Digital PCR-based Competitive Index for High-throughput Analysis of Fitness in Salmonella
Summary
This molecular-based approach for determining bacterial fitness facilitates precise and accurate detection of microorganisms using unique genomic DNA barcodes that are quantified via digital PCR. The protocol describes calculating the competitive index for Salmonella strains; however, the technology is readily adaptable to protocols requiring absolute quantification of any genetically-malleable organism.
Abstract
A competitive index is a common method used to assess bacterial fitness and/or virulence. The utility of this approach is exemplified by its ease to perform and its ability to standardize the fitness of many strains to a wild-type organism. The technique is limited, however, by available phenotypic markers and the number of strains that can be assessed simultaneously, creating the need for a great number of replicate experiments. Concurrent with large numbers of experiments, the labor and material costs for quantifying bacteria based on phenotypic markers are not insignificant. To overcome these negative aspects while retaining the positive aspects, we have developed a molecular-based approach to directly quantify microorganisms after engineering genetic markers onto bacterial chromosomes. Unique, 25 base pair DNA barcodes were inserted at an innocuous locus on the chromosome of wild-type and mutant strains of Salmonella. In vitro competition experiments were performed using inocula consisting of pooled strains. Following the competition, the absolute numbers of each strain were quantified using digital PCR and the competitive indices for each strain were calculated from those values. Our data indicate that this approach to quantifying Salmonella is extremely sensitive, accurate, and precise for detecting both highly abundant (high fitness) and rare (low fitness) microorganisms. Additionally, this technique is easily adaptable to nearly any organism with chromosomes capable of modification, as well as to various experimental designs that require absolute quantification of microorganisms.
Introduction
Assessing fitness and virulence of pathogenic organisms is a fundamental aspect of microbiology research. It enables comparisons to be made between strains or between mutated organisms, which allows researchers to determine the importance of certain genes under specific conditions. Traditionally, virulence assessment utilizes an animal model of infection using different bacterial strains and observing the outcome of the infected animal (e.g. Infectious Dose50, Lethal Dose50, time to death, symptom severity, lack of symptoms, etc.). This procedure provides valuable descriptions of virulence, but it requires strains to cause considerable differences in outcomes in order to detect variations from wild-type. Furthermore, results are semi-quantitative because while disease progression and symptom severity can be subjectively quantified over time, interpretation of virulence compared to wild-type is more qualitative (i.e. more, less, or equally virulent). A common alternative to performing animal infectivity assays is to generate competitive indices (CIs), values that directly compare fitness or virulence of a strain to a wild-type counterpart in a mixed infection1. This technique has numerous advantages over a traditional animal model of infection by standardizing virulence to a wild-type strain and determining a quantifiable value to reflect the degree of attenuation. This technique can also be adapted to analyze gene interactions in bacteria by determining a canceled out competitive index (COI)2. Calculating a COI for a group of mutated organisms allows researchers to determine whether two genes independently contribute to pathogenesis or if they are involved in the same virulence pathway and dependent on each other. Additionally, calculating a CI requires enumeration of bacteria which can provide valuable insights into the pathogenesis of organisms. CIs and COIs also allow researchers to asses avirulent strains that do not cause clinical disease but still have differences in fitness. This technique is limited by the use of traditional antibiotic resistance markers to identify strains, thereby limiting the number of input strains to only one or two at a time. Because of this limitation, large numbers of experimental groups and replicates are required, which in addition to adding to labor and material costs, also increases opportunities for variability in experimental conditions and inaccurate results. (For a thorough review of the benefits and applications of using mixed infections to study virulence, fitness, and gene interactions, see C.R. Beuzón and D.W. Holden 1)
Attempts have been made to overcome this limitation, such as the use of fluorescently-labeled cells quantified via flow cytometry3,4,5. This technique quantifies cells using either 1) labeled antibodies to phenotypic markers or 2) endogenously produced fluorescent proteins. The use of labeled antibodies has a limit of detection of 1,000 cells/mL, and therefore requires a high number of cells to analyze3. Cells expressing fluorescent proteins have an altered physiology and are susceptible to fitness changes resulting from high protein expression6. Both methods are limited by the number of fluorescent markers detectable using flow cytometry. An advancement in molecular quantification was achieved through the development of a microarray technique that detected attenuation in 120 strains from an initial mixed infection of over 1,000 strains in a murine model7. This technique utilized a microarray analysis of RNA from mutated strains, which lead to considerable variability in the outcome. Nevertheless, it established that large pools of mixed infections can be a useful tool and that by utilizing sensitive detection techniques, differences in bacterial virulence can be identified. With the development of the next generation sequencing, Tn-seq expanded the utility of transposon mutations, enabling a powerful method for quantifying bacteria that were randomly mutated8,9,10,11. An alternative protocol was recently developed that eliminates the need for transposons and instead uses DNA barcodes to more easily identify and track genomic changes and their impact on fitness12. This technology is a major advancement, but the insertion of the genomic barcodes is still a random process. To overcome the randomness of previous experiments, Yoon et al. developed a method to calculate the CIs of Salmonella strains using unique DNA barcodes inserted at precise locations on the chromosomes of bacteria13. Unique barcoded strains were detected using a qPCR-based method with SYBR green and primers specific to each unique barcode. The technique was limited by constraints imposed by qPCR, including differences in primer efficiencies and low sensitivity, evidenced by the need for nested-PCR prior to qPCR. Nevertheless, this approach demonstrated that targeted genomic modifications could be exploited for detecting and potentially quantifying pools of multiple bacterial strains.
In the following protocol, we describe a novel methodology to perform bacterial competition experiments with large pools of mixed inocula followed by accurate quantification using a highly sensitive digital PCR technique. The protocol involves genetically-labeling bacterial strains with a unique DNA barcode inserted on an innocuous region of the chromosome. This modification allows strains to be quickly and accurately quantified using modern molecular technology instead of traditional serial dilutions, replica plating, and counting colony forming units that rely on phenotypic markers (i.e. antibiotic resistance). The modifications allow for simultaneous assessment of many strains in a single pooled inoculum, substantially reducing the possibility of experimental variability because all strains are exposed to the exact same conditions. Furthermore, while this technique was developed in Salmonella enterica serovar Typhimurium, it is highly adaptable to any genetically malleable organism and nearly any experimental design where accurate bacterial counts are required, providing a new tool to increase accuracy and throughput in microbiology laboratories without the constraints imposed by previous methods.
Protocol
1. Incorporate Unique DNA Barcodes onto a Plasmid Containing the Necessary Components for Allelic Exchange
NOTE: A new plasmid, named pSKAP, with a high copy number and increased transformation efficiency compared to the existing pKD13 allelic exchange plasmid was created. This is described in steps 1.1-1.12 (Figure 1). The finalized plasmids containing unique DNA barcodes and components for allelic exchange are available through a plasmid repository (Table of Materials).
- Using a commercial plasmid miniprep kit, purify pKD1314 and pPCR Script Cam SK+ from overnight bacterial cultures grown in Luria-Bertani (LB) broth supplemented with 50 μg/mL kanamycin or 25 μg/mL chloramphenicol (for pKD13 and pPCR Script Cam SK+, respectively) (Table 1).
- Perform restriction digestions on both plasmids using commercial restriction enzymes HindIII and BamHI according to the manufacturer’s specifications.
- Remove restriction enzymes and excised DNA from the pPCR Script Cam SK+ reaction and purify the 3,370-base pair (bp) plasmid backbone using a commercially available DNA cleanup kit according to the manufacturer’s specifications.
- Separate the fragments from the pKD13 restriction digestion on a 1% agarose gel using an electrophoresis chamber.
- Visualize bands using a blue light transilluminator and excise the 1,333 bp fragment from the gel (Figure 1C).
NOTE: This fragment contains the FRT-flanked kanamycin resistance gene required for chromosomal allelic replacement. - Purify the excised DNA from step 1.5 using a commercial gel extraction kit.
- To create pSKAP, ligate the purified fragment from pKD13 (from step 1.6) into pPCR Script Cam SK+ (from step 1.3) using a commercial T4 DNA ligase according to the manufacturer’s specifications.
- Transform chemically competent DH5α cells with the ligated pSKAP plasmid following the manufacturer’s protocol (Table 1).
- Spread transformants onto LB agar plates supplemented with 50 μg/mL kanamycin and incubate at 37 °C overnight.
- Pick a colony from the plate and streak it on a new LB agar plate supplemented with 50 μg/mL kanamycin and incubate at 37 °C overnight. Pick a colony from this plate and use to inoculate LB broth supplemented with 50 μg/mL kanamycin. Incubate the culture overnight at 37 °C with constant agitation.
- Use a commercial plasmid miniprep kit to purify pSKAP from the overnight bacterial culture.
- Perform a diagnostic restriction digestion of the plasmid from step 1.11 using Hind III and Bam HI according to manufacturer’s specifications. Visualize fragments on a 1% agarose gel as in steps 1.4-1.5.
NOTE: The pSKAP total size should be 4,703 bp. Fragments after step 1.12 should be 3,370 and 1,333 bp. - Design PCR primers for insertional site-directed mutagenesis (SDM) (Table 2 and S1) in such a way as to insert a unique 25-basepair DNA sequence at position 725 of pSKAP (Figure 2).
NOTE: The barcode DNA is inserted into the plasmid just outside of the FRT-flanked kanamycin resistance gene, so the barcode is not lost during subsequent removal of the kanamycin resistance cassette. If generating new barcode sequences, use online tools to ensure that the fluorescently-labeled target-specific PCR probes (hereafter referred to simply as “probes”) will efficiently bind to the new sequence. Insertion sequences designed to-date, along with the necessary primers to create them, are provided in Tables 2 and S1. - Prepare SDM reactions using a commercial high-fidelity DNA polymerase, the desired primer pairs (Table 2), and pSKAP template. Set the thermocycler to perform the following: 1) 98 °C for 30 s, 2) 98 °C for 10 s, 56 °C for 15 s, 72 °C for 2 min, 3) repeat steps 2, 24 times, 4) 72 °C for 5 min, 5) hold at 4 °C.
- After completion of PCR, deplete pSKAP template by adding the restriction enzyme Dpn I to the reaction. Incubate at 37 °C for 20 min.
NOTE: Products from PCR should be visualized on an agarose gel to verify the size and purity of the product. A control Dpn I digestion consisting of the unmodified pSKAP template can be performed and used in subsequent steps to ensure template DNA is completely digested. - Use 5 μL of the product from step 1.15 to transform 100 μL of commercial chemically competent DH5α cells according to the manufacturer’s recommendations.
- Spread transformants onto LB agar plates supplemented with 25 μg/mL chloramphenicol and incubate at 37 °C overnight.
- Select a colony (or colonies) from overnight plates and streak onto individual LB agar plates supplemented with 25 μg/mL chloramphenicol and incubate at 37 °C overnight. Select a colony from overnight plates and use to inoculate 5 mL of LB broth supplemented with 25 μg/mL chloramphenicol. Incubate culture(s) overnight at 37 °C with constant agitation.
- Use a commercial plasmid miniprep kit to purify plasmids from the overnight culture(s).
- Sanger sequence purified plasmids using the M13 Forward sequencing primer (Table 2). Compare mutated region to the original plasmid and assess for SDM insertional accuracy.
- After confirming the barcode insertion and accuracy, assign each barcode and plasmid a name.
NOTE: Barcodes generated to-date have been assigned a two-letter designation: AA, AB, AC, …, BA, BB, BC, etc. Barcoded plasmids are denoted as pSKAP_AA, pSKAP_AB, pSKAP_AC, …, pSKAP_BA, pSKAP_BB, pSKAP_BC, etc. - Repeat steps 1.14-1.21 to generate the desired number of DNA barcodes.
2. Introduce DNA Barcode onto the Chromosome of S. Typhimurium
NOTE: Insertion of DNA barcodes onto the S. Typhimurium chromosome is achieved by using an allelic exchange method described by Datsenko and Wanner14 that has been modified for use in S. Typhimurium.
- Determine the locus on the S. Typhimurium genome at which to insert the DNA barcode (Figure 3).
NOTE: Select a large, intergenic region of the chromosome. Avoid regions that produce non-coding RNA. This study utilized a locus downstream of put P between residues 1,213,840 and 1,213,861 (determined from genome assembly GCA_000022165.1). This region has previously been genetically manipulated for in trans complementation of genes15. Alternatively, a DNA barcode could be introduced while simultaneously disrupting a gene of interest. Doing so would require minimal alterations to this protocol and streamline mutant creation. - Design PCR primers to amplify the unique barcode and FRT-flanked kanamycin resistance gene from the desired pSKAP barcode-containing plasmid from step 1.21 (Table 2). Add 40-nucleotide extensions that are homologous to the region selected in step 2.1 to the 5’ end of each primer (Table 2).
- Perform the amplification using a commercial high-fidelity polymerase, the primers from step 2.2, and the desired pSKAP barcode-containing plasmid as template. Set the thermocycler to perform the following: 1) 98 °C for 30 s, 2) 98 °C for 10 s, 3) 56 °C for 15 s, 4) 72 °C for 60 s, repeat steps 2-4 29 times, 5) 72 °C for 5 min, 6) hold at 4 °C.
- After completion of PCR, deplete template DNA using DpnI as described in step 1.15. Purify and concentrate the DNA using a commercial DNA cleanup kit according to the manufacturer’s specifications.
- Refer to the previously published protocol for generating mutants in S. Typhimurium strain 14028s from PCR products14,16,17,18.
NOTE: It is not essential that the kanamycin resistance gene is excised from the chromosome. However, excision of the gene is minimally disruptive to the bacterial chromosome as it results in a 129 bp scar that would leave potential downstream genes in-frame. It is recommended that the strain containing the kanamycin resistance gene is retained as it can be used to move barcodes between strains via P22-mediated transduction. - Repeat steps 2.3 – 2.5 to create the desired strains with the appropriate barcodes.
NOTE: Barcodes can be introduced into wild-type S. Typhimurium that can then be subjected to further genetic manipulation, or barcodes can be introduced into strains that have been previously genetically altered.
3. Bacterial Growth Conditions and In Vitro Competition Assays
- From bacteria stock, streak desired S. Typhimurium strains that each harbor a unique DNA barcode onto LB agar plates. Incubate plates overnight at 37 °C.
- Select a single colony from each strain and inoculate 5 mL of LB broth. Incubate for 20 h at 37 °C with constant agitation.
NOTE: Using the overnight culture, proceed to step 3.5 to collect pure genomic DNA (gDNA) from each barcoded strain. This is necessary for subsequent validation and control experiments in section 6. - For each competition assay, transfer an equal volume of each overnight culture into an appropriately sized sterile tube. Thoroughly mix strains together by vortexing vigorously for at least 5 s.
NOTE: The volume of each overnight culture to transfer should be sufficient for each condition and replicate, as well as for isolating gDNA to quantify input. While it is not entirely necessary to measure optical densities of cultures because the absolute number of input microorganisms will be quantified using digital PCR, the number of input bacteria for each strain should be approximately equal to avoid bottlenecks or unequal competition early in the experiment. Representative competition assays in this protocol compared growth rates of 8 strains simultaneously. Additional or fewer strains may be necessary for individual experimental designs. - Transfer 100 μL of the mixed inoculum into 4.9 mL sterile LB broth. Incubate at 37 °C for the desired time or to a desired optical density.
- Harvest 500 μL of the inoculum by centrifugation at >12,000 x g for 1 min. Remove and discard the supernatant. Proceed immediately to section 4 with the cells.
- At desired time points, remove 500 μL aliquots of culture and harvest cells by centrifugation at >12,000 x g for 1 min. Remove and discard supernatant.
NOTE: If collecting aliquots at multiple timepoints, freeze pellets at -20 °C or immediately proceed to step 4.1 after each collection.
4. Collecting and Quantifying gDNA from S. Typhimurium (from Steps 3.5 and 3.6)
- Harvest gDNA from cells using a commercial gDNA purification kit. If available, perform the optional RNA depletion step.
NOTE: RNA depletion is not necessary; however, the presence of RNA will artificially increase the DNA concentration, leading to aberrant calculations in subsequent steps. If using a commercial gDNA purification kit, follow the manufacturer’s recommendations to ensure that the column is not overloaded with DNA. There is no minimum DNA concentration required if the sample is quantifiable in subsequent steps. - Use a spectrophotometer to quantify DNA in each sample.
NOTE: DNA can be quantified using any reliable method. - Calculate gDNA copy number based on the bacterial genome size using the following equation where: X is the amount of DNA in ng and N is the length of a double-stranded DNA molecule (the genome size).
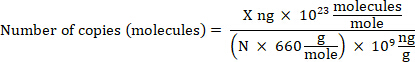
NOTE: 660 g/mole is used as the average mass of 1 DNA bp. Small variations may exist depending on the organism’s nucleotide composition. Numerous calculators are available online to perform the calculation.
5. Design Primers and Probes for Quantitative Detection of DNA Barcodes via dDigital PCR
- Design primers to amplify the barcoded region of the S. Typhimurium chromosome downstream of putP (Figure 3C and Table 2).
NOTE: Primer and probe designs can be facilitated by numerous online programs (Table of Materials). If barcodes are all inserted at the same loci, a single set of amplification primers is universal for all barcodes. - Design 6-carboxyfluorescein (FAM)-based and/or hexachlorofluorescein (HEX)-based probes specific to each barcode (Table 2 and S1).
NOTE: The droplet reader used in this experiment is capable of detecting both FAM- and HEX-based probes simultaneously in a multiplex reaction. Design ½ of the probes to utilize FAM and ½ to utilize HEX. This is not a necessary step but will reduce the reagent use and experimental costs if implemented. - Make 20x primer-probe master mixes containing 1) 20 mM of each forward and reverse amplification primer, 2) 10 mM of a single FAM probe, and 3) 10 mM of a single HEX probe (if multiplexing).
6. Validate the Sensitivity and Specificity of Each Pprimer-probe Set for Each Genomic Barcode Using Digital PCR
NOTE: This protocol uses validating eight unique barcodes with eight unique probes as an example. The number of barcodes utilized can be increased or decreased to accommodate various experimental designs.
- Create a pool of gDNA that contains every barcode except for one. Use this pool as the diluent to perform a dilution series with gDNA containing the single remaining barcode (sample dilution scheme is provided in Figure 4).
NOTE: Using pooled gDNA as a diluent ensures a consistent background while ascertaining sensitivity. Using the copy numbers determined in step 4.3, dilute gDNA to a copy number within the recommended digital PCR range (1-100,000 copies per 20 μL reaction). Keep in mind that the range is set for each unique target (barcode), not the total gDNA. - Prepare reactions for digital PCR in duplicate according to the manufacturer’s recommendations for using a digital PCR supermix designed for probe-based chemistry. Use the mixtures from step 6.1 as template DNA. Use the 20x primer-probe master mix from step 5.3 that contains the probe for the diluted barcode in step 6.1.
- Prepare replicate control reactions for digital PCR according to the manufacturer’s recommendations for using a digital PCR supermix designed for probe-based chemistry. Control reactions for each probe mix must consist of 1) no template controls (NTCs), 2) negative controls, and 3) positive controls.
NOTE: The minimum number of replicate control reactions is two. This example protocol uses four NTCs, six negative controls, and six positive controls for each barcode. Negative controls should contain gDNA with each barcode except for the barcode corresponding to the probe being tested. This will validate the specificity of each probe. - Repeat steps 6.1-6.3 to create digital PCR reactions for each barcoded gDNA sample. A sample plate arrangement is presented in Figure 5.
NOTE: This and subsequent steps are described based on a specific digital PCR platform that utilizes droplets and flow-based technology. Alternative digital PCR platforms that utilize chip-based technology can easily be substituted with slight modifications to this protocol. Step 6.4 may require more than one 96-well plate to validate all primer sets. In contrast to qPCR, separate plates analyzed by digital PCR can be readily compared without the need for standardized reference wells between plates. - Generate droplets for each reaction condition using a droplet generator according to the manufacturer’s instructions.
- Transfer newly created droplets into the appropriate 96-well plate. Use 200 μL pipette tips on a 5-50 μL multichannel pipette.
NOTE: When pipetting droplets, pipette slowly and smoothly! The digital PCR equipment manufacturer recommends using only pipettes and pipette tips from a particular manufacturer (e.g., Ranin). These pipette tips have a smooth opening with no microscopic plastic fragments that can destroy droplets or damage the microfluidics of the droplet reader. Numerous brands of tips were examined and observed to have a spectrum of manufacturing quality. Equivalent results have been achieved using pipette tip alternatives; however, caution should be used when deviating from the manufacturer’s recommendations. - After all the droplets have been generated and transferred, seal the plate with a foil plate sealer.
- Use the manufacturer-recommended thermocycler to perform the following cycling conditions: 1) 94 °C for 10 min; 2) 94 °C for 1 min, ramp rate set at 1 °C/s; 3) 55 °C for 2 min, ramp rate set at 1 °C/s; 4) repeat steps 2 and 3 49 times; 5) 98 °C for 10 min; 6) hold at 4 °C up to 24 h.
NOTE: Thermal transfer in a droplet reaction is not the same as standard PCR. Reaction conditions may require modification. - While thermocycling is being performed, program the data analysis software with the plate setup information such as sample name, experiment type (absolute quantification), supermix used, target 1 name (FAM barcode name), target 1 type (NTC, positive control, negative control, or unknown), target 2 name (HEX barcode name), target 2 type (blank, positive control, negative control, or unknown). The final plate setup information is shown in Figure 5.
- After thermocycling is complete, transfer the completed reactions to the droplet reader and start the reading process according to the manufacturer’s instructions.
7. Quantify the Number of Bacteria in a Competitive Index Experiment
- Dilute gDNA isolated and quantified from section 4 to an appropriate concentration as described above.
- Prepare reactions for digital PCR according to the manufacturer’s recommendations for using the appropriate supermix. Use DNA from step 7.1 as the template. Use one 20x primer-probe master mix that contains the probe (or probes if detecting both FAM and HEX) for possible barcodes present in the experiment.
- Prepare additional digital PCR reactions as in step 7.2 using different 20x primer-probe master mixes until all barcodes utilized in the experimental design can be detected.
- Include controls for each condition as described in 6.3. This includes 1) no template controls (NTCs), 2) negative controls, and 3) positive controls.
- Continue with the protocol as described in steps 6.5-6.10.
8. Analyze Digital PCR Data and Calculate Absolute Copy Numbers
- When all wells have been read and the run is complete, open the .qlp data file using the data analysis software.
NOTE: The file types and data analysis procedures described here are specific to one digital PCR manufacturer. If using alternative digital PCR platforms, file types and data analysis procedures will be specific to the platform used and should be performed according to the manufacturer’s recommended specifications. - Select all wells that utilize the same primer-probe master mix.
- On the Droplets tab, examine the number of droplets analyzed in each well (both positive and negative droplets). Exclude from analysis any well that has fewer than 10,000 total droplets.
- Move to the 1D Amplitude tab and examine the amplitudes of positive and negative droplets. Ensure they comprise two distinct populations.
- Within the software, use the thresholding feature to make a cutoff between positive and negative droplets for each probe that was utilized (Figure 6).
NOTE: All wells that use the same primer-probe master mix from step 5.3 should have the same thresholds. - Once appropriate thresholds have been applied to all wells, the software will calculate the number of DNA copies in each reaction. Export data to a spreadsheet to facilitate further analysis.
NOTE: The data analysis software uses the number of positive and negative droplets that are fit to a Poisson distribution to determine the copy number. - Using the values from step 8.6, calculate the initial copy number of each unique genomic barcode in the sample. Determine the mean false-positive rate from the negative control reactions and subtract this value from the values obtained in experimental reactions. Multiply values as necessary based on the dilutions that were performed when setting up each experiment (Table 3).
9. Determine Relative Fitness of an Organism by Calculating the CI or COI from Digital PCR-based Quantification of Barcoded Strains
- Calculate the CI of a barcoded strain using the following formulas where: AOutput is the absolute quantification of the barcoded strain at a given timepoint, WTOutput is the absolute quantification of barcoded wild-type bacteria at the same timepoint, AInput is the absolute quantification of the input inoculum of the barcoded strain, WTInput is the absolute quantification of the input inoculum of barcoded wild-type bacteria, XOutput is the summation of all strains at the same timepoint, XInput is the summation of the total input inoculum of all barcoded strains.
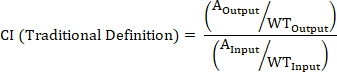
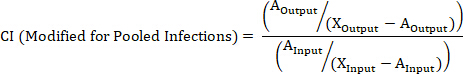
NOTE: See the discussion for advantages, disadvantages, and the most appropriate use of each formula.
- Repeat step 9.1 for each barcoded strain at all timepoints.
- If applicable, calculate the COI using the following formulas where: AOutput is the absolute quantification of a barcoded strain with mutated gene A at a given timepoint, ABOutput is the absolute quantification of a barcoded strain with mutated genes A and B at the same timepoint, AInput is the absolute quantification of the input inoculum of the barcoded strain with mutated gene A, ABInput is the absolute quantification of the input inoculum of barcoded strain with mutated genes A and B, BOutput is the absolute quantification of a barcoded strain with mutated gene B at a given timepoint, and BInput is the absolute quantification of the input inoculum of the barcoded strain with mutated gene B.
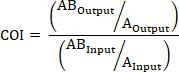
AND/OR
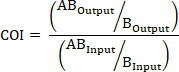
Representative Results
The use of this methodology requires that appropriate control reactions are performed to validate the sensitivity and specificity of each probe used to identify target DNA. In this representative experiment, we validated eight unique DNA barcodes with the eight corresponding probes for identification. All eight probes had a low rate of false positives in both NTC and negative control reactions (Table 3), highlighting their specificity even among highly similar DNA sequences. To assess the sensitivity of each condition, gDNA containing a unique barcode was serially diluted in a constant background of gDNA containing each of the seven remaining barcode sequences. With the approach outlined above, digital PCR could distinguish as few as 2 copies of gDNA in a background of nearly 2,000,000 similar DNA sequences (Table 3).
In addition to determining sensitivity and specificity of each probe and DNA barcode sequence, the dilutions performed in the validation study allowed us to calculate a simulated competitive index from the resulting data. While there was no true input or output for this experiment, the data can be analyzed as though a competition experiment has been performed. To do so, we consider each mixture in the serial dilution as an output (AOutput) for the diluted barcode, while the total output (XOutput), input (AInput), and the total input (XInput) of each strain is calculated from the quantification in the positive controls where all barcodes are included. Using the dilution factor for each mixture, the theoretical CI was determined and is reported in Table 3. In each of the dilution series that was performed for each barcode, the average simulated CI is reported along with the standard deviations for each duplicate dilution series. In all cases, the simulated CI that was calculated is similar to the theoretical CI. The majority of calculated CIs deviate from the theoretical CIs by less than 25%. In cases of lower theoretical CIs, the deviation of the calculated value was upwards of 2-fold. For example, this represented a change from a theoretical CI of 0.000625 to a calculated CI of 0.001220. These data highlight that the described method is both highly accurate and highly precise. The combination of high sensitivity, specificity, accuracy, and precision enable this system to reliably detect differences in fitness that may otherwise go unnoticed.
After validating that genomic barcodes could be accurately detected and quantified, we performed in vitro competition experiments (Table 4). The first competition experiment utilized eight wild-type S. Typhimurium strains that each contained a unique DNA barcode. Each strain was grown overnight, and the eight cultures were mixed together in equal amounts. 100 µL of this mixed inoculum was used to inoculate 4.9 mL of sterile LB broth and the resulting culture was incubated at 37 °C with constant agitation. gDNA was harvested from the inoculum to calculate the exact input of each strain. The growth of the culture was monitored by measuring the absorbance at 600 nm (OD600). At OD600 = 0.5 (logarithmic phase), a sample was collected from each culture and gDNA was harvested. The remaining culture was returned to 37 °C with constant agitation until 8 hours post-inoculation when a final sample was collected and gDNA harvested (stationary). Results were calculated using the CI formula modified for pooled infections. As expected, all wild-type strains had CI values nearly equal to 1 (Table 4). A similar competition experiment was performed using eight mutant S. Typhimurium strains that each had unique barcodes in addition to a single-, double-, or triple-transketolase deficiency18. As shown previously, the strains all grew similarly in LB broth, with only a slight lag observed in the triple-transketolase-deficient strain. However, when the growth of each strain was assessed by analyzing the CI, a much more profound defect was observed for the transketolase-deficient strain (CI was compared to growth curves in Shaw et al.18). Furthermore, this experiment allowed us to assign a quantifiable value to each strain’s growth characteristics instead of merely qualitatively describing the growth patterns. CIs for each strain were calculated using both the traditional formula where each strain was only compared to wild-type and the modified formula where all input strains were considered. While the changes were small, the CI of the triple-transketolase-deficient strain was artificially low in the traditional formula because it does not account for the other six competing strains that all exhibited near-wild-type fitness.
| Strains | Genotype | Source or reference |
| S. Typhimurium ATCC 14028s | wild-type | ATCC |
| TT22236 | LT2 Salmonella carrying pTP2223 | (27) |
| DH5α | F– φ80lacZΔM15 Δ(lacZYA-argF)U169 recA1 endA1 hsdR17(rK–, mK+) phoA supE44 λ– thi-1 gyrA96 relA1 | (28) |
| JAS18077 | putP::AA::FRT | This study |
| JAS18080 | putP::AD::FRT | This study |
| JAS18083 | putP::AG::FRT | This study |
| JAS18088 | putP::AL::FRT | This study |
| JAS18091 | putP::AO::FRT | This study |
| JAS18096 | putP::AT::FRT | This study |
| JAS18099 | putP::AW::FRT | This study |
| JAS18100 | putP::AX::FRT | This study |
| JAS18122 | ΔtktA::FRT putP::AD::FRT | This study |
| JAS18130 | ΔtktB::FRT putP::AL::FRT | This study |
| JAS18138 | ΔtktC::FRT putP::AT::FRT | This study |
| JAS18125 | ΔtktA::FRT ΔtktB::FRT putP::AG::FRT | This study |
| JAS18133 | ΔtktA::FRT ΔtktC::FRT putP::AO::FRT | This study |
| JAS18141 | ΔtktB::FRT ΔtktC::FRT putP::AW::FRT | This study |
| JAS18142 | ΔtktA::FRT ΔtktB::FRT ΔtktC::FRT putP::AX::FRT | This study |
| Plasmids | ||
| pKD13 | bla FRT ahp FRT PS1 PS4 oriR6K | (14) |
| pPCR Script Cam SK+ | ColE1 ori; CmR | Stratagene/Aligent |
| pTP2223 | Plac lam bet exo tetR | (16) |
| pCP20 | bla cat cI857 PRflp pSC101 oriTS | (29) |
| pSKAP | ColE1 ori; CmR; bla FRT ahp FRT | This study |
| pSKAP_AA | ColE1 ori; CmR; bla FRT ahp FRT; AA | This study |
| pSKAP_AD | ColE1 ori; CmR; bla FRT ahp FRT; AD | This study |
| pSKAP_AG | ColE1 ori; CmR; bla FRT ahp FRT; AG | This study |
| pSKAP_AL | ColE1 ori; CmR; bla FRT ahp FRT; AL | This study |
| pSKAP_AO | ColE1 ori; CmR; bla FRT ahp FRT; AO | This study |
| pSKAP_AT | ColE1 ori; CmR; bla FRT ahp FRT; AT | This study |
| pSKAP_AW | ColE1 ori; CmR; bla FRT ahp FRT; AW | This study |
| pSKAP_AX | ColE1 ori; CmR; bla FRT ahp FRT; AX | This study |
Table 1: Strains and plasmids used in this study.
| Name | Sequence (5' – 3')1,2,3 |
| pSKAP SDM AA – F | AGAAGTCTCCTGCTGGTGCTTGAGTCGATTGTGTAGGCTGGAGC |
| pSKAP SDM AA – R | ACTCAAGCACCAGCAGGAGACTTCTCTCAAGACGTGTAATGCTG |
| pSKAP SDM AD – F | AAGAGCACGGTGAGGTGATAGTAGGCGATTGTGTAGGCTGGAGC |
| pSKAP SDM AD – R | CCTACTATCACCTCACCGTGCTCTTCTCAAGACGTGTAATGCTG |
| pSKAP SDM AG – F | AGTAGTGTCCTGGAGGAGCATGTGACGATTGTGTAGGCTGGAGC |
| pSKAP SDM AG – R | TCACATGCTCCTCCAGGACACTACTCTCAAGACGTGTAATGCTG |
| pSKAP SDM AL – F | ACCACACATCGAAGGCACTAGCTCTCTCAAGACGTGTAATGCTG |
| pSKAP SDM AL – R | AGAGCTAGTGCCTTCGATGTGTGGTCGATTGTGTAGGCTGGAGC |
| pSKAP SDM AO – F | GTCCACAACCACACTCAGTGATACTCTCAAGACGTGTAATGCTG |
| pSKAP SDM AO – R | AGTATCACTGAGTGTGGTTGTGGACCGATTGTGTAGGCTGGAGC |
| pSKAP SDM AT – F | ACCAGTGTCCGTGACATGGCTAGACCGATTGTGTAGGCTGGAGC |
| pSKAP SDM AT – R | GTCTAGCCATGTCACGGACACTGGTCTCAAGACGTGTAATGCTG |
| pSKAP SDM AW – F | ACGACTGAGTGATGTGGATGTGACGCGATTGTGTAGGCTGGAGC |
| pSKAP SDM AW – R | CGTCACATCCACATCACTCAGTCGTCTCAAGACGTGTAATGCTG |
| pSKAP SDM AX – F | ACTATCGTGGTGTAACGACAGGCTGCGATTGTGTAGGCTGGAGC |
| pSKAP SDM AX – R | CAGCCTGTCGTTACACCACGATAGTCTCAAGACGTGTAATGCTG |
| M13 – F | GTAAAACGACGGCCAG |
| putP Recombination – F | TAGCGATGGGAGAGAGGACACGTTAATTATTCCATTTTAA TGCAGCATTACACGTC |
| putP Recombination – R | TACTGCGGGTATTAATGCTGAAAACATCCATAACCCATTG CCTGCAGTTCGAAGTTCC |
| qPCR Barcode Region Amplify – F1 | TGCAGCATTACACGTCTTG |
| qPCR Barcode Region Amplify – R2 | TAGGAACTTCGAAGCAGC |
| Barcode AA Probe – FAM | 6-FAM/AGAAGTCTC/ZEN/CTGCTGGTGCTTGAGTC/IBFQ |
| Barcode AD Probe – FAM | 6-FAM/AAGAGCACG/ZEN/GTGAGGTGATAGTAGGC/IBFQ |
| Barcode AG Probe – FAM | 6-FAM/AGTAGTGTC/ZEN/CTGGAGGAGCATGTGAC/IBFQ |
| Barcode AL Probe – FAM | 6-FAM/AGAGCTAGT/ZEN/GCCTTCGATGTGTGGTC/IBFQ |
| Barcode AO Probe – HEX | HEX/AGTATCACT/ZEN/GAGTGTGGTTGTGGACC/IBFQ |
| Barcode AT Probe – HEX | HEX/ACCAGTGTC/ZEN/CGTGACATGGCTAGACC/IBFQ |
| Barcode AW Probe – HEX | HEX/ACGACTGAG/ZEN/TGATGTGGATGTGACGC/IBFQ |
| Barcode AX Probe – HEX | HEX/ACTATCGTG/ZEN/GTGTAACGACAGGCTGC/IBFQ |
| 1Underlined nucleotides indicate complementary sequences for each DNA barcode that is inserted onto pSKAP after SDM. 2Double underlined nucleotides indicate a complementary region on the S. Typhimurium chromosome used for allelic replacement. 3PrimeTime qPCR Probes are hybridization oligos labelled with a 5' fluorescent dye, either 6-carboxyfluorescein (6-FAM) or hexachlorofluorescein (HEX), an internal quencher (ZEN), and the 3' quencer Iowa Black® FQ (IBFQ). |
|
Table 2: Primers and probes used in this study.
| Quantification (copies/20µL reaction) | ||||||||
| Description | AA | AD | AG | AL | AO | AT | AW | AX |
| NTC | N/A | 0.000 | 0.000 | 3.510 | N/A | 0.000 | 0.000 | 0.000 |
| NTC | 3.600 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| NTC | 3.510 | 0.000 | 1.280 | 1.180 | 2.340 | 0.000 | 0.000 | 0.000 |
| NTC | 2.290 | 0.000 | 0.000 | 1.200 | 1.150 | 1.160 | 0.000 | 0.000 |
| Mean | 3.133 | 0.000 | 0.320 | 1.473 | 1.163 | 0.290 | 0.000 | 0.000 |
| Negative | 5.130 | 1.156 | 0.000 | 1.124 | 3.745 | 1.281 | 7.354 | 7.142 |
| Negative | 5.270 | 0.000 | 0.000 | 1.087 | 1.666 | 1.643 | 7.746 | 2.269 |
| Negative | 2.660 | 0.000 | 1.361 | 1.451 | 8.974 | 0.000 | N/A | 0.000 |
| Negative | 6.090 | 0.000 | 0.000 | 2.251 | 0.000 | 2.531 | 8.700 | 3.495 |
| Negative | 1.740 | 0.000 | 1.086 | 2.130 | 4.171 | 0.000 | 7.113 | 5.522 |
| Negative | 6.220 | 3.581 | 0.000 | 4.022 | 1.175 | 1.341 | N/A | 5.950 |
| Mean | 4.518 | 0.789 | 0.408 | 2.011 | 3.288 | 1.133 | 7.728 | 4.063 |
| Positive | 22281.540 | 42673.039 | 46442.242 | 45359.180 | 47885.625 | 15708.027 | 45325.906 | 20810.559 |
| Positive | 23989.676 | 44625.523 | 47356.438 | 45790.660 | 47456.973 | 15096.601 | 47929.840 | 22455.234 |
| Positive | 17846.824 | 38980.133 | 45633.809 | 44174.820 | 33875.039 | 15156.063 | 42536.270 | 21467.840 |
| Positive | 21047.588 | 40140.848 | 41672.648 | 46028.496 | 47426.527 | 16718.000 | 46978.664 | 19876.473 |
| Positive | 20218.238 | 44660.602 | 41718.707 | 45799.375 | 46495.602 | 14590.264 | 54741.023 | 22011.938 |
| Positive | 18531.740 | 41620.801 | N/A | 48082.313 | 35645.199 | 15341.382 | 48950.992 | 21117.559 |
| Mean | 20652.601 | 42116.824 | 44564.769 | 45872.474 | 43130.827 | 15435.056 | 47743.783 | 21289.934 |
| Blank Subtraction | 20648.083 | 42116.035 | 44564.361 | 45870.463 | 43127.539 | 15433.923 | 47736.054 | 21285.871 |
| Undiluted A | 23024.961 | 44448.875 | 58897.510 | 51120.948 | 55450.191 | 18155.305 | 62844.068 | 27567.828 |
| Undiluted B | 18278.174 | 35252.586 | 54409.510 | 66022.396 | 43101.148 | 15732.609 | 60761.328 | 26581.979 |
| 1/50 A | 521.670 | 755.035 | 1066.898 | 1287.187 | 1053.339 | 181.324 | 1278.336 | 580.961 |
| 1/50 B | 435.326 | 634.215 | 1168.087 | 1383.537 | 991.040 | 165.443 | 1180.445 | 596.461 |
| 1/100 A | 228.028 | 598.848 | 603.911 | 631.116 | 507.956 | 258.405 | 665.647 | 331.590 |
| 1/100 B | 256.330 | 585.834 | 583.325 | 670.875 | 459.325 | 289.207 | 638.916 | 307.948 |
| 1/200 A | 121.283 | 305.293 | 258.247 | 346.965 | 234.774 | 114.163 | 169.055 | 172.553 |
| 1/200 B | 114.638 | 313.040 | 253.685 | 297.216 | 191.637 | 179.895 | 280.989 | 147.297 |
| 1/400 A | 42.829 | 141.343 | 127.337 | 163.605 | N/A | 71.241 | 157.697 | 85.976 |
| 1/400 B | 59.544 | 180.543 | 162.080 | 162.108 | 115.508 | 104.682 | 151.141 | 87.804 |
| 1/800 A | 34.304 | 67.934 | 65.939 | 83.857 | 66.134 | 31.784 | 82.722 | 45.616 |
| 1/800 B | 20.390 | 80.222 | 81.453 | 85.325 | 53.102 | 38.034 | 55.460 | 29.660 |
| 1/1600 A | 15.405 | 44.505 | 47.672 | 39.613 | 33.027 | 18.006 | 37.655 | 20.988 |
| 1/1600 B | 22.091 | 48.828 | 46.781 | 37.388 | 30.245 | 30.138 | 34.553 | 19.795 |
| 1/3200 A | 12.333 | 22.104 | 16.850 | 18.403 | 11.460 | 10.535 | 21.245 | 9.218 |
| 1/3200 B | 6.796 | 32.555 | 15.742 | 26.249 | 15.111 | 9.908 | 23.393 | 10.175 |
| Blank Subtraction | ||||||||
| Undiluted A | 23020.443 | 44448.086 | 58897.103 | 51118.937 | 55446.903 | 18154.172 | 62836.339 | 27563.765 |
| Undiluted B | 18273.655 | 35251.797 | 54409.103 | 66020.385 | 43097.860 | 15731.477 | 60753.600 | 26577.916 |
| 1/50 A | 517.152 | 754.246 | 1066.490 | 1285.176 | 1050.051 | 180.192 | 1270.607 | 576.898 |
| 1/50 B | 430.808 | 633.426 | 1167.679 | 1381.526 | 987.751 | 164.311 | 1172.717 | 592.398 |
| 1/100 A | 223.509 | 598.059 | 603.503 | 629.105 | 504.668 | 257.272 | 657.918 | 327.527 |
| 1/100 B | 251.812 | 585.044 | 582.918 | 668.864 | 456.036 | 288.074 | 631.187 | 303.885 |
| 1/200 A | 116.765 | 304.503 | 257.840 | 344.954 | 231.486 | 113.030 | 161.327 | 168.490 |
| 1/200 B | 110.120 | 312.251 | 253.277 | 295.205 | 188.348 | 178.762 | 273.261 | 143.234 |
| 1/400 A | 38.310 | 140.554 | 126.929 | 161.594 | N/A | 70.108 | 149.968 | 81.913 |
| 1/400 B | 55.026 | 179.753 | 161.672 | 160.097 | 112.219 | 103.549 | 143.413 | 83.741 |
| 1/800 A | 29.786 | 67.145 | 65.531 | 81.847 | 62.846 | 30.651 | 74.994 | 41.553 |
| 1/800 B | 15.872 | 79.433 | 81.045 | 83.314 | 49.813 | 36.901 | 47.732 | 25.596 |
| 1/1600 A | 10.886 | 43.716 | 47.264 | 37.602 | 29.739 | 16.874 | 29.927 | 16.925 |
| 1/1600 B | 17.573 | 48.039 | 46.373 | 35.377 | 26.957 | 29.005 | 26.825 | 15.732 |
| 1/3200 A | 7.815 | 21.314 | 16.442 | 16.392 | 8.172 | 9.402 | 13.517 | 5.155 |
| 1/3200 B | 2.278 | 31.765 | 15.334 | 24.238 | 11.822 | 8.776 | 15.664 | 6.112 |
| Simulated CI | ||||||||
| Undiluted A | 1.114895 | 1.055372 | 1.321619 | 1.114419 | 1.285650 | 1.176251 | 1.316329 | 1.294932 |
| Undiluted B | 0.885005 | 0.837016 | 1.220911 | 1.439279 | 0.999312 | 1.019279 | 1.272698 | 1.248618 |
| 1/50 A | 0.025046 | 0.017909 | 0.023931 | 0.028018 | 0.024348 | 0.011675 | 0.026617 | 0.027102 |
| 1/50 B | 0.020864 | 0.015040 | 0.026202 | 0.030118 | 0.022903 | 0.010646 | 0.024567 | 0.027831 |
| 1/100 A | 0.010825 | 0.014200 | 0.013542 | 0.013715 | 0.011702 | 0.016669 | 0.013782 | 0.015387 |
| 1/100 B | 0.012195 | 0.013891 | 0.013080 | 0.014582 | 0.010574 | 0.018665 | 0.013222 | 0.014276 |
| 1/200 A | 0.005655 | 0.007230 | 0.005786 | 0.007520 | 0.005367 | 0.007323 | 0.003380 | 0.007916 |
| 1/200 B | 0.005333 | 0.007414 | 0.005683 | 0.006436 | 0.004367 | 0.011582 | 0.005724 | 0.006729 |
| 1/400 A | 0.001855 | 0.003337 | 0.002848 | 0.003523 | N/A | 0.004542 | 0.003142 | 0.003848 |
| 1/400 B | 0.002665 | 0.004268 | 0.003628 | 0.003490 | 0.002602 | 0.006709 | 0.003004 | 0.003934 |
| 1/800 A | 0.001443 | 0.001594 | 0.001470 | 0.001784 | 0.001457 | 0.001986 | 0.001571 | 0.001952 |
| 1/800 B | 0.000769 | 0.001886 | 0.001819 | 0.001816 | 0.001155 | 0.002391 | 0.001000 | 0.001203 |
| 1/1,600 A | 0.000527 | 0.001038 | 0.001061 | 0.000820 | 0.000690 | 0.001093 | 0.000627 | 0.000795 |
| 1/1,600 B | 0.000851 | 0.001141 | 0.001041 | 0.000771 | 0.000625 | 0.001879 | 0.000562 | 0.000739 |
| 1/3,200 A | 0.000378 | 0.000506 | 0.000369 | 0.000357 | 0.000189 | 0.000609 | 0.000283 | 0.000242 |
| 1/3,200 B | 0.000110 | 0.000754 | 0.000344 | 0.000528 | 0.000274 | 0.000569 | 0.000328 | 0.000287 |
| Average CI (Theoretical) | ||||||||
| Undiluted (1) | 0.999950 | 0.946194 | 1.271265 | 1.276849 | 1.142481 | 1.097765 | 1.294514 | 1.271775 |
| 1/50 (0.02) | 0.022955 | 0.016474 | 0.025067 | 0.029068 | 0.023625 | 0.011161 | 0.025592 | 0.027466 |
| 1/100 (0.01) | 0.011510 | 0.014046 | 0.013311 | 0.014148 | 0.011138 | 0.017667 | 0.013502 | 0.014832 |
| 1/200 (0.005) | 0.005494 | 0.007322 | 0.005735 | 0.006978 | 0.004867 | 0.009453 | 0.004552 | 0.007322 |
| 1/400 (0.0025) | 0.002260 | 0.003803 | 0.003238 | 0.003507 | 0.00260* | 0.005626 | 0.003073 | 0.003891 |
| 1/800 (0.00125) | 0.001106 | 0.001740 | 0.001645 | 0.001800 | 0.001306 | 0.002188 | 0.001285 | 0.001577 |
| 1/1,600 (0.000625) | 0.000689 | 0.001089 | 0.001051 | 0.000795 | 0.000657 | 0.001486 | 0.000594 | 0.000767 |
| 1/3,200 (0.000313) | 0.000244 | 0.000630 | 0.000357 | 0.000443 | 0.000232 | 0.000589 | 0.000306 | 0.000265 |
| Standard Deviation | ||||||||
| Undiluted | 0.11494 | 0.10918 | 0.05035 | 0.16243 | 0.14317 | 0.07849 | 0.02182 | 0.02316 |
| 1/50 | 0.00209 | 0.00143 | 0.00114 | 0.00105 | 0.00072 | 0.00051 | 0.00103 | 0.00036 |
| 1/100 | 0.00069 | 0.00015 | 0.00023 | 0.00043 | 0.00056 | 0.00100 | 0.00028 | 0.00056 |
| 1/200 | 0.00016 | 0.00009 | 0.00005 | 0.00054 | 0.00050 | 0.00213 | 0.00117 | 0.00059 |
| 1/400 | 0.00040 | 0.00047 | 0.00039 | 0.00002 | 0* | 0.00108 | 0.00007 | 0.00004 |
| 1/800 | 0.00034 | 0.00015 | 0.00017 | 0.00002 | 0.00015 | 0.00020 | 0.00029 | 0.00037 |
| 1/1,600 | 0.00016 | 0.00005 | 0.00001 | 0.00002 | 0.00003 | 0.00039 | 0.00003 | 0.00003 |
| 1/3,200 | 0.00013 | 0.00012 | 0.00001 | 0.00009 | 0.00004 | 0.00002 | 0.00002 | 0.00002 |
| *Represents results from a single experiment. | ||||||||
Table 3: Absolute quantification and simulated CI calculation.
| Condition | Competitive Index1 | |||||||
| Experiment 1 | ||||||||
| WTAA | WTAD | WTAG | WTAL | WTAO | WTAT | WTAW | WTAX | |
| Logarithmic | 0.927 ± 0.033 | 0.992 ± 0.031 | 1.068 ± 0.025 | 0.921 ± 0.02 | 1.044 ± 0.03 | 1.051 ± 0.057 | 1.094 ± 0.027 | 0.929 ± 0.005 |
| Stationary | 1.1 ± 0.021 | 1.071 ± 0.053 | 1.079 ± 0.065 | 0.948 ± 0.02 | 0.98 ± 0.02 | 0.873 ± 0.044 | 0.97 ± 0.056 | 1.021 ± 0.007 |
| Experiment 2 | ||||||||
| CI (Traditional) | WTAA | ΔAAD | ΔBAL | ΔCAT | ΔABAG | ΔACAO | ΔBCAW | ΔABCAX |
| Logarithmic | 1 ± 0 | 0.802 ± 0.084 | 0.957 ± 0.02 | 0.989 ± 0.073 | 0.581 ± 0.153 | 0.86 ± 0.053 | 0.995 ± 0.011 | 0.695 ± 0.061 |
| Stationary | 1 ± 0 | 0.97 ± 0.063 | 1.043 ± 0.058 | 0.99 ± 0.036 | 1.625 ± 0.589 | 0.835 ± 0.051 | 0.912 ± 0.047 | 0.477 ± 0.049 |
| CI (Pooled Inoculum) | ||||||||
| Logarithmic | 1.114 ± 0.039 | 0.864 ± 0.074 | 1.073 ± 0.032 | 1.1 ± 0.068 | 0.633 ± 0.152 | 0.938 ± 0.056 | 1.111 ± 0.043 | 0.746 ± 0.06 |
| Stationary | 1.078 ± 0.039 | 1.039 ± 0.049 | 1.166 ± 0.093 | 1.066 ± 0.01 | 1.735 ± 0.613 | 0.876 ± 0.035 | 0.97 ± 0.045 | 0.49 ± 0.047 |
| 1Values represent mean CI ± standard deviation for three or four replicate experiments. | ||||||||
Table 4: Representative results from in vitro competition between S. Typhimurium strains.
Table S1. Optional primers for creating additional barcode sequences and corresponding fluorescent probes for their detection. Please click here to download this file.
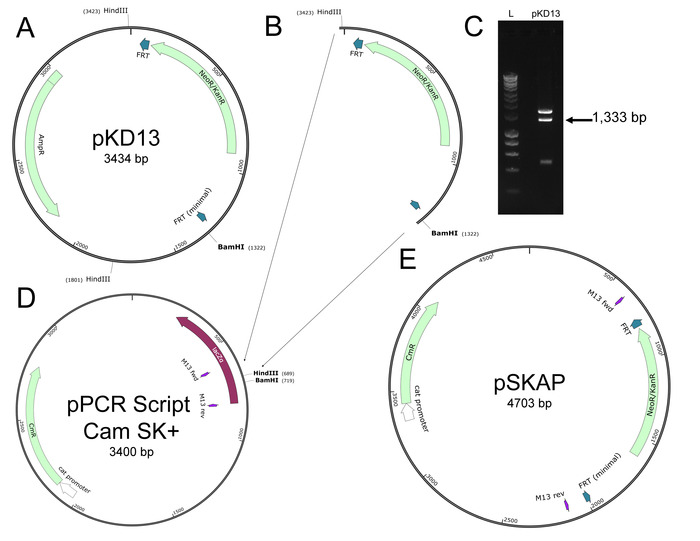
Figure 1. Generation of pSKAP. (A) Purified pKD13 was subjected to restriction digestion with HindIII and BamHI. (B, C) The 1,333 bp fragment of interest containing an FRT-flanked kanamycin resistance gene was purified. (D) pPCR Script Cam SK+ was also digested with HindIII and BamHI and the fragment from pKD13 (B) was ligated in to generate (E) pSKAP. Please click here to view a larger version of this figure.

Figure 2: Insertional site-directed mutagenesis to pSKAP. (A) Insertion of 25 bp DNA barcodes at position 725 was performed using PCR. Forward and reverse primers specific to that location were designed with complementary 25-nucleotide 5’ extensions (denoted in the primer as lowercase “a”). (B) A generic pSKAP_Barcode plasmid resulting from SDM is shown with the location of the inserted DNA barcode highlighted orange. Please click here to view a larger version of this figure.
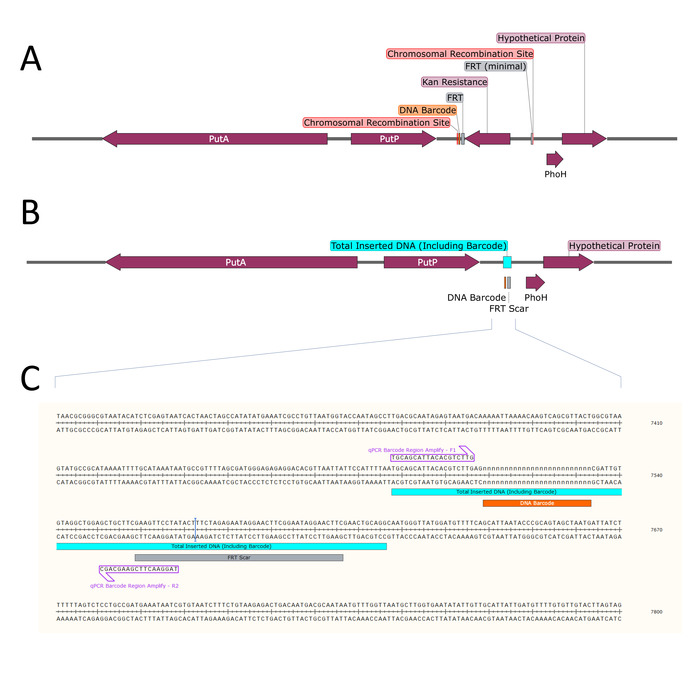
Figure 3: Chromosomal rearrangement downstream of putP. (A) After λ-Red mediated recombination, the selectable kanamycin resistance gene (dark purple) flanked by FRT sites (grey) is inserted on the chromosome between the loci indicated (Chromosomal Recombination Site, red). The unique DNA barcode (orange) is inserted just outside the FRT site. (B) The kanamycin resistance gene is removed by FRT-mediated excision, leaving a remnant of inserted DNA on the chromosome (Total Inserted DNA, blue) consisting of the DNA barcode and an FRT scar. (C) The modified chromosomal DNA sequence surrounding the inserted DNA is shown, along with the amplification priming sites (light purple) used for digital PCR. Please click here to view a larger version of this figure.

Figure 4: Dilution scheme for validating fluorescent probe sensitivity and specificity. (A) Purified gDNA from seven barcoded strains is mixed together in equal amounts to create the diluent for diluting the omitted barcoded gDNA (AX in the example above). (B) Perform a serial dilution of the omitted barcoded gDNA (AX in the example above) using the prepared diluent described previously. Thoroughly mix the contents of each tube before transferring to the next tube. Figure 4 was created with BioRender. Please click here to view a larger version of this figure.
Figure 5: Plate layout for analyzing sensitivity and specificity of primer-probe sets 1 and 2. The digital PCR experiment includes NTCs, positive controls, negative controls, and the dilution schemes for each of the tested barcodes. The plate for validating primer-probe sets 3 and 4 is laid out in the same pattern using the appropriate barcoded gDNA. Please click here to view a larger version of this figure.
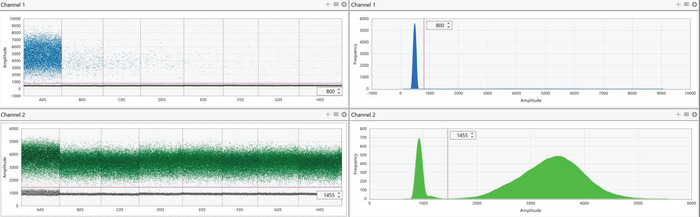
Figure 6: Representative digital PCR results of diluted AA-barcoded gDNA. gDNA containing the AA barcode was diluted in a background of all other barcoded gDNA as described in Figure 4. Channel 1 represents the FAM probe for the AA barcode (top panels) while channel 2 represents the HEX probe for the AO barcode (bottom panels). Results of each probe are presented as both individual droplet fluorescent amplitude (left panels) and a histogram representing the frequency of fluorescent intensity of all droplets in the selected wells (right panels). For each condition, positive (high fluorescence) and negative (low fluorescence) droplets should form two distinct populations. In the case of AA that was diluted, and most droplets were negative, the histogram (top right panel) appears to only depict a single population. This is because positive droplets are substantially outnumbered by negative droplets; however, two distinct populations are still visible by examining droplet fluorescent amplitude in the left panels. The populations should be separated using the threshold feature to define positive and negative droplets (visualized by the pink line). Threshold values will vary depending on the probes that were used, but all wells that utilize the same probe mix should have identical thresholds. As the AA-barcoded gDNA was diluted, there is a decrease in positive (high fluorescent) droplets while the number of positive AO droplets remains constant in the background. Please click here to view a larger version of this figure.
Discussion
The ability to accurately quantify microorganisms is of paramount importance to microbiology research, and the ability to enumerate unique strains from an initial mixed population has proved to be an invaluable tool for assessing fitness and virulence traits in bacteria. However, the techniques for accomplishing this have not progressed in pace with modern developments in molecular biology. The technology to easily modify the chromosomes of many bacteria, including S. Typhimurium, has been available for nearly two decades14, yet this ability has been rarely utilized for molecularly tagging strains with unique DNA sequences. By exploiting the ability to create readily identifiable strains based on unique, minimally-disruptive DNA barcodes inserted onto the bacterial chromosome, coupled with the most state-of-the-art technology to detect and quantify these molecular identifiers (i.e. digital PCR), we have created a system that provides exquisite sensitivity, specificity, accuracy, and precision for easily quantifying individual strains within a diverse population of bacteria.
Calculating CIs and COIs as described above relies on the ability to accurately make the appropriate modifications to bacterial chromosomes. All modifications should be verified by Sanger sequencing to ensure that no random mutations occurred. The use of a high-fidelity polymerase will minimize these errors, but any such mutations that do occur will impair the ability to detect the strain, which is another critical aspect of this protocol. Although we have demonstrated that digital PCR can detect as few as two gDNA copies in a background of nearly 2 million, strains outside of this range may require additional dilutions for their accurate quantification. Furthermore, DNA barcode sequences must be designed to facilitate the use of high-quality probe sequences. Probe sequences should be analyzed for tendencies to self-dimerize, form hairpins, or ineffectively bind their targets. The importance of using quality sequences and probes cannot be minimalized, a fact that is evidenced by the careful validation experiments that must be performed with each probe. Efforts to create optimal DNA barcodes will create optimal digital PCR quantification results.
While carefully designed molecular tags are important for obtaining quality results, interpreting the results is another critical aspect of this protocol. The CI is defined as the ratio between the mutant strain and the wild-type strain in the output divided by the ratio of the two strains in the input1,19,20. This traditional CI presented in section 9 is useful when the mixed infection consists of only one strain versus wild-type. However, when using large pools of strains to inoculate media or animals, strains are not only competing against wild-type, but also against every other strain present in the inoculum. Previous studies that performed competition experiments using multiple infecting strains have failed to take this into account in their calculations7,13. To account for this feature of mixed infections, we have introduced a formula to calculate CIs that has been modified for pooled infections. It is unlikely that all strains will provide the same level of competition a wild-type strain would. However, because most bacterial genes have little impact on virulence, as the number of strains used in competitive index experiment increases, the likelihood that overall virulence will tend toward the wild-type strain increases. This may not necessarily be the case for certain experimental designs using pools of many strains all with known virulence defects. However, this is accounted for in the modified equation because less abundant strains (less fit) in the outcome will have a smaller effect on Xoutput. Depending on the specific experimental design, there may be cases in which one or the other formula is preferred. In most instances involving pooled infections, however, it is important to consider that in a mixed infection, all strains compete against each other, not just against wild-type. When analyzing results, it is critical that the rationale behind each formula is well-understood to make the most accurate interpretations of strain fitness. When reporting results, it is equally important to accurately disclose how data were analyzed.
Section 9 of the protocol also includes formulas to help determine gene interactions using a COI. With this analysis, predictions can be made to determine whether two virulence genes operate independently or together. COI is defined as the ratio of the double mutant to single mutant strain in the output divided by the ratio of the two strains in the input1. The formula is designed to detect phenotypic additivity of gene disruptions. If genes function independently to enhance virulence, a disruption of both genes should cause a greater decrease in fitness compared to a single disruption of either gene alone. If genes function together to enhance virulence (such as genes encoding two enzymes in a pathway), a disruption of either gene should have the same effect on virulence as disrupting both genes. Detecting phenotypic additivity can be difficult in cases where the level of attenuation caused by a single gene is either very high or very low. Nevertheless, direct comparison of strains within the same animal system provides less variability and a more reliable account of the functional relationship between genes, and this calculation can be performed from two strains within a larger mixed population.
A final critical aspect for interpreting results is to consider the effects of population dynamics. In some mixed infections that have multiple strains, a single strain may emerge as either more dominant or less fit because of random population drift. This phenomenon can be amplified when bottleneck events occur. This can be caused from using a very large number of input strains, a very small number of total bacteria in the inoculum, or a combination of both. Another interfering aspect that arises from mixed infections is the possibility of in trans complementation. This occurs when a fit strain, such as the wild-type, artificially enhances the virulence of a less fit strain. A hypothetical example of this would be to compare the fitness of an S. Typhimurium Pathogenicity Island 2 (SPI2)-knockout strain co-infected with a wild-type strain. SPI2 enables S. Typhimurium to survive intracellularly by secreting effectors into the host cytosol that modify the phagosome within a macrophage. Disruption of this system makes S. Typhimurium susceptible to intracellular killing. However, because macrophages are capable of engulfing two or more bacteria at once, the SPI2-knockout could receive a considerable increase in fitness if it is residing in the same macrophage as a wild-type S. Typhimurium that is secreting effectors into the host cytosol. Random population dynamics and the possibility of in trans complementation is a limitation of any competition experiment. If in trans complementation is suspected, phenotypes should be confirmed using other complementary methods to assess fitness. To overcome random population dynamics, increasing the number of replicate experiments increases the likelihood of identifying outliers in results. Fortunately, the protocol described above makes it easier to have a greater number of identical replicate experiments because the number of experimental conditions is drastically reduced.
A key element to the CI technique described above is its ability to be adapted to almost any organism and any experimental design that requires accurate quantification of microorganisms. It does, however, require the genetic manipulation of an organism to incorporate a unique DNA sequence on the chromosome. The adaptability of the technique requires the species to be genetically-malleable and will rely on the generation of alternative protocols for modifying the genome (Steps 1 and 2). The DNA barcodes listed in Tables 2 and S1 should be enough for most bacteria; however, as in all quantitative PCR experiments, it is pertinent to analyze the bacterial genome to ensure minimal potential for off-target binding of primers and probes by a simple BLAST analysis. The DNA barcode sequences used in this study differ by only 3-4 bases in some cases, highlighting the exquisite specificity of the probes that minimizes the potential for non-specific binding. Regardless of fluorescent probes’ specificity, all new barcode sequences must be appropriately validated for sensitivity and specificity as described in step 6. After creating barcoded strains, it is possible to adapt this protocol to many types of experiments besides in vitro competition assays as described above. The strains are suitable for in vivo competition assays in mice or other animal model systems. Only minimal modifications are required to extract gDNA from animal organs and tissues, and these modifications are well described by manufacturers of kits for such purposes. Further, large pools of mixed infections for in vivo competition have been successfully utilized previously7, offering the potential to reduce the number of animals necessary for a single experiment, which not only decreases costs of those experiments but also decreases the potential for animal-to-animal variability. Similarly, barcoded strains could be used in other in vitro assays that examine the susceptibility of strains to various treatment conditions (e.g. antibiotics, acid susceptibility, killing by reactive oxygen or nitrogen species, etc.). For these experiments, assessing growth inhibition could be achieved by adding the desired chemical to the growth medium in step 3.4 and proceeding as described. To assess bacterial death, a large pool of strains could be mixed, and the input quantified as described above. After exposure to the desired treatment, live cells could be selectively quantified by coupling the digital PCR quantification procedure with Viability PCR to differentiate between live and dead cells21,22,23,24,25,26. Throughput for such experiments would be dramatically increased because dilutions and replica plates for each strain are replaced by more streamlined molecular techniques. Lastly, although analyzing evolutionary biology and population genetics is beyond the scope of this paper, barcoded organisms are highly adaptable for such studies. Ultimately, the purpose of this protocol was to develop a powerful technique for quantifying bacteria that is highly adaptable for its use in many diverse species and in many types of experiments.
Declarações
The authors have nothing to disclose.
Acknowledgements
Research reported in this publication was supported by the George F. Haddix President’s Faculty Research Fund and the National Institute of General Medical Science of the National Institutes of Health (NIH) under award number GM103427. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Materials
| 1.5 mL microcentrifuge tubes | Eppendorf | 22600028 | Procure from any manufacturer |
| 16 mL culture tubes | MidSci | 8599 | Procure from any manufacturer |
| 5-200 μL pipette tips | RAININ | 30389241 | Procure alternative tip brands with caution based on manufacturing quality |
| 5-50 μL multichannel pipette | RAININ | 17013804 | Use alternative multichannel pipettes with caution |
| Agarose | ThermoFisher Scientific | BP160-500 | Procure from any manufacturer |
| BLAST Analysis | NCBI | N/A | https://blast.ncbi.nlm.nih.gov/Blast.cgi |
| C1000 Touch Thermocycler with 96-Deep Well Reaction Module | Bio Rad | 1851197 | Must procure ddPCR supplies from Bio Rad. Alternatives are not yet available. |
| Chemically competent DH5α | Invitrogen | 18258012 | Procure from any manufacturer or prepare yourself |
| Chloramphenicol | ThermoFisher Scientific | BP904-100 | Procure from any manufacturer |
| Cytation5 Microplate reader | BioTek | CYT5MF | Procure from any manufacturer, use any system capable of accurately quantifying DNA |
| Data Analysis Software (QuantaSoft and QuantaSoft Data Analysis Pro) | Bio Rad | N/A | Must procure ddPCR supplies from Bio Rad. Alternatives are not yet available. |
| ddPCR 96-Well Plates | Bio Rad | 12001925 | Must procure ddPCR supplies from Bio Rad. Alternatives are not yet available. |
| ddPCR Droplet Reader Oil | Bio Rad | 1863004 | Must procure ddPCR supplies from Bio Rad. Alternatives are not yet available. |
| ddPCR Supermix for Probes (No dUTP) | Bio Rad | 1863024 | Must procure ddPCR supplies from Bio Rad. Alternatives are not yet available. |
| DG8 Cartridges for QX200/QX100 Droplet Generator | Bio Rad | 1864008 | Must procure ddPCR supplies from Bio Rad. Alternatives are not yet available. |
| DG8 Gaskets for QX200/QX100 Droplet Generator | Bio Rad | 1863009 | Must procure ddPCR supplies from Bio Rad. Alternatives are not yet available. |
| Droplet Generation Oil for Probes | Bio Rad | 1863005 | Must procure ddPCR supplies from Bio Rad. Alternatives are not yet available. |
| Kanamycin | ThermoFisher Scientific | BP906-5 | Procure from any manufacturer |
| Luria-Bertani agar | ThermoFisher Scientific | BP1425-2 | Procure from any manufacturer or make it yourself from agar, tryptone, yeast digest, and NaCl |
| Luria-Bertani broth | ThermoFisher Scientific | BP1426-2 | Procure from any manufacturer or make it yourself from tryptone, yeast digest, and NaCl |
| PCR Plate Heat Seal, foil, pierceable | Bio Rad | 1814040 | Must procure ddPCR supplies from Bio Rad. Alternatives are not yet available. |
| PCR Tubes | Eppendorf | 951010022 | Procure from any manufacturer |
| Petri dishes | ThermoFisher Scientific | FB0875712 | Procure from any manufacturer |
| pPCR Script Cam SK+ | Stratagene/Agilent | 211192 | No longer available commercially |
| Primer/Probe Design | IDT | N/A | https://www.idtdna.com/Primerquest/Home/Index |
| pSKAP and pSKAP_Barcodes | Addgene | Plasmid numbers 122702-122726 | www.addgene.org |
| PX1 PCR Plate Sealer | Bio Rad | 1814000 | Must procure ddPCR supplies from Bio Rad. Alternatives are not yet available. |
| QX200 Droplet Generator | Bio Rad | 1864002 | Must procure ddPCR supplies from Bio Rad. Alternatives are not yet available. |
| QX200 Droplet Reader | Bio Rad | 1864003 | Must procure ddPCR supplies from Bio Rad. Alternatives are not yet available. |
| S. Typhimurium strain ATCC 14028s | ATCC | ATCC 14028s | www.atcc.org |
| Take3 Micro-Volume Plate | BioTek | TAKE3 | Procure from any manufacturer, use any system capable of accurately quantifying DNA |
| Thermo Scientific FastDigest BamHI | ThermoFisher Scientific | FERFD0054 | Procure from any manufacturer |
| Thermo Scientific FastDigest DpnI | ThermoFisher Scientific | FERFD1704 | Procure from any manufacturer |
| Thermo Scientific FastDigest HindIII | ThermoFisher Scientific | FERFD0504 | Procure from any manufacturer |
| Thermo Scientific GeneJet Gel Extraction and DNA Cleanup Micro Kit | ThermoFisher Scientific | FERK0832 | Procure from any manufacturer |
| Thermo Scientific GeneJet Miniprep Kit | ThermoFisher Scientific | FERK0503 | Procure from any manufacturer |
| Thermo Scientific Phusion High-Fidelity DNA Polymerase | ThermoFisher Scientific | F534L | Procure from any manufacturer |
| Thermo Scientific T4 DNA Ligase | ThermoFisher Scientific | FEREL0011 | Procure from any manufacturer |
| Thermocycler | Bio Rad | 1861096 | Procure from any manufacturer |
| UVP Visi-Blue Transilluminator | ThermoFisher Scientific | UV95043301 | Or other transiluminator that allows visualization of DNA |
| Water, Molecular Biology Grade | ThermoFisher Scientific | BP28191 | Procure from any manufacturer |
Referências
- Beuzón, C. R., Holden, D. W. Use of mixed infections with Salmonella strains to study virulence genes and their interactions in vivo. Microbes and Infection. 3 (14-15), 1345-1352 (2001).
- Bäumler, A. J., Tsolis, R. M., Valentine, P. J., Ficht, T. A., Heffron, F. Synergistic effect of mutations in invA and lpfC on the ability of Salmonella typhimurium to cause murine typhoid. Infection and Immunity. 65 (6), 2254-2259 (1997).
- Vital, M., Hammes, F., Egli, T. Competition of Escherichia coli O157 with a drinking water bacterial community at low nutrient concentrations. Water Research. 46 (19), 6279-6290 (2012).
- Schellenberg, J., Smoragiewicz, W., Karska-Wysocki, B. A rapid method combining immunofluorescence and flow cytometry for improved understanding of competitive interactions between lactic acid bacteria (LAB) and methicillin-resistant S. aureus (MRSA) in mixed culture. Journal of Microbiological Methods. 65 (1), 1-9 (2006).
- Becker, D., et al. Robust Salmonella metabolism limits possibilities for new antimicrobials. Nature. 440 (7082), 303 (2006).
- Rang, C., Galen, J. E., Kaper, J. B., Chao, L. Fitness cost of the green fluorescent protein in gastrointestinal bacteria. Canadian Journal of Microbiology. 49 (9), 531-537 (2003).
- Santiviago, C. A., et al. Analysis of pools of targeted Salmonella deletion mutants identifies novel genes affecting fitness during competitive infection in mice. PLoS Pathogens. 5 (7), e1000477 (2009).
- Ahmer, B. M., Gunn, J. S. Interaction of Salmonella spp. with the intestinal microbiota. Frontiers in Microbiology. 2, 101 (2011).
- Gallagher, L. A., Shendure, J., Manoil, C. Genome-scale identification of resistance functions in Pseudomonas aeruginosa using Tn-seq. MBio. 2 (1), (2011).
- Van Opijnen, T., Bodi, K. L., Camilli, A. Tn-seq: high-throughput parallel sequencing for fitness and genetic interaction studies in microorganisms. Nature Methods. 6 (10), 767 (2009).
- van Opijnen, T., Lazinski, D. W., Camilli, A. Genome‐wide fitness and genetic interactions determined by Tn‐seq, a high‐throughput massively parallel sequencing method for microorganisms. Current Protocols in Microbiology. 36 (1), (2017).
- Mutalik, V. K., et al. Dual-barcoded shotgun expression library sequencing for high-throughput characterization of functional traits in bacteria. Nature Communications. 10 (1), 308 (2019).
- Yoon, H., Gros, P., Heffron, F. Quantitative PCR-based competitive index for high-throughput screening of Salmonella virulence factors. Infection and Immunity. 79 (1), 360-368 (2011).
- Datsenko, K. A., Wanner, B. L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proceedings of the National Academy of Sciences. 97 (12), 6640-6645 (2000).
- Henard, C. A., Bourret, T. J., Song, M., Vázquez-Torres, A. Control of redox balance by the stringent response regulatory protein promotes antioxidant defenses of Salmonella. Journal of Biological Chemistry. 285 (47), 36785-36793 (2010).
- Poteete, A. R., Fenton, A. C. λ red-dependent growth and recombination of phage P22. Virology. 134 (1), 161-167 (1984).
- McCollister, B. D., Bourret, T. J., Gill, R., Jones-Carson, J., Vázquez-Torres, A. Repression of SPI2 transcription by nitric oxide-producing, IFNγ-activated macrophages promotes maturation of Salmonella phagosomes. Journal of Experimental Medicine. 202 (5), 625-635 (2005).
- Shaw, J. A., et al. Salmonella enterica serovar Typhimurium has three transketolase enzymes contributing to the pentose phosphate pathway. Journal of Biological Chemistry. 293 (29), 11271-11282 (2018).
- Freter, R., O’Brien, P., Macsai, M. S. Role of chemotaxis in the association of motile bacteria with intestinal mucosa: in vivo studies. Infection and immunity. 34 (1), 234-240 (1981).
- Taylor, R. K., Miller, V. L., Furlong, D. B., Mekalanos, J. J. Use of phoA gene fusions to identify a pilus colonization factor coordinately regulated with cholera toxin. Proceedings of the National Academy of Sciences. 84 (9), 2833-2837 (1987).
- Nogva, H. K., Dromtorp, S., Nissen, H., Rudi, K. Ethidium monoazide for DNA-based differentiation of viable and dead bacteria by 5′-nuclease PCR. Biotechniques. 34 (4), 804-813 (2003).
- Nocker, A., Camper, A. K. Novel approaches toward preferential detection of viable cells using nucleic acid amplification techniques. FEMS Microbiology Letters. 291 (2), 137-142 (2009).
- Nocker, A., Cheung, C. -. Y., Camper, A. K. Comparison of propidium monoazide with ethidium monoazide for differentiation of live vs. dead bacteria by selective removal of DNA from dead cells. Journal of Microbiological Methods. 67 (2), 310-320 (2006).
- Nocker, A., Mazza, A., Masson, L., Camper, A. K., Brousseau, R. Selective detection of live bacteria combining propidium monoazide sample treatment with microarray technology. Journal of Microbiological Methods. 76 (3), 253-261 (2009).
- Nocker, A., Sossa, K. E., Camper, A. K. Molecular monitoring of disinfection efficacy using propidium monoazide in combination with quantitative PCR. Journal of Microbiological Methods. 70 (2), 252-260 (2007).
- Nocker, A., Sossa-Fernandez, P., Burr, M. D., Camper, A. K. Use of propidium monoazide for live/dead distinction in microbial ecology. Applied and Environmental Microbiology. 73 (16), 5111-5117 (2007).
- Price-Carter, M., Tingey, J., Bobik, T. A., Roth, J. R. The Alternative Electron Acceptor Tetrathionate Supports B12-Dependent Anaerobic Growth ofSalmonella enterica Serovar Typhimurium on Ethanolamine or 1, 2-Propanediol. Journal of Bacteriology. 183 (8), 2463-2475 (2001).
- Taylor, R. G., Walker, D. C., McInnes, R. E. coli host strains significantly affect the quality of small scale plasmid DNA preparations used for sequencing. Nucleic Acids Research. 21 (7), 1677 (1993).
- Cherepanov, P. P., Wackernagel, W. Gene disruption in Escherichia coli: Tc R and Km R cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene. 158 (1), 9-14 (1995).

