic reporter mouse. Proc Natl Acad Sci USA. 110, 9547-9552 (2013).
- Condeelis, J., Weissleder, R. In Vivo Imaging in Cancer. Cold Spring Harbor Perspectives in Biology. , (2010).
- Madero-Visbal, R. A., et al. Bioluminescence imaging correlates with tumor progression in an orthotopic mouse model of lung cancer. Surgical Oncology. 21, 23-29 (2012).
- Thalhofer, C. J., et al. In vivo imaging of transgenic Leishmania parasites in a live host. J Vis Exp. 10, (2010).
- Troy, T., Jekic-McMullen, D., Sambucetti, L., Rice, B. Quantitative comparison of the sensitivity of detection of fluorescent and bioluminescent reporters in animal models. Molecular Imaging: Official Journal of the Society for Molecular Imaging. 3, 9-23 (2004).
- Michelini, E., Cevenini, L., Mezzanotte, L., Roda, A. Luminescent probes and visualization of bioluminescence. Methods Mol Biol. 574, 1-13 (2009).
- Branchini, B. R., Ablamsky, D. M., Rosenberg, J. C. Chemically Modified Firefly Luciferase Is an Efficient Source of Near-Infrared Light. Bioconjugate Chemistry. 21, 2023-2030 (2010).
- Saito, K., et al. Luminescent proteins for high-speed single-cell and whole-body imaging. Nat Commun. 3, 1262 (2012).
- Dragulescu-Andrasi, A., Chan, C. T., De, A., Massoud, T. F., Gambhir, S. S. Bioluminescence resonance energy transfer (BRET) imaging of protein-protein interactions within deep tissues of living subjects. Proc Natl Acad Sci USA. 108, 12060-12065 (2011).
- Bacart, J., Corbel, C., Jockers, R., Bach, S., Couturier, C. The BRET technology and its application to screening assays. Biotechnology Journal. 3, 311-324 (2008).
- So, M. -. K., Xu, C., Loening, A. M., Gambhir, S. S., Rao, J. Self-illuminating quantum dot conjugates for in vivo imaging. Nat Biotechnol. 24, 339-343 (2006).
- Wu, Q., Chu, M. Self-illuminating quantum dots for highly sensitive in vivo real-time luminescent mapping of sentinel lymph nodes. Int J Nanomedicine. 7, 3433-3443 (2012).
- Wu, C., et al. In vivo far-red luminescence imaging of a biomarker based on BRET from Cypridina bioluminescence to an organic dye. Proc Natl Acad Sci USA. 106, 15599-15603 (2009).
- Piston, D. W., Kremers, G. -. J. Fluorescent protein FRET: the good, the bad and the ugly. Trends Biochem Sci. 32, 407-414 (2007).
- Dragavon, J., et al. in Imaging, Manipulation, and Analysis of Biomolecules, Cells, and Tissues. , 790210-790219 (2011).
- Dragavon, J., et al. In vivo excitation of nanoparticles using luminescent bacteria. Proceedings of the National Academy of Sciences. 109, 8890-8895 (2012).
- Holland, A. D., et al. In vitro characterization of fluorescence by unbound excitation from luminescence: Broadening the scope of energy transfer. Methods. , (2013).
- Choi, K. -. H., Schweizer, H. P. mini-Tn7 insertion in bacteria with single attTn7 sites: example Pseudomonas aeruginosa. Nat. Prot. 1, 153-161 (2006).
- Riottot, M. M., Fournier, J. M., Jouin, H. Direct evidence for the involvement of capsular polysaccharide in the immunoprotective activity of Klebsiella pneumoniae ribosomal preparations. Infect Immun. 31, 71-77 (1981).
- Becker, W. Fluorescence lifetime imaging–techniques and applications. J Microsc. 247, 119-136 (2012).
- Rogers, K. L., et al. Visualization of local Ca2+ dynamics with genetically encoded bioluminescent reporters. Eur J Neurosci. 21, 597-610 (2005).
- Dubochet, J., McDowall, A. W., Menge, B., Schmid, E. N., Lickfeld, K. G. Electron microscopy of frozen-hydrated bacteria. Journal of Bacteriology. 155, 381-390 (1983).
- Graham, L. L., Harris, R., Villiger, W., Beveridge, T. J. Freeze-substitution of gram-negative eubacteria: general cell morphology and envelope profiles. Journal of Bacteriology. 173, 1623-1633 (1991).
- Hobot, J. A., Carlemalm, E., Villiger, W., Kellenberger, E. Periplasmic gel: new concept resulting from the reinvestigation of bacterial cell envelope ultrastructure by new methods. Journal of Bacteriology. 160, 143-152 (1984).

