Pattern-based Search of Epigenomic Data Using GeNemo
Summary
Unlike DNA sequence data, epigenomic data are not readily subjected to text-based searches. Presented here are the procedures to use an upgraded version of GeNemo, a web-based bioinformatics tool, to conduct pattern-based searches for similarities in epigenomic data comparing available online databases including Encyclopedia of DNA Elements with user's data.
Abstract
Compared with the robust text-based search tools for genomic or RNA sequencing data, current methodologies for pattern-based searches of epigenomic and other functional genomic data are very limited. GeNemo is the first online search tool that accomplishes this goal. Users input their functional genomic data in the Browser Extensible Data (BED), Peaks, and bigWig formats, and may search for data in any of the three formats. Users may specify which types of datasets to search against, choosing from a variety of online datasets, with the Encyclopedia of DNA Elements (ENCODE) representing different epigenomic marks, transcriptional factor binding sites, and chromatin hypersensitivities or accessibilities in specific cell types, and developmental stages or species (mouse or human). GeNemo returns a list of genomic regions with matching patterns to the input data, which may be viewed in the browser as well as downloaded in the BED file format. The upgraded GeNemo has improved graphical display, has more robust interface, and is no longer prone to errors due to changes in the University of California, Santa Cruz (UCSC) genome browser. Troubleshooting steps for common problems are discussed. As the amount of functional genomic data is expanding exponentially, there is a critical need to develop and refine new bioinformatic tools such as GeNemo for data analyses and interpretation.
Introduction
Recent technological advances have allowed for a rapid expansion of epigenomic or functional genomic data depositories, which have outpaced the development of relevant analytic tools to extract biological insights. One important way to analyze epigenomic data is to search user-generated data against data depositories and especially those from the Encyclopedia of DNA Elements (ENCODE)1 projects for matching patterns that could lead to new knowledge. For instance, identifying similarities in the patterns of two different epigenomic marks at defined loci across the genome may indicate coordinated action by different molecular players on chromatin conformation and transcriptional regulation2,3,4.
Conventional text-based search engines are ineffective in this regard because, unlike DNA sequence, epigenomic data predominantly exist in the format of intensities or functional genomic regions. GeNemo, standing for Gene Nemo (as in Finding Nemo), was developed to address this unmet need using pattern-based searches5. Its algorithm utilizes a Markov Chain Monte Carlo maximization process5. Users take their own data or a dataset downloaded from depositories and search an array of online epigenomic data to identify similarities in patterns.
The current version of GeNemo has an updated display, interfaces more robustly with the University of California, Santa Cruz (UCSC) genome browser6, and is less susceptible to issues caused by changes in the latter. In particular, while GeNemo's Results page used to be based on the UCSC genome browser interface, the current version of GeNemo supports its own Results page and consequently is no longer adversely affected by structural changes to the UCSC genome browser. GeNemo can use any genomic signal, including protein-binding, histone modification, chromatin accessibility, topological domains, and so on, as a query to find colocalized/similar segments among known data sets from large consortia. Therefore, it is an important tool to study the relationship between different epigenomic data of interest and known data generated in large scale genomic projects.
Protocol
NOTE: The protocol can be paused anywhere.
1. Basic Setup
- Obtain a BED, Peaks format, or BigWig7 file containing the data to be input into Genome. The file should have extension name "bed", "broadpeaks""narrowpeaks", or "bigWig" respectively.
NOTE: Zipped versions of these type of files will also work. - Use an internet browser to go to genemo.org. Any operating system capable of running most common internet browsers should be able to use GeNemo.
- Choose which species to search against using the dropdown menu. Currently available species include human and mouse.
- Upload user file using a url or a direct upload. BigWig files only work with the url upload method. BED and Peaks format files work with both methods (wiggle files cannot be uploaded as the main data as of now).
2. Optional Setup
- Provide an email address in the corresponding box in order to receive the search results by email when the search is done.
NOTE: When searching a large part of the genome and/or against a large number of tracks (see below), it is recommended that the user provides his/her email, since the search may take a long time. For example, a 100 megabase search takes around 15 s. A link to the search results will be sent to the email address provided when the search is completed. The link will expire in 7 days after the completion of a search. - Provide a bigwig file or the wiggle display file may be from a url. This display file will not affect the results; it will only be shown alongside the results.
- Specify a search range (including the chromosome and base pair positions) in the corresponding box.
- List the chromosome, start base pair, and end base pair.
- Use 'chrN' for the chromosome format, where 'N' is the chromosome number/letter (1, 2, … X, or Y). For the base pairs, just type in the numbers.
- Include spaces between all three entries, or include a colon (:) between chromosome number and the first base pair, and/or a hyphen between the two base pairs. For example: chr1:1000000-2000000, chr1 1000000 2000000, chr1 1000000-2000000, chr1:1000000 2000000.
NOTE: Steps 2.1 – 2.3 are optional.
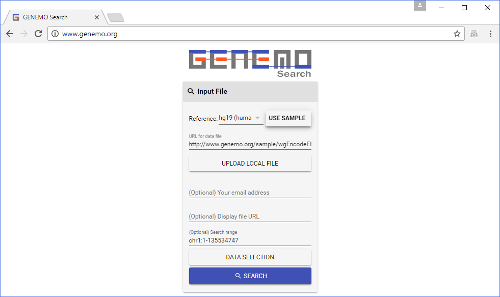
Figure 1: GeNemo's front page with the necessary areas filled out. A user needs to input the species, search file and search range, and select tracks he/she wishes to search against. Email address and display file are optional. Please click here to view a larger version of this figure.
3. Data Selection
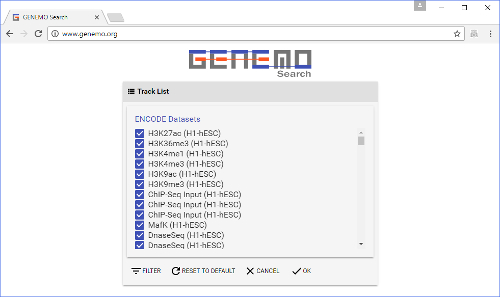
Figure 2: Track selection window. This is brought up by clicking the "DATA SELECTION" button on the front page. Here, users select tracks to search the input file against. Some of the tracks are already selected by default. Please click here to view a larger version of this figure.
- After clicking the data selection button, choose which types of tracks to search against (i.e., to add to the query). The track collection includes many different datasets from labs around the world.
- As the list of tracks is quite long, users may want to use the filter button (on the top) to facilitate track selections. Tracks may be filtered by Experiment, Tissue, Cell Line and/or Lab.
- There are five buttons on the bottom to help execute track selection: Select All, Select None, Add, Filter, Exclude.
- Select All" and "Select None" are self-explanatory.
- The "Add" button adds currently selected tracks to the query. It serves as the logic gate "OR". Note that selecting the filter(s) above (e.g., certain Experiments, Tissues, Cell Lines or Labs) does not automatically add corresponding tracks to the search query. Users must first select tracks (e.g., brain, liver under tissue), and then click the "Add" button to add them to the query. When selecting tracks, note that only the filters specified in the opened tab in the filter window will be applied to the search query. Selections on other tabs will be saved in the filter window, but not applied to the search query.
- The "Filter" button retains only the types of tracks currently selected in the filter window in the query and removes all other types of tracks. It serves as the logic gate "AND". Essentially, "Filter" allows the selection of the interaction between two categories of tracks (e.g., certain tissues with certain Labs). Note that "Filter" does not add the selected types of tracks to the query if they are not already in the query.
- The "Exclude" button removes all types of tracks that are currently selected in the filter window from the query. It serves as the logic gate "NOT", in opposition to the "Filter" function. Again, "Exclude" does not add any tracks currently not selected in the filter window to the query.
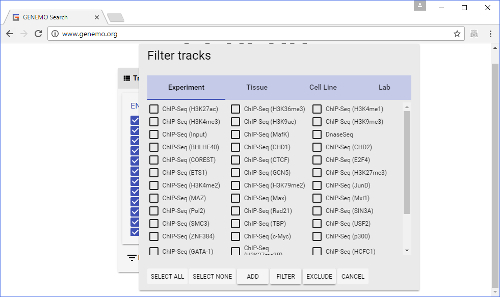
Figure 3: Filter window. This is brought up by clicking the "FILTER" button on the Track selection window. Here, users can select many tracks at the same time, with relative ease. Please click here to view a larger version of this figure.
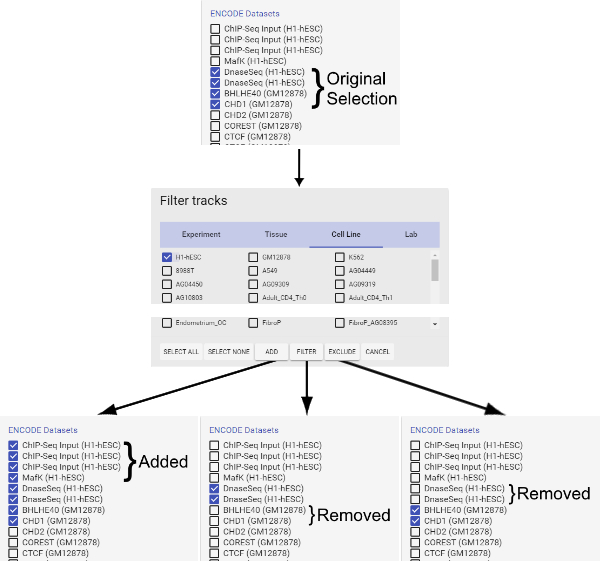
Figure 4: How to use the filter function. Please click here to view a larger version of this figure.
- After adding the desired tracks to the query, click the "Update" button on the bottom right. This is needed in order to accommodate two ways to select data: selecting individual data tracks or filtering/excluding. The "Reset View" button resets the query to the default tracks related to gene expression regulation in human/mouse embryonic stem cells.
NOTE: Selecting tracks to be searched against via "Data selection" is optional but recommended because the default search tracks are most likely not suited to the user's needs.
4. Search and Results
- Click the "Search" button after data selection. The search may take some time.
- Once the search is completed, users will see various boxes in the Results page. Each box represents a section of the genome where a user's data file has a closely matched pattern with one or more of the tracks the user has queried.
- If there are no boxes visible, try searching more types of tracks or making the search range bigger with the same input file. An easy way to do this without redoing everything is clicking the "☰" button next to the logo. This will open up a sidebar that allows the user to modify the search.
- The results may be exported as a BED file by clicking on the "DOWNLOAD BED FILE" button on the bottom of the Results page.
- Click the Visualize button on the top right of each box to visualize the results.
- In the Visualization panel on the right, multiple things are displayed including the data, which incorporates the user input file, the display file if one was inputted, matching tracks, and some default tracks. From the results, the user may compare known ENCODE datasets against the provided dataset for further investigation. The user may also refer to UCSC genes to see the context of the query results. If tracks from multiple cell lines/tissues are selected, the user may use such results to gain insights about the tissue specificity of the similarities between the given dataset and ENCODE datasets.
- On the Results page, user may drag on any tracks to move upstream or downstream of the genome; when the mouse cursor is on the coordinates, the user may use the mouse wheel and/or zoom in and out.
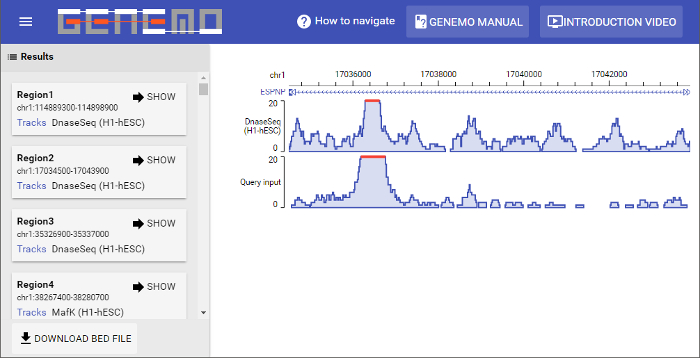
Figure 5: Results page. This particular search returned 363 matching regions. Displaying the first matching region can be done by clicking the "SHOW" button on the bottom left of each resulting region box. On the left part of the display window it can be seen that the two data files (input and selected track) are similar in signal strength pattern. Please click here to view a larger version of this figure.
Representative Results
Shown here in Figure 5 is a simulated search. The human species was selected, and the corresponding sample file was used as the input data file. In addition, the default tracks, as seen in Figure 3, were selected. There were a total of 363 matching regions, and the first region is shown in the display page. It can be seen that the intensity pattern from base 17036000 to 17038000 on chromosome 1 for the input file and one of the selected tracks is very similar.
Discussion
A thorough understanding of the epigenome is required to achieve the full potential of human genome sequencing in providing new biological insights8. Currently there are only ways to search online epigenomic datasets by their data description and title (i.e., metadata)1. This severely limits the types of search one can do with epigenomic data. Pattern-based search tools for epigenomic data are essential for exploring the relationship between different epigenomic marks, which may lead to new biological insights. GeNemo, which searches by the content of the data and not metadata, is the first service of its kind to compare patterns in epigenomic data from published depositories such as the ENCODE database with a user-generated or downloaded dataset5. This marks the beginning of the availability of an epigenomic search tool that is widely accessible to researchers around the world just as text-based sequence search tool became widely available in the 1990s. Currently, there are no alternatives for pattern-based online search tools for epigenomic data other than GeNemo.
One potential example of using GeNemo is to search the co-appearing histone modifications and other epigenetic marks with the transcriptional factor E2F6 in human embryonic stem cells (an example E2F6 binding signal file is available at ENCODE data portal or at https://sysbio.ucsd.edu/public/xcao3/ENCODESample/ENCFF001UBC.bed). By using this file as query to search against all ENCODE datasets in H1-hESC, GeNemo will show that E2F6 binding signal is heavily enriched with H3K4me1, H3K4me2, H3K4me3, and H3K27me3, which agrees with existing research showing that E2F6 regulates some genes via methylation of H3K279. On the other hand, there appears to be colocalization of E2F6 and CtBP2 binding sites, which is known to interact with a factor in the same family, E2F710. These results for the entire genome against a large number of epigenetic marks, transcriptional factor binding signals, and other signals included in ENCODE can be fairly easily obtained with GeNemo, which can provide all potential targets for further analysis.
Since the first publication5 of GeNemo as a web-based epigenomic data search tool, the Results section of GeNemo has been updated to have a matching appearance with GeNemo's front page. The old Results section closely mirrored the UCSC genome browser results section, and was largely dependent on the remote UCSC server for display. With the new interface, GeNemo is more user-friendly and no longer dependent on the UCSC genome server (even though data are still fetched remotely). This makes GeNemo more robust and less susceptible to problems due to code changes at the UCSC server. Furthermore, the new, faster polymer interface of GeNemo gives the user more tools to visualize and analyze patterns in the data.
Critical steps include providing the appropriate input file and selecting data tracks to search against. Users are strongly encouraged to experiment with various track selection functions to become familiar with the selection process and how different commands can be combined to achieve the intended outcome. In particular, note that the "Add" function is required to add desired tracks selected to the query, while "Filter" or "Exclude" can be used as logic gate commands "AND" and "OR", respectively. The "Update" function is required to affect all the selections before implementing the search. When no results are returned, a user may check the input data file, search more tracks or increase the search range. Whenever there is an error, there will be a window popping up defining what exactly the error is. There are some ambiguous errors, though. For example, when the window says that 'no file was uploaded,' either no file was uploaded, or the uploaded file was not of an acceptable format and, consequently, the program was not able to read it correctly. Acceptable file formats for file upload include BED and Peaks format file for both upload methods, and bigWig for online link upload only. The zipped versions of these file formats are also acceptable.
Current limitations of this approach include the yet-to-be-optimized algorithms and functions employed in GeNemo. GeNemo cannot yet provide any guidance on the interpretation of any datasets returned. This task is up to the users, which requires significant knowledge and expertise in the biology of the genome and epigenome. In addition, another current limitation is that users cannot change the sensitivity and noise level of the searches. We expect to continue to improve and expand GeNemo on its pattern searching capabilities and dataset collection in future.
Declarações
The authors have nothing to disclose.
Acknowledgements
This work was supported by NIH grants including DP1HD087990 from NICHD, R01HG008135 from NHGRI. We thank members of the Zhong lab for valuable feedback.
Author Contributions:
X.C. and A.T.Z. updated GeNemo by coding new interface and features; A.T.Z. produced the in-house sample video; A.T.Z., X.C and S.Z. wrote the paper.
Referências
- The ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature. 489, 57-74 (2012).
- Barski, A., et al. High-Resolution Profiling of Histone Methylations in the Human Genome. Cell. 129 (4), 823-837 (2007).
- Meaney, M. J., Ferguson-Smith, A. C. Epigenetic regulation of the neural transcriptome: the meaning of the marks. Nature Neuroscience. 13, 1313-1318 (2010).
- Roh, T. -. Y., Cuddapah, S., Cui, K., Zhao, K. The genomic landscape of histone modifications in human T cells. PNAS. 103 (43), 15782-15787 (2006).
- Zhang, Y., Cao, X., Zhong, S. GeNemo: a search engine for web-based functional genomic data. Nucleic Acids Res. 44, W122-W127 (2016).
- Fujita, P. A., Rhead, B., Zweig, A. S., Hinrichs, A. S., Karolchik, D., Cline, M. S., Goldman, M., Barber, G. P., Clawson, H., Coelho, A., et al. The UCSC Genome Browser database: update 2011. Nucleic Acids Res. 39, 876-882 (2011).
- Neph, S., Vierstra, J., Stergachis, A. B., Reynolds, A. P., Haugen, E., Vernot, B., Thurman, R. E., John, S., Sandstrom, R., Johnson, A. K., et al. An expansive human regulatory lexicon encoded in transcription factor footprints. Nature. 489, 83-90 (2012).
- Sarda, S., Hannenhalli, S. Next-generation sequencing and epigenomics research: a hammer in search of nails. Genomics Inform. 12 (1), 2-11 (2014).
- Storre, J., et al. Silencing of the Meiotic Genes SMC1β and STAG3 in Somatic Cells by E2F6. J Biol Chem. 280, 41380-41386 (2005).
- Liu, B., Shats, I., Angus, S. P., Gatza, M. L., Nevins, J. R. Interaction of E2F7 Transcription Factor with E2F1 and C-terminal-binding Protein (CtBP) Provides a Mechanism for E2F7-dependent Transcription Repression. J Biol Chem. 288, 24581-24589 (2013).

