Visualization of IL-22-expressing Lymphocytes Using Reporter Mice
Summary
We describe here a transgenic reporter mouse model to visualize the IL-22-producing cells inside different mouse tissues. This method can be used to track the location of other cytokines or secretary proteins in the mouse.
Abstract
Reporter mice have been widely used to observe the localization of expression of targeted genes. This protocol focuses on a strategy to establish a new transgenic reporter mouse model. We chose to visualize interleukin (IL) 22 gene expression because this cytokine has important activities in the intestine, where it contributes to repair tissues damaged by inflammation. Reporter systems offer considerable advantages over other methods of identifying products in vivo. In the case of IL-22, other studies had first isolated cells from tissues and then re-stimulated the cells in vitro. IL-22, which is normally secreted, was trapped inside cells using a drug, and intracellular staining was used to visualize it. This method identifies cells capable of producing IL-22, but it does not determine whether they were doing so in vivo. The reporter design includes inserting a gene for a fluorescent protein (tdTomato) into the IL-22 gene in such a way that the fluorescent protein cannot be secreted and therefore remains trapped inside the producing cells in vivo. Fluorescent producers can then be visualized in tissue sections or by ex vivo analysis through flow cytometry. The actual construction process for the reporter included recombineering a bacterial artificial chromosome that contained the IL-22 gene. This engineered chromosome was then introduced into the mouse genome. Homeostatic IL-22 reporter expression was observed in different mouse tissues, including the spleen, thymus, lymph nodes, Peyer's patch, and intestine, by flow cytometry analysis. Colitis was induced by T-cell (CD4+CD45RBhigh) transfer, and reporter expression was visualized. Positive T cells were first present in the mesenteric lymph nodes, and then they accumulated inside the lamina propria of the distal small intestine and colon tissues. The strategy using BACs gave good-fidelity reporter expression compared to IL-22 expression, and it is simpler than knock-in procedures.
Introduction
Cell type-specific expression of reporter genes is useful to identify cells actively expressing the target in tissues under homeostatic and perturbed states. It also allows for the purification of these cells, which remain viable, to study their other properties. Reporter mice have been utilized in elucidating the mechanism of action for specific cytokines, transcription factors, and regulatory elements. Previous strategies1,2,3 have largely relied on knocking the reporter into the target locus in the mouse chromosome, a time-consuming and costly procedure. Thus, a simpler method for the generation of reporter mice is desirable.
Cytokines are a broad class of small, secreted proteins/peptides that regulate immune responses through intercellular signaling. Interleukin 22 (IL-22) is a cytokine with many reported activities, including barrier function, tissue repair, and inflammation4. Although IL-22 was initially discovered as a T-cell product5, subsequent reports demonstrated its expression in natural killer (NK) cells in humans6 and mice7 and in other classes of innate lymphocytes8. Despite extensive observation of IL-22-producing cells, visualization of IL-22 previously required ex vivo stimulation and permeabilization to stains with antibodies. Therefore, novel IL-22 reporter mice would be a very useful tool to investigate the function of IL-22 in homeostatic and pathogenic processes.
Here, we developed a simplified transgenic reporter mouse model to observe the IL-22-producing cells in vivo and in vitro. Using a BAC recombineering method9, we inserted the tdTomato cDNA sequence with Poly A signal fragments into the IL-22 locus and replaced exon 1. The other untranslated regions, exons, and regulatory elements were not perturbed, since we would like to mimic the natural regulation of IL-22 as much as possible. The site of reporter insertion disrupts the signal sequence, resulting in the accumulation of the reporter inside the producing cells, unlike IL-22 itself, which is rapidly secreted. This new method can also be applied to the generation of reporter mice for other secreted proteins.
Protocol
All animals received proper care in accordance with the experimental procedures outlined in the 2011 Guide for Care and Use of Laboratory Animals Committee of Frederick National Laboratory for Cancer Research.
1. Generation of IL-22-tdTomato Reporter Mice by BAC Recombineering
NOTE: The mice should be unconscious and do not move in response to a noxious stimulus. Sterilize the surgical area with 70% ethanol and sterilize all surgical tools using a glass bead sterilizer.
- Purify BAC DNA (RP23-401E11, clone length 228391 bps) using an alkaline extraction method10,11. Characterize the BAC clone by SpeI digestion of 5 µg of RP23-401E11 to confirm the correct size of the target gene. NOTE: The BAC DNA was digested into 14 fragments.
- Insert the TdTomato reporter gene into the Not I/Sal I site of the PLD53SC-A-GFP-B shuttle vector11 and replace the GFP gene in the same site. Then, using BAC RP23-401E11 as a template, amplify the homology boxes (A and B) to the first translated exon of Il-22 (exon 1) by PCR (95 °C 2 min; 30 cycles of 95 °C for 30 sec, 55 °C for 30 sec, and 72°C for 30 sec; and a final extension at 72 °C for 10 min). In a 50 µl PCR reaction system, add 50 ng (1 µl) of BAC DNA, 0.5 µl of primers (10 µM), 40 µl of PCR superMix high fidelity, and 8 µl of H2O.
NOTE: The primers for the A and B boxes are as follows: A box Forward: 5'- TGGCGCGCCGGAGCTGTGAAGAAAG and Reverse: 5'- CAGAGATCGCACAAGTGTCAAC; B box Forward: 5'- GTTAATTAACTGCCCGTCAACA and Reverse: 5'- TGGCCGGCCTGAGCACCTGCTT CATC10. The sites of the restriction enzymes (Asc I and Pac I) are underlined. - Ligate 500 ng of Asc I-digested A and 500 ng of Pac I-digested B fragments into 50 ng of PLD53SC-ATB (tdTomato) vector at 16 °C for 1 hr.
- Transform the purified PLD53SC-ATB vector into BAC-competent cells11 and culture them on Luria-Bertani(LB) agar plates with chloramphenicol (20 µg/ml) and ampicillin (30 µg/ml) at 37 °C. Pick two to three colonies of co-integrate from the Ch/Amp master plates.
- Inoculate each colony into 1 ml of LB supplemented with 20 µg/ml chloramphenicol and incubate them for 1 hr at 37 °C and 225 rpm. Spread 100 µl of each mixture on a LB-agar plate (10 g of Peptone 140, 5 g of yeast extract, 12 g of agar, and 1 L of water; pH 7.5) supplemented with chloramphenicol and 4-4.5% of sucrose. Incubate the culture overnight at 37 °C.
- Pick both big and small colonies and replate them on two LB-agar plates with chloramphenicol. Then, expose the plates either with or without UV light for 30 sec and pick the UV-sensitive colonies for further analysis.
- Identify the modified BAC DNA by PCR using tdTomato/IL-22 heterozygous primer (Forward: 5' ACT TgT gCg ATC TCT gAT ggT; Reverse: 5' TgT AAT Cgg ggA TgT Cgg C). The thermocycler conditions are as follows: 95 °C for 2 min; 30 cycles of 95 °C for 30 sec, 56 °C for 30 sec, and 72 °C for 30 sec; and a final extension at 72 °C for 10 min.
- Confirm the sequence of the modified area of the BAC by direct BAC sequencing of large-prep BAC DNA12.
- Anesthetize a recipient mouse with a dose of 0.25% tribromoethanol into the peritoneal cavity when implanting the microinjected zygotes into pseudo-pregnant C57BL/6c mice.
- Linearize the IL-22-tdTomato BAC construct (10 µg) by digestion with 5 µl of restriction endonucleasePi-SceI in 100 µl of the reaction system and microinject the linearized BAC DNA into fertilized eggs of C57BL/6 females at the pronuclear stage13. Screen the transgenic mice by Southern blot analysis of DNA isolated from tail biopsies using standard procedures.
2. Single-cell Preparation from Spleens, Thymus, Lymph Nodes, and Peyer's Patch
NOTE: IL-22-tdTomato reporter mice were maintained at the National Cancer Institute (NCI, Frederick, MD). The euthanasia method conforms to the most recent AVMA Guidelines on Euthanasia. All mice were euthanized using CO2 inhalation14.
- Euthanize an IL-22-tdTomato mouse in a CO2 chamber and put it on a clean pad. Sterilize the abdomen skin by spraying 70% alcohol and cut it open. Remove the spleen in the left flank and the thymus behind the sternum.
- To harvest the mesentericlymph nodes (located in the mesenteric tissue) and Peyer's patches (small lymphoidnodules throughout the ileum region of the small intestine), take the pearly white mesenteric nodes close to the wall of the small intestine using dissect forceps with fine-point serrated tips first and then remove the whole small intestine and colon out of the body. Find the extended oval patches (Peyer's patches)along the gut and cut them with scissors. Grind these lymphoid tissues (lymph nodes and Peyer's patches) individually between two frosted slides into a single-cell suspension in PBS/1% fetal bovine serum (FBS) buffer on ice.
- Mince the spleen or thymus tissues using the same method as in step 2.2. Centrifuge the splenic suspension for 8 min at 500 x g and 4 °C. Add 1 ml of ACK lysing buffer per spleen to the cell pellets.
- Mix well and let them stand for 1 min. Add 9 ml of sterile phosphate-buffered saline (PBS) buffer to each sample. Spin down by centrifugation for 8 min at 500 x g and 4 °C to remove the red blood cells.
NOTE: Do not add ACK lysing buffer to the thymus cells. - Resuspend the resulting leukocytes in 5 ml of PBS and pass the cells through a 100 µm cell strainer. Collect the cell pellets by centrifugation at 500 x g for 8 min.
- Resuspend the cells in 5 ml of RPMI media or FACS buffer and count the viable cells using 0.4% trypan blue dye/PBS solution.
3. Isolation of Intraepithelial Lymphocytes and Lamina Propria Cells from the Intestines
- Open the abdominal skin as in step 2.1, cut the small intestine, from duodenum (next to the stomach) to ileum (close to the appendix), and the whole colon, right above the anus. Remove the extra adipose and mesenteric tissue using forceps. Put the intestines immediately in ice-cold PBS.
- Look for Peyer's patches (small, rough nodules) along the distal region of the small intestine. Remove the masses using forceps andopen the intestine lengthwise using scissors. Thoroughly flush the intestine with 10 ml of ice-cold PBS.
- Incubate the tissues in 20 ml of pre-digestion buffer (5 mM EDTA and 1 mM dithiothreitol in Hank's Balanced Salt Solution (HBSS)) for 30 min at 37 °C with slow rotation (50 rpm) in a shaker. After each of 2 incubations, remove the epithelial cell layer, containing the intraepithelial lymphocytes (IELs), by intense vortexing (30 sec at 12,000 rpm) and pass all the tissues through a 100 µm cell strainer.
- Add 20 ml of new EDTA solution to the tissue for second incubation for 30 min at 37 °C. Pass the IEL fractions through a 100 µm cell strainer and pool them with the previous fractions. Keep the remaining intestine fragments on ice to be treated in step 3.9.
- Centrifuge the pooled IEL fractions at 800 x g for 5 min at 4 °C. Wash the pooled IEL fractions with 10 ml of HBSS/5% FBS and spin them down for 5 min at 800 x g and 4 °C.
- Resuspend the cells in 8 mL of 44% colloidal silica medium15 and overlay them with 5 ml of 67% colloidal silica medium. Centrifuge the 44%/67.5% cell mixture gradient for 20 min at room temperature and 1,500 x g.
- Collect the IEL cells from the band at the interface. First, remove the top layer of the supernatant (about 5 ml) using 1 ml pipette. Then, harvest IELs at the interphase of the colloidal silica medium gradient.
- Wash the IELs by adding 10 ml of RPMI medium. Spin the cells down for 5 min at 800 x g and 4 °C. Resuspend the IELs in 5 ml of RPMI for step 3.15.
- For the isolation of the lamina propria lymphocytes (LPLs), mince the intestine tissue fragments from step 3.4 into small pieces using scissors (1 mm) and digest the pieces with 10 ml of digestion buffer (0.05 g of collagenase, 0.05 g of DNaseI, and 0.3 g of dispase II in RPMI/5% FBS) for 15-20 min at 37 °C.
- Filter the mixture through a 70 µm cell strainer. The flow-through supernatant contains the released LPL.
- Fully digest the intestine fragments by repeating steps 3.9-3.10 (normally, they must be repeated 3 times). Each time, pool the supernatants containing the LPLs.
- Collect the cells by centrifugation for 5 min at 1,500 x g and room temperature and resuspend the pellets in RPMI/5% FBS. Pass them through a 40 µm cell strainer.
- Resuspend the cell pellets in 10 ml of the 40% fraction of a 40:80 colloidal silica medium gradient and overlay them on 5 ml of the 80% fraction in a 15 ml tube.
- Perform the density gradient separation by centrifugation for 20 min at 1,500 x g and room temperature15.
- Collect the LPLs, as described in step 3.7. Wash them once with 10 ml of PBS and resuspend the pellets in 1 ml of PBS/1% FBS buffer or RPMI medium.
- Count the viable cells (both IELs and LPLs) using 0.4% trypan blue dye/PBS solution.
4. Expression of IL-22-tdTomato in Colitis
- Prepare single spleen cell suspensions from IL-22-TdTomato mice at a concentration of 1 x 107 cells/ml in sterile PBS, as described in steps 2.1-2.4.
- Purify the CD4+ T cells using the Mouse CD4 Cell Negative Isolation method10. Label them with anti-CD4-APC (clone RM4-5, 0.5 µl/1 x 106 cells in PBS/1% FBS buffer) and anti-CD45RB-FITC (clone 16A, 0.5 µl/1 x 106 cells in PBS/1% FBS buffer) by incubation at 4 °C for 30 min.
- Spin the cells down for 5 min at 500 x g and 4 °C. Wash the cells with 2 ml of PBS/1% FBS buffer and resuspend them in 1 ml of PBS/1% FBS staining buffer.
- Run the labeled T cells on a flow cytometry machine16. Gate the cells on a T-cell population and sort CD4+CD45RBhigh double-positive cells (the wavelength detected at 488 nm and 633 nm lasers).
- Harvest the sorted cells by centrifugation for 5 min at 500 x g and 4 °C. Resuspend the cell pellets in sterile PBS buffer at 1 x 106 cells/ml. Inject the cells (0.5 x 106) into the peritoneum ofeach Rag1-/- recipient mouse.
- Sacrifice the recipient mice under CO2 at various time points. Harvest the mesenteric lymph nodes and the small and large intestines, as described in steps 2.1 and 2.2. Put each mesenteric lymph node and cut-open intestine (paper/intestine/paper sandwich) in 4% paraformaldehyde (PFA; handle under a fume hood) overnight to fix the tissues. Replace the PFA with 18% sucrose for 16-24 hr and then freeze the tissues for sectioning.
- Use the cryostat machine to cut the tissue17,18,19. Warm the tissue sections to room temperature for approximately 60 min and place them in acetone for 10 min at room temperature. Dry them at room temperature for approximately 5 min.
- Add FBS in excess, remove it by pipette, and add anti-RFP at a 1:200 dilution in 0.1% BSA/1x PBS. Incubate the antibody at 37 °C for 60 min.
- Rinse the sections in 1x PBS for 10 min and apply the corresponding secondary antibody (goat anti-rabbit 568) to the tissue, diluted 1:300 in 1% BSA/1x PBS, for 30 min at room temperature.
- Rinse slides in 1x PBS for 10 min, wipe them dry, and add coverslips.
- Visualize all samples with a fluorescent microscope under consistent illumination (RFP: excitation filter 540-580 nm) using the 40X objective.
Representative Results
A murine IL-22 reporter transgene was created using recombineering to modify a bacterial artificial chromosome carrying the IL-22 locus. Figure 1 shows a diagram of pBACe3.6 vector containing the sacBII gene, a positive-selection marker, and chloramphenicol antibiotic resistance gene11. After introducing tdTomato into exon 1, the signal peptide sequence was disrupted, as shown in Figure 2. Thus, the tdTomato reporter was trapped inside the IL-22-expressing cells, enabling their detection and isolation by flow cytometry. The homozygous mice were bred from founder lines and screened by PCR, and they did not require backcrossing in order to achieve offspring with a genetic identity, as is required in the usual knock-in strategy. To test the fidelity of the IL-22 reporters, in vitro-generated splenocytes were cultured under Th22 or neutral conditions. Figure 3 demonstrates that IL-22-tdtomato was well expressed in Th22 cells in vitro, detected either by flow cytometry or by fluorescent microscopy. In vivo, as shown in Figure 4, the IL-22 reporter was detected in different mouse tissues under homeostatic condition. Most of the tdTomato signal was found in the lamina propria (LP) cells from the gut, but not in the axillary lymph node (ALN), the spleen, or the thymus. To visualize the tdTomato reporter in inflamed tissue, we used a mouse colitis model induced by the transfer of reporter CD4+CD45Rbhi T cells into Rag1-/- mice. Figure 5 demonstrates that the reporters were first present in the mesenteric lymph nodes and then accumulated inside the lamina propria of distal small intestine and colon tissues. Taken together, this novel method for the generation of IL-22 reporter mice is effective and time-saving.
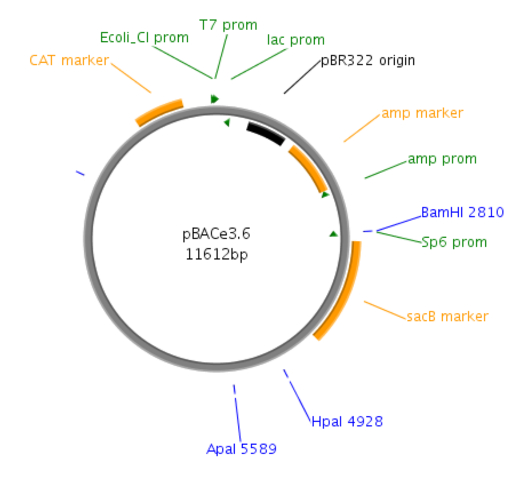
Figure 1: Site map of the pBACe3.6 vector. pBACe3.6 is characterized by containing chloramphenicol antibiotic resistance and a sacBII gene as a positive-selection marker. During recombination, the desired clones are sucrose-resistant through their expressing the sacBII gene and are allowed to grow on sucrose-containing media, while the negative BAC colonies are sucrose-sensitive.
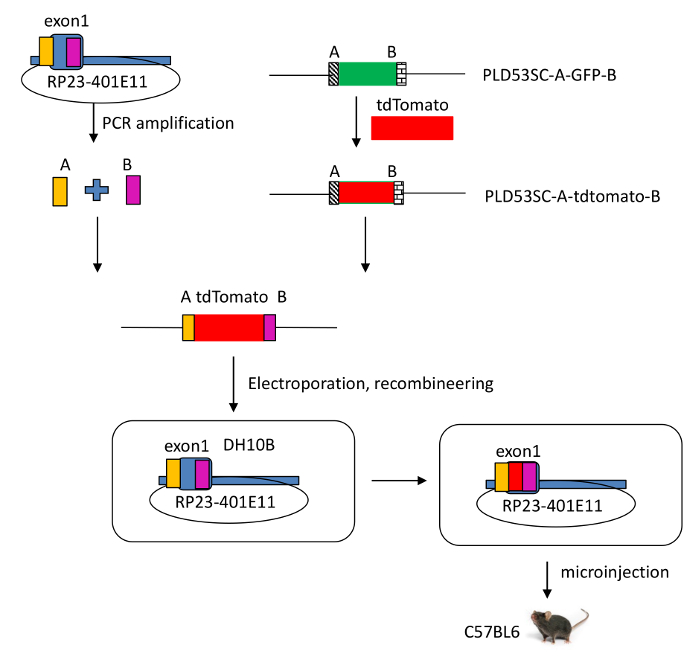
Figure 2: Schematic view of the modified RP23-401E11 BAC clone. A tdTomato reporter gene was introduced into the Il-22 locus, which includes IL-22 and regulatory elements in a murine bacterial artificial chromosome (BAC RP23-401E11), using recombineering technology. By homologous recombination, the sequence of the signal peptide of Il-22 in the BAC was disrupted and exon 1 of Il-22 was replaced by the tdTomato reporter gene. Please click here to view a larger version of this figure.
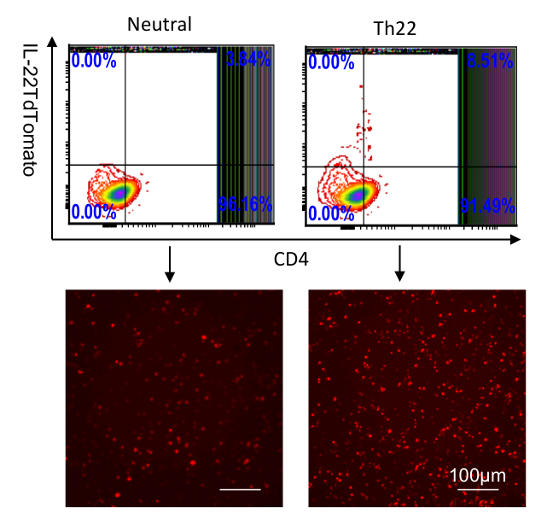
Figure 3: Identification of IL-22 reporters in splenocytes cultured in vitro. CD4 T cells were purified from the spleen and then stimulated with anti-CD28; anti-CD3 (neutral condition); or anti-CD3, anti-CD28, anti-IFNƳ, anti-IL4, IL-6, TGF-β, and 6-Formylindolo (3,2-b)carbazole (FICZ) for 5 days. IL-22-tdTomato was analyzed by flow cytometry (top two) and fluorescent microscopy (bottom two, scale bar: 100 µm). The numbers in the quadrants indicate the percent of CD45+ cells. Please click here to view a larger version of this figure.
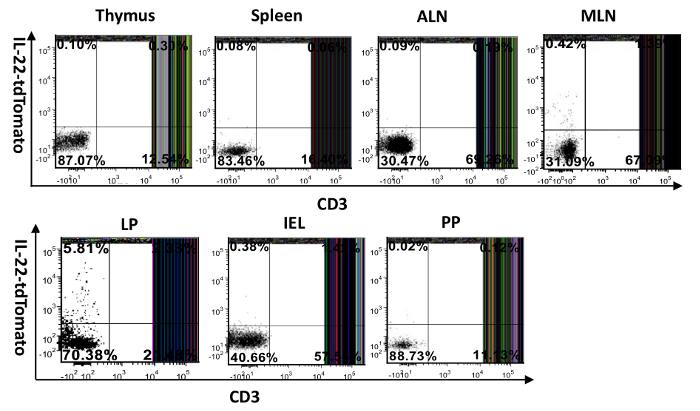
Figure 4: Visualization of IL-22-producing cells in different mouse tissues. Single-cell suspensions were prepared from the spleen, thymus, lymph nodes, intestines (IEL and LP), and Peyer's patch. They were then surface-stained with FITC-anti-CD3 and examined for tdTomato expression. MLN: mesenteric lymph node, ALN: axillary lymph node, PP: Peyer's patch, IEL: intraepithelial cells isolated from the small intestine, LP: lamina propria cells purified from the small intestine. The numbers in the quadrants indicate the percent of CD45+ cells. Please click here to view a larger version of this figure.
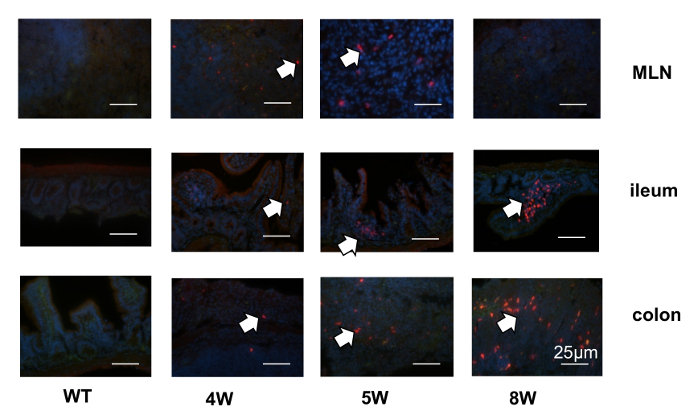
Figure 5: Localization of IL-22 reporters during the development of T-cell transfer colitis. CD4CD45RBHigh T cells from IL-22 reporter mice were transferred into Rag1-/- mice, and the tdTomato signals were evaluated by immunohistochemistry in the different tissues at different time points during the development of colitis. The arrows point to the IL-22-tdTomato signals. Scale bars = 25 µm. Please click here to view a larger version of this figure.
Discussion
IL-22 plays an essential role in innate host defense and tissue remodeling. IL-22-producing cells have been identified ex vivo by intracellular staining. However, it still remains difficult to track IL-22 expression in situ, either in the normal state or in inflammatory conditions. This protocol describes a novel method to develop an IL-22 reporter mouse model, which enables us to localize the reporter-expressing cells in vivo. The reporter gene encoding TdTomato was inserted into the IL-22 locus at a site that disrupts the signal sequence. This strategy results in the reporter being trapped in the producing cell, unlike IL-22 which is rapidly secreted, enabling visualization of the producing cell. The engineered IL-22-reporter was contained within a large BAC, with the aim of preserving the regulatory elements, thus conferring fidelity of expression of the reporter corresponding to that of IL-22. The intestinal tissues of transgenic mice displayed homeostatic reporter expression in CD4 T cells and innate lymphocytes in the lamina propria of the small intestine. In induced colitis, CD4 T cells expressed the reporter in the colon.
The role of IL-22 in inflammatory bowel disease has been unclear. By participating in the intestinal mucosal defense and tissue regeneration, IL-22 is capable of eliciting the production of both protective20,21,22 and proinflammatory mediators (e.g., IL-8)23,24. Two models of T cell-driven colitis showed increases in IL-22 in the colon, including TCR α -/- mice25,26 and the CD45RBhi transfer model27. Comparing inflammatory bowel disease models, IL-22 expression in the intestine was higher in a Crohn's disease modelthan in a model of ulcerative colitis21,24.Here, we transfer CD4CD45RBhi T cells from the reporter mice into Rag1-/- recipients and mimic the pathogenic process of Crohn's disease. We found that, preceding colitis, IL-22 reporters were visualized in intestinal draining lymph nodes (MLN). The signals then diminished in the MLN and moved to the intestines, especially the distal small intestines and colons, with the development of colitis. Most tdTomato reporters were localized inside the lamina propria mucosa of the intestine.
A different type of reporter for IL-22 has been previously described, in which cells are permanently marked such that they and their daughters continue to express the reporter, whether or not transcription of IL-22 has continued28. As in our study, gut innate lymphocytes were marked under homeostatic conditions, and in colitis, the reporter was localized in T cells from the mesenteric lymph nodes to the intestinal tissue.
The procedures described in this protocol would be applicable for the establishment of other transgene reporter mice, such as IL-17, IL-25, and so on. However, it is crucial to choose suitable BAC clones carrying distal regulatory elements to assure the fidelity of reporter gene expression, since the transgenes are randomly integrated into the mouse genome. The reporter mouse allows for the identification of cells expressing in vivo, as well as for the purification and characterization of live cells. We used tdTomato, an exceptionally bright red fluorescent protein, as a fluorescent reporter gene to make the reporter suitable for live animal imaging studies.
Disclosures
The authors have nothing to disclose.
Acknowledgements
We thank Kelli Czarra and Megan Karwan for animal technical assistance, Kathleen Noer and Roberta Matthai for flow cytometry assistance, and Donna Butcher andMiriam R. Anver for pathology analysis. This project was supported by a grant from the Ely and Edythe Broad Foundation (to Scott Durum) and has been funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under Contract No. HHSN261200800001E (MRA).
Materials
| RP23-401E11 BAC | Thermo Fisher Scientific | RPCI23.C | Need gene ID: 50929 |
| NucleoBond BAC 100 | Takara Clontech | 740579 | |
| PCR SuperMix High Fidelity | Thermo Fisher Scientific | 10790020 | |
| PI-SceI | New England Biolabs | R0696S | |
| SpeI | New England Biolabs | R0133S | |
| LB Broth | Thermo Fisher Scientific | 10855-001 | 1L: 10 g SELECT Peptone 140, 5 g SELECT Yeast Extract, 5 g sodium chloride |
| Anti-mouse CD3 | eBioscience | 11-0031 | |
| Anti-mouse CD4 | eBioscience | 17-0041 | |
| Anti-mouse CD45 | Thermo Fisher Scientific | MCD4530 | |
| Anti-mouse CD45RB | eBioscience | 11-0455 | |
| Anti-mouse RFP | Abcam | Ab62341 | |
| HBSS, no calcium, no magnesium, no phenol red | Thermo Fisher Scientific | 14175145 | KCl, KH2PO4, Na2HPO4, NaHCO3, NaCl, D-Glucose |
| Dnase I | Roche | 10104159001 | |
| ACK lysing buffer | Thermo Fisher Scientific | A1049201 | |
| Percoll | GE healthcare life sciences | 17-0891-01 | |
| Collagenase D | Roche | 11088858001 | |
| Dispase II (neutral protease, grade II) | Roche | 4942078001 | |
| IX70 inverted fluorescence microscope | Olympus | Ask for quote | |
| Nikon Eclipse 80i microscope | Nikon | Ask for quote | |
| Dynal shaker | Electron Microscopy Science | 61050-10 | |
| FACSAria | BD Bioscience | Ask for quote | |
| LSRII SORP/flow cytometry | Becton, Dickinson and Company | Ask for quote |
References
- Awasthi, A., et al. Cutting edge: IL-23 receptor gfp reporter mice reveal distinct populations of IL-17-producing cells. J Immunol. 182, 5904-5908 (2009).
- Price, A. E., Reinhardt, R. L., Liang, H. E., Locksley, R. M. Marking and quantifying IL-17A-producing cells in vivo. PLoS One. 7, e39750 (2012).
- Kamanaka, M., et al. Expression of interleukin-10 in intestinal lymphocytes detected by an interleukin-10 reporter knockin tiger mouse. Immunity. 25, 941-952 (2006).
- Sonnenberg, G. F., Fouser, L. A., Artis, D. Border patrol: regulation of immunity, inflammation and tissue homeostasis at barrier surfaces by IL-22. Nat Immunol. 12, 383-390 (2011).
- Dumoutier, L., Louahed, J., Renauld, J. C. Cloning and characterization of IL-10-related T cell-derived inducible factor (IL-TIF), a novel cytokine structurally related to IL-10 and inducible by IL-9. J Immunol. 164, 1814-1819 (2000).
- Cella, M., et al. A human natural killer cell subset provides an innate source of IL-22 for mucosal immunity. Nature. 457, 722-725 (2009).
- Sanos, S. L., et al. RORgammat and commensal microflora are required for the differentiation of mucosal interleukin 22-producing NKp46+ cells. Nat Immunol. 10, 83-91 (2009).
- Spits, H., et al. Innate lymphoid cells–a proposal for uniform nomenclature. Nat Rev Immunol. 13, 145-149 (2013).
- Mazzucchelli, R. I., et al. Visualization and identification of IL-7 producing cells in reporter mice. PLoS One. 4, e7637 (2009).
- Shen, W., Hixon, J. A., McLean, M. H., Li, W. Q., Durum, S. K. IL-22-Expressing Murine Lymphocytes Display Plasticity and Pathogenicity in Reporter Mice. Front Immunol. 6, 662 (2015).
- Gong, S., Yang, X. W., Li, C., Heintz, N. Highly efficient modification of bacterial artificial chromosomes (BACs) using novel shuttle vectors containing the R6Kgamma origin of replication. Genome Res. 12, 1992-1998 (2002).
- Shen, W., Huang, Y., Tang, Y., Liu, D. P., Liang, C. C. A general method to modify BACs to generate large recombinant DNA fragments. Mol Biotechnol. 31, 181-186 (2005).
- Cho, A., Haruyama, N., Kulkarni, A. B. Generation of transgenic mice. Curr Protoc Cell Biol. Chapter. Chapter 19, Unit 19 11 (2009).
- Danneman, P. J., Stein, S., Walshaw, S. O. Humane and practical implications of using carbon dioxide mixed with oxygen for anesthesia or euthanasia of rats. Lab Anim Sci. 47, 376-385 (1997).
- Omata, Y., et al. Isolation of coccidian enteroepithelial stages of Toxoplasma gondii from the intestinal mucosa of cats by Percoll density-gradient centrifugation. Parasitol Res. 83, 574-577 (1997).
- Kumar, N., Borth, N. Flow-cytometry and cell sorting: an efficient approach to investigate productivity and cell physiology in mammalian cell factories. Methods. 56, 366-374 (2012).
- Fischer, A. H., Jacobson, K. A., Rose, J., Zeller, R. Cryosectioning tissues. CSH Protoc. 2008, pdb.prot4991 (2008).
- Hilbe, W., et al. Comparison of automated cellular imaging system and manual microscopy for immunohistochemically stained cryostat sections of lung cancer specimens applying p53, ki-67 and p120. Oncol Rep. 10, 15-20 (2003).
- Suzuki, Y., Furukawa, M., Abe, J., Kashiwagi, M., Hirose, S. Localization of porcine trappin-2 (SKALP/elafin) in trachea and large intestine by in situ hybridization and immunohistochemistry. Histochem Cell Biol. 114, 15-20 (2000).
- Wolk, K., et al. IL-22 increases the innate immunity of tissues. Immunity. 21, 241-254 (2004).
- Brand, S., et al. IL-22 is increased in active Crohn’s disease and promotes proinflammatory gene expression and intestinal epithelial cell migration. Am J Physiol Gastrointest Liver Physiol. 290, G827-G838 (2006).
- Nagalakshmi, M. L., Rascle, A., Zurawski, S., Menon, S., Waal Malefyt, d. e., R, Interleukin-22 activates STAT3 and induces IL-10 by colon epithelial cells. Int Immunopharmacol. 4, 679-691 (2004).
- Maloy, K. J., Powrie, F. Intestinal homeostasis and its breakdown in inflammatory bowel disease. Nature. 474, 298-306 (2011).
- Andoh, A., et al. Interleukin-22, a member of the IL-10 subfamily, induces inflammatory responses in colonic subepithelial myofibroblasts. Gastroenterology. 129, 969-984 (2005).
- Bhan, A. K., Mizoguchi, E., Smith, R. N., Mizoguchi, A. Colitis in transgenic and knockout animals as models of human inflammatory bowel disease. Immunol Rev. 169, 195-207 (1999).
- Mombaerts, P., et al. Spontaneous development of inflammatory bowel disease in T cell receptor mutant mice. Cell. 75, 274-282 (1993).
- Powrie, F. Immune regulation in the intestine: a balancing act between effector and regulatory T cell responses. Ann N Y Acad Sci. 1029, 132-141 (2004).
- Ahlfors, H., et al. IL-22 fate reporter reveals origin and control of IL-22 production in homeostasis and infection. J Immunol. 193, 4602-4613 (2014).

