Preparation of Parasagittal Slices for the Investigation of Dorsal-ventral Organization of the Rodent Medial Entorhinal Cortex
Summary
We describe procedures for preparation and electrophysiological recording from brain slices that maintain the dorsal-ventral axis of the medial entorhinal cortex (MEC). Because neural encoding of location follows a dorsal-ventral organization within the MEC, these procedures facilitate investigation of cellular mechanisms important for navigation and memory.
Abstract
Computation in the brain relies on neurons responding appropriately to their synaptic inputs. Neurons differ in their complement and distribution of membrane ion channels that determine how they respond to synaptic inputs. However, the relationship between these cellular properties and neuronal function in behaving animals is not well understood. One approach to this problem is to investigate topographically organized neural circuits in which the position of individual neurons maps onto information they encode or computations they carry out1. Experiments using this approach suggest principles for tuning of synaptic responses underlying information encoding in sensory and cognitive circuits2,3.
The topographical organization of spatial representations along the dorsal-ventral axis of the medial entorhinal cortex (MEC) provides an opportunity to establish relationships between cellular mechanisms and computations important for spatial cognition. Neurons in layer II of the rodent MEC encode location using grid-like firing fields4-6. For neurons found at dorsal positions in the MEC the distance between the individual firing fields that form a grid is on the order of 30 cm, whereas for neurons at progressively more ventral positions this distance increases to greater than 1 m. Several studies have revealed cellular properties of neurons in layer II of the MEC that, like the spacing between grid firing fields, also differ according to their dorsal-ventral position, suggesting that these cellular properties are important for spatial computation2,7-10.
Here we describe procedures for preparation and electrophysiological recording from brain slices that maintain the dorsal-ventral extent of the MEC enabling investigation of the topographical organization of biophysical and anatomical properties of MEC neurons. The dorsal-ventral position of identified neurons relative to anatomical landmarks is difficult to establish accurately with protocols that use horizontal slices of MEC7,8,11,12, as it is difficult to establish reference points for the exact dorsal-ventral location of the slice. The procedures we describe enable accurate and consistent measurement of location of recorded cells along the dorsal-ventral axis of the MEC as well as visualization of molecular gradients2,10. The procedures have been developed for use with adult mice (> 28 days) and have been successfully employed with mice up to 1.5 years old. With adjustments they could be used with younger mice or other rodent species. A standardized system of preparation and measurement will aid systematic investigation of the cellular and microcircuit properties of this area.
Protocol
1. Parasagittal Slice Preparation
1.1 Dissect out cerebral hemispheres
All animal experimentation should follow local ethical review and national regulations. In the case of the experiments described here, the work conforms to the United Kingdom Animals (Scientific Procedures) Act 1986. We routinely use cervical dislocation without anesthetic to euthanize the mouse before removing the brain. Alternatively the mouse can be terminally anesthetized, but in this case it may be necessary to determine if the choice of anesthetic influences neuronal properties.
Remove the brain from the mouse and immediately place in cold (4-8 °C) cutting artificial cerebrospinal fluid (ACSF) (see Table 1 for solution compositions) bubbled to saturation with carbogen (95% O2, 5% CO2).
After a maximum of three minutes, carefully remove the brain from the cutting ACSF using a spatula and gently place it in an upright position (dorsal side facing upwards) onto filter paper that has been moistened with the cutting ACSF (Figure 1A).
To facilitate accurate mounting of the hemispheres, use a razor or scalpel to remove as much of the cerebellum as possible without impacting the MEC (located at the caudal extreme of the cerebrum) and remove the rostral third of the cerebrum by sectioning in the coronal plane (Figure 1B).
Hemisect the brain, taking care that the section is exactly along the vertical plane of the midline (Figure 1C).
Return the hemispheres to the bubbled cutting ACSF for one and a half minutes.
1.2 Mount the hemispheres on a vibratome
Before mounting, ensure that the cutting edge of the vibratome blade is angled at 20 degrees from horizontal (Figure 1D). On the mounting surface make a shallow strip of superglue parallel to the vibratome blade, approximately the width of a hemisphere and long enough to accommodate the two hemispheres end-to-end.
Taking care to minimize physical impact, remove each hemisphere from the cutting ACSF with a spatula and position so that its medial surface rests on the spatula and its dorsal extent faces towards the microtome blade. Gently slide each hemisphere onto the strip of superglue, taking care to ensure that the medial surface of each hemisphere is parallel to the microtome base. We have found that for best results the dorsal surface of each hemisphere should be parallel to and face the vibratome blade (Figure 1D).
1.3 Maintenance of the preparation during slicing
Following mounting immediately submerge the hemispheres in cold cutting ACSF (4 – 8 °C). Maintain the temperature and carbogen saturation throughout the slicing procedure. If direct cooling and bubbling of the solution in the cutting chamber is not practical, periodically replenish the cutting ACSF solution in the chamber with fresh chilled and bubbled solution.
1.4 Cut sections
Using a vibratome, remove cortex from both hemispheres in the sagittal plane until you identify the lateral most extent of the MEC (typically ~1mm down from the lateral surface) (Figure 1F).
Between cuts lift the vibratome blade up ~200 μm to avoid dragging the blade back over and damaging the exposed tissue. Simultaneously cut 400 μm parasagittal sections from both hemispheres (if they are lined up as shown in Figure 1D they will both be cut at the same time) until the medial extent of the MEC is reached. This can be identified by the absence of the thick white band around the ventro-caudal curve of the hippocampus (the external capsule), the non-convex shape of its rostral boundary and the angular dorso-caudal ‘corner’. Figure 1E illustrates the circular appearance of the hippocampus in the parasagittal plane lateral to the MEC. Figures 1F-G illustrate how sections within the MEC appear at different lateral-medial positions when slicing. Note the progressively more bean-shaped appearance of the hippocampus at more medial positions. Each hemisphere typically yields two or three 400 μm thick slices containing the MEC.
1.5 Incubate slices
After each cut immediately place the slices in carbogen-saturated standard ACSF maintained in a water bath at 35 °C. Allow slices to incubate at 35 °C for approximately 15 minutes in the ACSF after slicing is complete.
Remove the slice holder from the water bath and continue bubbling with carbogen at room temperature for at least 45 minutes.
2. Example Parasagittal Slice Experiment
A typical experiment using this preparation is to make electrophysiological recordings from stellate cells in layer II of the MEC.
2.1 Optimize optics
Before recording, ensure that the condenser is in focus on the slice plane (Koehler illumination) and is centered under the high magnification (e.g. 40x) objective.
2.2 Identify cells of interest
Use a low magnification (e.g. 4x) objective to identify an approximate recording region within the MEC (Figure 2A-B). Switch to a high magnification objective to identify viable cells within this region (Figure 2C-D).
As an example, putative layer II stellate cells are visually identified by their polygonal or ovoid shape and multiple primary dendrites with similar diameter, and absence of a single large diameter apical dendrite2,13,14. They are reliably identified on the edge of the Layer I/II border, where they are abundant and often appear in small groups2,9 (Figure 2C-D). Other cell types, such as interneurons and pyramidal cells should also be identifiable.
At this point experiments can be carried out, for example using whole-cell patch-clamp to record membrane potential or membrane current from identified neurons, and electrical or optogenetic methods to activate synaptic inputs to the recorded neuron. Neuron identity can be verified from the electrophysiological properties of the recorded neuron and by including fluorescent labels within the intracellular solution.
3. Measure Location Along the Dorso-ventral Axis
3.1 Image location of interest and surrounding slice
To determine the position of a recorded neuron along the dorsal-ventral axis, first use a low magnification objective to image the MEC region of the slice and surrounding areas. Mark the location of interest by, for example, including the recording electrode in an image (Figure 3A) or by stepping down the field iris diaphragm to leave a bright circle around the location of interest in a duplicate image. To pinpoint the recording location within the image the duplicate can then be superimposed on the initial image of the recording location and its surroundings (Figure 3B). Up to 3 separate low magnification (4x) images may be required to cover the area from a ventral recording location to the dorsal border of the MEC. These images can then be stitched together using image-manipulation software to provide an image that distance measurements can be taken from (Figure 3). Further verification of neuron identity and location can be carried out following recording by including inactive markers such as biocytin or Alexa dyes within the intracellular solution and then using appropriate processing of the tissue following recording2.
3.2 Establish the dorsal border of MEC
The dorsal border of the MEC provides a convenient landmark from which to measure dorsal-ventral position. The ventral border of the MEC is not as well defined.
Figure 4 illustrates how the MEC cellular landmarks defined in Nissl stained slices at different medial-lateral positions appear under DIC illumination.
Figure 4 A shows the circular hippocampus (Figure 4 (i)) and lack of a parasubicular protrusion into layer I (Figure 4 (iv)) associated with parasagittal slices containing Lateral Entorhinal Cortex (LEC).
Figure 4 B-C show typical slices that contain the MEC. The dorsal part of the prominent dark dorsal Entorhinal/Parasubicular boundary region in the DIC illuminated slices contains an area of the parasubiculum. The parasubiculum area is clearly revealed by Nissl staining in the corresponding images in column (iii). The dorsal border of the MEC (black arrows in 4B and 4C column (iv)) is ventral to the group of parasubicular cells that protrudes far out into layer I (red arrows in 4B and 4C column (iv)).
Comparison of Figures 4B and 4C columns (ii) and (iii) shows that the dorsal border of the MEC in the Nissl sections corresponds to a location that is ventral to the dorsal edge of the dark Entorhinal/Parasubicular boundary region in the DIC slice (black arrows). The location of the border can be estimated from DIC images and comparison to reference images (see also ref. 14). Future validation of the dorsal MEC border with molecular markers will improve the accuracy of this estimate. We note that stained reference sections, and sections that are processed for morphological identification of labeled neurons, may be subject to considerable shrinkage. Comparison of absolute distances relative to landmarks in DIC and reference sections therefore first requires measurement and correction for shrinkage (see e.g. ref 2).
3.3 Calibrate and measure distances
To facilitate easy measurement of distance in images, use the same low magnification objective to image a reference grid to establish a pixel:distance conversion ratio. Measure the pixel distance from the dorsal border of the MEC to the location of interest along the contour of the MEC using a graphics program and convert pixel distance to distance in μm (Figure 5A). If neurons are filled with a marker such as biocytin, then location of the recorded neuron can then also be recovered by appropriate processing (see e.g. ref 2).
4. Representative Results
Figure 5A shows an example low magnification (4x) composite image of a parasagittal slice after recording, with the locations of recorded neurons and measurement guides superimposed. Recordings from the marked dorsal and ventral stellate cells are shown in Figure 5B. These recordings help to establish the identity of the cells and illustrate how electrical properties of MEC layer II stellate cells differ at dorsal and ventral locations.
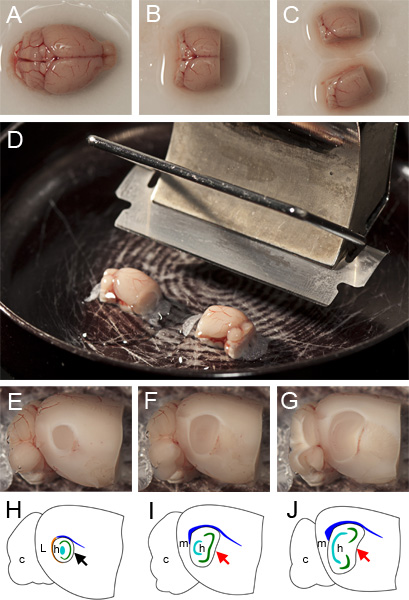
Figure 1. Preparation of parasagittal slices. A Whole brain resting with dorsal side facing upwards on filter paper moistened with cutting ACSF. B Brain after removal of cerebellum and rostral third of frontal cortex. C Hemisected brain ready for mounting. D mounted hemispheres on glue strip parallel to cutting edge of vibratome blade prior to submersion in cutting ACSF and slicing. E Appearance of section containing LEC during the slicing procedure after the removal of lateral tissue and not yet sufficiently medial for a standard parasagittal MEC slice. F Appearance of ‘lateral’ part of the MEC after removing a further 400 μm of tissue (the surface is 400 μm medial to that shown in (F)). G Appearance of ‘medial’ part of the MEC after a 400 μm parasagittal slice has been cut. H-I Schematics of E-F respectively indicating anatomical landmarks, c: cerebellum, L: lateral entorhinal cortex (LEC), m: Medial Entorhinal Cortex (MEC), h: hippocampus, orange: external capsule, blue: corpus callosum, cyan: dentate gyrus, green: CA3 & CA1. The appearance of rostral boundary of the hippocampus in slices that contain LEC is concave (black arrow in H) whereas in slices that contain only MEC, the boundary can be linear (red arrow in I) or concave (red arrow in J). The approximate shapes of colored anatomical landmarks were obtained from the annotations in the Allen Brain Atlas (http://mouse.brain-map.org/atlas/ARA/Sagittal/browser.html).

Figure 2. Identifying MEC layer II cells under DIC illumination. A Example composite image of a parasagittal brain slice at low magnification (4x). Separate images have been aligned and blended to remove distracting edges and vignetting. Important landmark areas h: hippocampus, m: MEC. B Crop from an individual low magnification (4x) image that contains layer II of the MEC, visible as the dark vertical stripe near the right hand edge. C MEC layer II at high magnification (40x) with DIC illumination. The image is an aligned and blended composite of several images. The dense central ‘column’ of cells is Layer II. D Single high magnification DIC image showing a group of healthy putative stellate cells and interneurons in layer II. As shown here, stellate cells often occur in groups on the border of layer I/II. Pyramidal cells tend to be found closer to the Layer II/III border. The dotted outlines in A-C indicate the extent of B-D respectively. Scale bars: in A and B 500 μm, in C and D 100 μm.
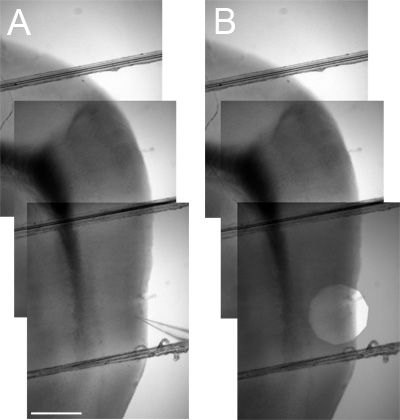
Figure 3. Measuring the dorsal-ventral position of locations of interest. A Aligned low magnification (4x) images that include the entire dorso-ventral extent of the MEC between the location of interest (marked by the tip of a recording electrode) and the dark Entorhinal / Parasubicular boundary region landmark (used to establish the dorsal border of the MEC – see Figure 4). B As in A but including a superimposed image with a stopped down field iris diaphragm (overlaid with reduced opacity) instead of a recording electrode to mark the location of interest.
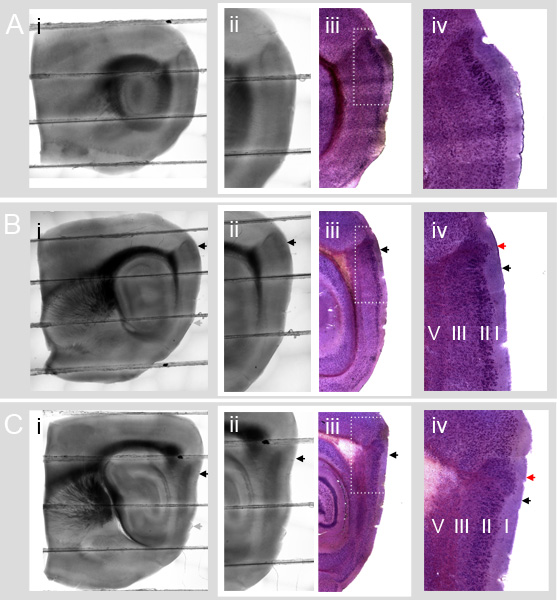
Figure 4. Estimating the dorsal border of the MEC from DIC images of parasagittal slices. A-C Parasagittal sections (lateral to medial) from the Entorhinal Cortex. DIC and Nissl sections are from different mice. A-C column (i): Whole parasagittal slices (400 μm thick) (aligned and blended composites of low magnification (4x) images). A-C column (ii): Close up of entorhinal cortex with characteristic dark Entorhinal / Parasubicular boundary region in the top area of each image. A-C column (iii) Nissl stained slices (40 μm thick) from a different mouse aligned with images in A-C (ii). The darker, dense layer of cells is layer II. Comparing columns (ii) and (iii) shows how the Nissl stained cells appear in DIC images. A-C column (iv): Detail of Nissl stained slices. B and C column (iv) include cells from the parasubiculum (dorsal part of the dark Entorhinal/Parasubicular boundary region in the DIC images) and cells from dorsal MEC. In B (iv) and C (iv) the large dorsal patch of parasubicular cells that extends deep into layer I is easily visible (red arrows). The ventral edge of these patches corresponds to the dorsal border of the MEC (black arrows). In A (iv) the parasubicular patch is absent, indicating that the slice is too lateral for a standard parasagittal MEC slice preparation. In all panels, black arrows indicate the dorsal border of MEC. Grey arrows indicate the approximate ventral border of the MEC.
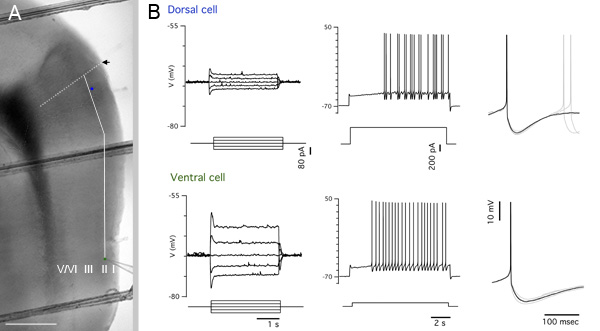
Figure 5. Representative results. A Blended and cropped portion of the image in Figure 3A. The positions of a dorsal cell and a ventral cell are indicated by blue and green filled circles respectively. The black arrow marks the estimated dorsal border of the MEC and is extended into the deep layers by the white dotted line. The solid white line is a guide showing the contour path along which the pixel measurement of distance from the dorsal border of the MEC to the ventrally located cell was taken. Scale bar: 500 μm. B Electrophysiological traces from the cells indicated in A. From left to right – subthreshold responses to current steps used to calculate input resistance, spiking response to a large positive current step, spike detail.
Discussion
To facilitate investigation of MEC circuit properties that follow a dorsal-ventral organization we have described here in detail a procedure for producing a parasagittal slice preparation that preserves the dorsal-ventral extent of the MEC.
Critical steps
Removing the brain from the animal. Take particular care to avoid exerting pressure on the brain. This is more important than rapid removal of the brain.
Slicing. Slice should be firmly glued to the floor of the holding chamber. The vibratome should have z-axis vibration < 2 μm and should be used with high amplitude and low speed settings. The optimal settings can differ between models and must be established by trial and error.
Monitoring solution temperatures. We find that slice quality is sensitive to temperature during and after cutting. Outside the recommended temperature ranges the number of healthy neurons is reduced, neurons can become more difficult to patch and responses to synaptic input can be reduced.
Imaging. Koehler illumination and centering of the field iris diaphragm are important for visualization of cells before recording.
Troubleshooting
Slices contain few healthy cells. Replace solutions. Check z-axis displacement on the vibratome. Check temperature of solutions. Check orientation of the slice when cutting.
Neurons in the slice are difficult to see. Check Koehler illumination is correctly configured. Check microscope lenses are clean.
Difficulty forming gigaseals. Check the shape of recording electrodes. Examine slice for signs of cell death, e.g. round cell shapes, or high contrast cells.
Images of the slice are difficult to align. Ensure the slices are in focus to capture maximum detail. For automatic image alignment >40% image overlap is typically sufficient.
Limitations
One potential limitation of the parasagittal slice preparation is that very long range synaptic connections within the MEC may not run directly parallel to the parasagittal plane9, 15 and so may not be preserved. Maintaining these connections may require modifying the preparation procedure to change the angle of the slice cut by either hemisecting or mounting the brain hemispheres on the vibratome at an angle in the rostral-caudal plane. Similar considerations may apply for preservation of other connections, for example afferent inputs from the medial septum.
Comparison with horizontal MEC slice preparations
- Accurate measurement of dorsal-ventral location is important for establishing quantitative relationships between position along on the dorsal-ventral axis and physiological properties. In horizontal slice preparations7,8,11,12 it is difficult to determine the exact dorsal-ventral location of the cut and this complicates establishing a consistent reference location. In contrast, in the parasagittal slice preparation the dorsal border of the MEC is easy to establish as a reference point and knowledge of the depth of a neuron in a slice is unnecessary to measure its dorsal-ventral position. Parasagittal slices therefore greatly facilitate accurate measurement of dorsal-ventral location, allowing more rapid and precise investigation of quantitative relationships between dorsal-ventral location and cellular and circuit properties.
- Molecular gradients are important for topographical circuit organization in the MEC and can be visualized using fluorescence or other labeling techniques. In comparison to horizontal slices, in which the signal would have to be compared between slices at different dorsal-ventral locations, the parasagittal preparation allows for easier, more fine-grained and more reliable visualization and quantification of dorsal-ventral molecular gradients.
- Dorsal-ventral gradients in synaptic connectivity may play an important role in MEC function. The parasagittal slice preparation can preserve synaptic connections between dorsal and ventral areas within the MEC, which is important for investigating how connections differ within and between distinct locations along the dorsal-ventral axis of the MEC and for maintaining circuit integrity equally at different dorsal-ventral locations. If connectivity varies substantially along the dorsal-ventral axis, horizontal slices are less likely to equally include entire functional microcircuits at each extreme.
Future developments and applications
Knowledge of the molecular determinants of cellular identity in the MEC will enable definitive characterization of neuronal identity and permit more precise determination of dorsal-ventral location.
Using the parasagittal slice preparation, topographically organized responses of cell type- and location-specific activity of multiple neurons could be observed simultaneously (for example using voltage sensitive dyes or calcium imaging) within a single slice.
In combination with optogenetic tools, the parasagittal preparation permits selective activation at different positions along the dorsal-ventral axis. Asking how the microcircuitry at different dorsal-ventral locations responds to spatially targeted activation could deliver important insights into MEC microcircuit function.
We expect that this preparation will provide a simple, robust and standardized basis for investigating dorsal-ventral gradients in physiological properties and facilitate understanding of how these gradients contribute to the information processing properties of the MEC.
Disclosures
The authors have nothing to disclose.
Acknowledgements
We are thank the following for their support: Commonwealth Scholarship Commission UK funding (HP), EPSRC (HP), BBSRC (MFN) and European Union Marie Curie Actions (MFN).
Materials
| Cutting ACSF(mM) | Standard ACSF(mM) | Internal solution (mM) | CASNumber | Supplier Catalogue Number | |
| NaCl | 86 | 124 | 7647-14-5 | Sigma S9888 | |
| NaH2PO4 | 1.2 | 1.2 | 13472-35-0 | Sigma 71505 | |
| KCl | 2.5 | 2.5 | 10 | 7447-40-7 | Sigma P3911 |
| NaHCO3 | 25 | 25 | 144-55-8 | Fischer S/4240 | |
| Glucose | 25 | 20 | 50-99-7 | Sigma G5767 | |
| Sucrose | 75 | 57-50-1 | Sigma S5016 | ||
| CaCl2 | 0.5 | 2 | 10043-52-4 | VWR 190464K | |
| MgCl2 | 7 | 1 | 2 | 7786-30-3 | Sigma 63020 |
| K Gluconate | 130 | 299-27-4 | Sigma G4500 | ||
| HEPES | 10 | 7365-45-9 | Sigma H3375 | ||
| EGTA | 0.1 | 67-42-5 | Sigma E4378 | ||
| Na2ATP | 2 | 34369-07-8 | Sigma A7699 | ||
| Na2GTP | 0.3 | 36051-31-7 | Sigma G8877 | ||
| NaPhospho-Creatine | 10 | 19333-65-4 | Sigma P7936 | ||
| Biocytin (optional) | 2.7 | 576-19-2 | Sigma B4261 |
Table 1. Cutting ACSF, standard ACSF and K-Gluconate internal solution recipes.
References
- O’Donnell, C., Nolan, M. F. Tuning of synaptic responses: an organizing principle for optimization of neural circuits. Trends Neurosci. 34, 51-60 (2011).
- Garden, D. L. F., Dodson, P. D., O’Donnell, C., White, M. D., Nolan, M. F. Tuning of synaptic integration in the medial entorhinal cortex to the organization of grid cell firing fields. Neuron. 60, 875-889 (2008).
- Kuba, H., Yamada, R., Fukui, I., Ohmori, H. Tonotopic specialization of auditory coincidence detection in nucleus laminaris of the chick. Journal of Neuroscience. 25, 1924-1934 (2005).
- Hafting, T., Fyhn, M., Molden, S., Moser, M. -. B., Moser, E. I. Microstructure of a spatial map in the entorhinal cortex. Nature. 436, 801-806 (2005).
- Sargolini, F. Conjunctive representation of position, direction, and velocity in entorhinal cortex. Science. 312, 758-762 (2006).
- Fyhn, M., Hafting, T., Witter, M. P., Moser, E. I., Moser, M. -. B. Grid cells in mice. Hippocampus. 18, 1230-1238 (2008).
- Giocomo, L. M., Zilli, E. A., Fransén, E., Hasselmo, M. E. Temporal frequency of subthreshold oscillations scales with entorhinal grid cell field spacing. Science. 315, 1719-1722 (2007).
- Giocomo, L. M., Hasselmo, M. E. Time constants of h current in layer II stellate cells differ along the dorsal to ventral axis of medial entorhinal cortex. Journal of Neuroscience. 28, 9414-9425 (2008).
- Burgalossi, A. Microcircuits of functionally identified neurons in the rat medial entorhinal cortex. Neuron. 70, 773-786 (2011).
- Dodson, P. D., Pastoll, H., Nolan, M. F. Dorsal-ventral organization of theta-like activity intrinsic to entorhinal stellate neurons is mediated by differences in stochastic current fluctuations. J. Physiol. (Lond). 589, 2993-3008 (2011).
- Nolan, M., Dudman, J., Dodson, P., Santoro, B. HCN1 channels control resting and active integrative properties of stellate cells from layer II of the entorhinal cortex. Journal of Neuroscience. 27, (2007).
- Boehlen, A., Heinemann, U., Erchova, I. The range of intrinsic frequencies represented by medial entorhinal cortex stellate cells extends with age. Journal of Neuroscience. 30, 4585-4589 (2010).
- Klink, R., Alonso, A. Morphological characteristics of layer II projection neurons in the rat medial entorhinal cortex. Hippocampus. 7, 571-583 (1997).
- van Groen, T. Entorhinal cortex of the mouse: cytoarchitectonical organization. Hippocampus. 11, 397-407 (2001).
- Dolorfo, C. L., Amaral, D. G. Entorhinal cortex of the rat: organization of intrinsic connections. The Journal of Comparative Neurology. 398, 49-82 (1998).

