Label-free Super-resolution Imaging Enabled by Vibrational Imaging of Swelled Tissue and Analysis
概要
By combining sample-expansion hydrogel chemistry with label-free chemical-specific stimulated Raman scattering microscopy, the protocol describes how to achieve label-free super-resolution volumetric imaging in biological samples. With an additional machine learning image segmentation algorithm, protein-specific multi-component images in tissues without antibody labeling were obtained.
Abstract
The universal utilization of fluorescence microscopy, especially super-resolution microscopy, has greatly advanced knowledge about modern biology. Conversely, the requirement of fluorophore labeling in fluorescent techniques poses significant challenges, such as photobleaching and non-uniform labeling of fluorescent probes and prolonged sample processing. In this protocol, the detailed working procedures of vibrational imaging of swelled tissue and analysis (VISTA) are presented. VISTA circumvents obstacles associated with fluorophores and achieves label-free super-resolution volumetric imaging in biological samples with spatial resolution down to 78 nm. The procedure is established by embedding cells and tissues in hydrogel, isotropically expanding the hydrogel sample hybrid, and visualizing endogenous protein distributions by vibrational imaging with stimulated Raman scattering microscopy. The method is demonstrated on both cells and mouse brain tissues. Highly correlative VISTA and immunofluorescence images were observed, validating the protein origin of imaging specificities. Exploiting such correlation, a machine learning-based image-segmentation algorithm was trained to achieve multi-component prediction of nuclei, blood vessels, neuronal cells, and dendrites from label-free mouse brain images. The procedure was further adapted to investigate pathological poly-glutamine (polyQ) aggregates in cells and amyloid-beta (Aβ) plaques in brain tissues with high throughput, justifying its potential for large-scale clinical samples.
Introduction
The development of optical imaging methods has revolutionized the understanding of modern biology because they provide unprecedented spatial and temporal information of targets across different scales, from subcellular proteins to whole organs1. Among them, fluorescence microscopy is the most well-established, with a large palette of organic dyes with high extinction coefficients and quantum yields2, easy-to-use genetic-encoded fluorescent proteins3, and super-resolution methods such as STED, PALM, and STORM for imaging nanometer-scale structures4,5. In addition, recent advancements in sample engineering and preservation chemistry, which expand specimens embedded in swellable polymer hydrogels6,7,8, enable sub-diffraction limited resolution on conventional fluorescence microscopes. For instance, typical expansion microscopy (ExM) effectively enhances the image resolution by four times with fourfold isotropic sample expansion7.
Despite its advantages, super-resolution fluorescence microscopy shares limitations that originate from fluorophore labeling. First, photobleaching and inactivation of fluorophores compromise the capacity for repetitive and quantitative fluorescence evaluations. Photobleaching is an inevitable event when light keeps pumping electrons into electronically excited states9. Second, labeling the fluorophores to the desired targets is not always a straightforward task. For instance, immunostaining demands a long and laborious sample preparation process and hinders imaging throughput10. It could also introduce artifacts due to inhomogeneous antibody-labeling, especially deep inside tissues11. Moreover, proper labeling strategies that target fluorophores for the desired proteins might be underdeveloped. For example, extensive screenings were required to find effective antibodies for Aβ plaques12. Smaller organic dyes, such as Congo red, often have limited specificity, only staining the core of the Aβ plaque. It is, therefore, highly desirable to develop a label-free super-resolution modality that circumvents the drawbacks of fluorophore-labeling and provides complementary high-resolution imaging from cells to tissues, and even to large-scale human samples.
Raman microscopy provides label-free contrast for chemical-specific structures and maps out the distribution of otherwise invisible chemical bonds by looking at the excited vibrational transitions13. In particular, stimulated Raman scattering (SRS) imaging on label-free or tiny-labeled samples has been demonstrated to have similar speed and resolution to fluorescence microscopy14,15. For example, healthy brain region has been readily differentiated from tumor-infiltrated region in human and mouse tissues16,17. Aβ plaques were also clearly imaged by targeting protein CH3 vibration (2940 cm−1) and amide I (1660 cm−1) on a fresh-frozen brain slice without any labeling18. Raman scattering, therefore, offers robust label-free contrast that overcomes the limitations of fluorophores. The question then became how one can accomplish super-resolution capacity using Raman scattering, which could reveal nanoscale structural details and functional implications in biological samples.
Although extensive efforts have been made to achieve super-resolution for Raman microscopy with elegant optic instrumentations, the resolution enhancement on biological samples has been rather limited19,20,21. Here, based on the recent works22,23, we present a protocol that combines a sample-expansion strategy with stimulated Raman scattering for super-resolution label-free vibrational imaging, named Vibrational Imaging of Swelled Tissues and Analysis (VISTA). First, cells and tissues were embedded in hydrogel matrixes through an optimized protein-hydrogel hybridization protocol. The hydrogel tissue hybrids were then incubated in detergent-rich solutions for delipidation, followed by expansion in water. The expanded samples were then imaged by a regular SRS microscope by targeting CH3 vibrations from retained endogenous proteins. VISTA, owing to its label-free imaging feature, bypasses photobleaching and inhomogeneous labeling arising from fluorophore labeling, with much higher sample processing throughput. This is also the first sub-100 nm (down to 78 nm) label-free imaging reported. No additional optical instrumentation besides typical SRS setup22,24 is required, making it readily applicable. With correlative VISTA and immunofluorescence images, an established machine-learning image-segmentation algorithm was trained25,26 to generate protein-specific multiplex images from single-channel images. The method was further applied to investigate Aβ plaques in mouse brain tissues, providing a holistic image suited for sub-phenotyping based on the fine views of the plaque core and peripheral filaments surrounded by cell nuclei and blood vessels.
Protocol
All animal procedures performed in this study were approved by the California Institute of Technology Institutional Animal Care and Use Committee (IACUC), and the protocol procedures complied with all relevant ethical regulations.
1. Preparation of stock solutions for fixation and sample expansion
- Prepare 40 mL of fixation solution by first dissolving 12 g of acrylamide (30% w/v) solid in 26 mL of nuclease-free water. Then, add 10 mL of 16% PFA stock solution to the mixture. Finally, add 4 mL of 10x phosphate-buffered saline (PBS, pH 7.4). The prepared solution can be stored at 4 °C for up to 2 weeks.
NOTE: Acrylamide is hazardous so the step of dissolving acrylamide solid in water should be done in a fume hood. - Prepare gelation solution (stock X) by dissolving 70 mg of sodium acrylate (7% w/v), 200 mg of acryl amide (20% w/v), and 50 µL of N, N′-methylenebisacrylamide (0.1% w/v) in 420 µL of ultrapure water. Add 57 µL of sterile-filtered 10x PBS (pH 7.4) at the end and store this solution at −20 °C for up to 1 week.
NOTE: Avoid sodium acrylate solid that forms clumps; make sure the sodium acrylate used is in the form of dispersive powders. The stock X made will be a colorless liquid; if the liquid looks light yellow, obtain new sodium acrylate. - Prepare the polymerization initiator solution by dissolving 1 g of ammonium persulfate (APS, 10% w/w) in 9 mL of nuclease-free water. Similarly, prepare polymerization accelerator solution by dissolving 1 g of tetramethylethylenediamine (TEMED, 10% w/w) in 9 mL of nuclease-free water. Aliquot the resulting solutions and store at −20 °C.
- Prepare 50 mL of denaturing buffer by dissolving 2.88 g of sodium dodecyl sulfate (SDS, 200 mM), 0.584 g of sodium chloride (200 mM), and 2.5 mL of 1M Tris-HCl buffer (50 mM, pH 8) in nuclease-free water.
NOTE: The concentration of SDS is close to its saturation condition. Some crystallization in the buffer is normal. Slight warming it up to 37 °C can make the solution clear and ready to use.
2. Preparation of mammalian cell samples
- Seed 3.5 x 104 HeLa cells onto a 12 mm borosilicate coverslip (#1.5) in a 24-well plate and then culture the cells in 400 µL of Dulbecco's Modified Eagle Medium (DMEM) with 10% fetal bovine serum and 1% penicillin-streptomycin antibiotics (complete DMEM) under a humidified atmosphere with 5% CO2 at 37 °C.
- To obtain cells in different mitotic stages, incubate the cells in complete DMEM til 60% confluency. Then, treat the cells with DMEM without fetal bovine serum for 20 h to synchronize the cell stages. After synchronization, switch the medium for the cells to complete DMEM and incubate for another 2-6 h to target the various stages of mitosis.
- Wash the HeLa cells on coverslips with sterile PBS and incubate them with fixation solution (4% PFA 30% acrylamide [AA] in PBS) at 37 °C for 7-8 h before further processing.
NOTE: Higher fixation temperature together a with high concentration of acrylamide helps to quench the intermolecular crosslinking between proteins during the fixation processes and is reported to preserve the detailed ultra-structures of proteins8. - For cells with polyQ aggregates, grow the cells in complete DMEM until they reach 70%-90% confluency. After changing to new complete DMEM, transfect 1 µg of plasmid encoding mHtt97Q-GFP into the cells using transfection agent (details in Table of Materials). Perform transfection according to the manufacturer's protocol.
- After 24-48 h of transfection, remove the culture medium and add 800 µL of fixation solution (4% PFA, 30% AA) to the coverslips with cells. Incubate the coverslips at 37 °C for 6-8 h. Wash the coverslip with PBS 3x, and the resulting cell samples are ready for further processing.
3. Preparation of mouse brain samples
- Purchase C57BL/6J mice (6- to 8-week-old, male and female, six mice) and Alzheimer mice (9-month-old, female, three mice) from commercial sources (details in Table of Materials). Perform euthanasia via carbon dioxide narcosis according to standard protocol27. In brief, place mice in a chamber and fill the chamber with a flow of 100% CO2 in the order of 30%-70% of the volume of the chamber/min and maintain the flow for at least 1 min after clinical death.
- After confirming the humane euthanasia, quickly decapitate the mice using a scissor. Expose the skull with a large incision through the skin down the midline. Pull the skin toward the nose of the animal to fully expose the skull.
- Cut the skull open along the midline with fine scissors and peel the two halves of the skull away to the side. Use tweezers to scoop out the brain into a Petri dish on ice with PBS. Wash the fresh mouse brain with ice-cold PBS and transfer the brain to 10 mL of fixation buffer (4% PFA, 30% AA).
- Incubate the mouse brain in fixation buffer at 4 °C for 24-48 h and then transfer it to 37 °C overnight. After washing it with sterile PBS, cut the mouse brain into 150 µm sections using a vibratome and store in PBS at 4 °C.
4. Hydrogel embedding, denaturation, and expansion of cell and tissue samples
- Make sure the samples were incubated in the fixation buffer for the required amount of time, as indicated above. Thaw stock X, free-radical initiator, and accelerator stock solutions and keep them on ice during the whole process.
NOTE: Insufficient incubation time could cause a reduced number of proteins retained on the hydrogel hybrid and, therefore, cause a decrease in signal. - Pick up the coverslip with cells and place it on a glass slide using tweezers. Pick the brain slices (150 µm thick) and place them on another glass slide using a soft-wool paintbrush, ensuring that they are flat. Get rid of excess liquid that comes with the samples.
- Stack two coverslips (#1.5), by adding drops of water between them, on both sides of the sample on the glass slide to make a gelation chamber. Place the glass slide with the sample facing upward onto an ice-cold heat block and cool it to 4 °C for 1 min.
- Add 10% (w/w) TEMED to stock X and then 10% APS solution. Quickly vortex and, within 1 min after mixing, slowly drop the resulting solution into the chamber onto the samples to cover the surface of the sample, avoiding air bubbles. Keep the solution and samples on ice (or an ice-cold heat block) to avoid gel formation (high degree of polymerization) before adding to the sample.
- Once the sample is fully immersed in the solution, place a flat, transparent, film-covered coverslip on top of the sample as a lid for the chamber. Leave the chamber on ice or an ice-cold heat block for another 1 min before incubating it in a humidifying incubator at 37 °C for 30 min.
- Take out the gelation chamber on the glass slide from the 37 °C incubator. A non-transparent gel should be observed after removing the lid. Retrieve the gel by cutting with a razorblade or, alternatively, put the glass slide into denaturing buffer at room temperature for 15 min. The gel will separate itself from the glass slide.
- Incubate the isolated gel in denaturing buffer under heat conditions to achieve further denaturation and delipidation. For cell samples, denature at 95 °C for 1 h. For 150 µm thick brain slices, denature at 70 °C for 3 h, followed by 95 °C for 1 h.
NOTE: The gel could become curly when first incubated with denaturing buffer at room temperature. It is normal and will become flat after high-temperature denaturation. When working with thicker tissue samples, a longer denaturation time is needed. - After the heat treatment, wash the denatured sample 3x with PBS for 10 min. At this point, the gel should have expanded to about 1.5 times the original size. Incubate the washed gel with ultrapure water in a large container (at least 20 times the volume of the gel) to achieve a higher expansion ratio. Change the water every 1 h 3x and leave the sample in darkness overnight. The resulting gel is ready to image.
NOTE: The expansion ratio largely depends on the mechanical properties of the original sample. A 4.2 times expansion for the cell samples and 3.4 times expansion for the brain tissue sample were obtained.
5. Label-free imaging of endogenous protein distribution in expanded cell and tissue samples
- Keep the expanded gel samples in water during the imaging process. The gel is fragile after expansion so should be handled with caution. Place the expanded sample-bearing gel onto a microscope slide (1 mm thick) with a microscope spacer with appropriate opening sizes and depths, filled with water. Cover the spacer with a coverslip (#1.5) and ensure it is properly sealed to avoid sample movement.
- Place the sample with either the expanded cell or tissue onto the motorized stage with the coverslip facing the objective. Click Ocular on the software to change the light path to bright field and adjust the z position using a manual knob.
- Add immersion oil on the top of the sample and adjust the condenser to the correct position for Köhler illumination while watching under bright field with 25x magnification from the objective. Input 791.3 nm in the OPO wavelength window on the laser panel to target protein CH3 vibration at 2940 cm−1.
- Switch the light of the microscope from bright field (eye piece) to laser scanning mode by clicking the LSM button and open the SRS laser shutter by hitting the Shutter button on the laser control panel. Press the LIVE button in the microscope software to start real-time image acquisition.
- Find the proper focus by changing the z while looking at the SRS image under short pixel-dwell time (12.5 µs/pixel) and low image resolution (256 pixels x 256 pixels). Once the ideal z-position is found, change the scan size and pixel-dwell time by pulling the respective bar in the microscope software.
- Acquire the image by hitting the LSM button using the super-sampling condition (1024 pixels x 1024 pixels) with a longer pixel-dwell time (80 µs/pixel) that matches the time constant of the lock-in amplifier. Acquire volumetric images by collecting a z-stack with a step size of 1 µm in the z-direction. Process and analyze the saved OIR file from the software using ImageJ.
NOTE: The detailed SRS setup has recently been reported in Mutlu et al.24. Here, a tunable picosecond laser with an 80 MHz repetition rate and 2 ps bandwidth for pump (770-990 nm) and stokes (1031.2 nm) was routed into a laser scanning confocal microscope (detailed in Table of Materials). Optimize the temporal and spatial overlapping of the two beams for signals with pure D2O. It takes some effort to find the right z for the expanded sample under bright field because the refractive index is very homogenous throughout the gel. - Determine the resolution of VISTA by taking an SRS image of polystyrene beads (100 nm) using both a 25x objective and a 60x objective. Set the pump laser to 784.5 nm, corresponding to Raman shift of 3050 cm−1, characteristic of the aromatic C-H stretches vibrations of polystyrene.
NOTE: The pump laser wavelength used here is similar to the one used in VISTA for proteins. - With the 60x objective, the experimental full wave half maximum (FWHM, which is the width of the line shape at half of the amplitude) of the bead image was 276.17 nm. Model the function of the bead object as a half-circle.
NOTE: When the PSF Gaussian function has a c (σ) = 269 nm/2.35, the convoluted bead image would have a measured FWHM of 276.17 nm. As a result, the resolution of the SRS system is 269 nm × 1.22 = 328 nm by the Rayleigh Criterion. As VISTA has an average of 4.2 times expansion of cell samples, the effective resolution is down to 328 nm/4.2 = 78 nm.
6. Correlative VISTA and fluorescent imaging of immuno-labeled and expanded tissue samples
- After denaturation, pre-incubate the hydrogel embedded samples (e.g., 150 µm brain coronal section) in 1% (v/v) Triton X-100 (PBST) for 15 min. Switch the incubation buffer to PBST with primary antibody at a 1:100 dilution. If multiple protein targets are needed for multiplex imaging, dilute respective primary antibodies for different targets to proper concentrations in the same cocktail and incubate with the samples simultaneously.
- Incubate the gels with diluted primary antibodies at 37 °C with gentle shaking at 80 rpm for 16-18 h, followed by extensive washing 3x with PBST for 1-2 h at 37 °C. Flip over the gel during the incubation to prevent inhomogeneous antibody labeling here and in all subsequent incubations.
- Incubate the samples with secondary antibodies of corresponding species targets at 1:100 dilutions with PBST at 37 °C for 12-16 h, protected from the light. Wash labeled hydrogel samples 3x with PBST for 1-2 h at 37 °C with gentle shaking.
- Dilute DAPI to 3 µM final concentration in PBS. Add a sufficient volume of DAPI solution to the sample hydrogel so that the gel is submerged in the solution. Incubate for 1-2 h at room temperature with gentle shaking at 80 rpm. Wash the sample 3x with PBS.
- Expand the immuno-labeled gel samples by incubating with a large volume of double deionized H2O. Change the water every 1 h 3x and incubate the samples in H2O overnight, protected from light. During labeling, the gel should expand 1.5 times compared to the PBS buffer.
- Prepare the imaging sample as described in step 5.1. Place the imaging sample onto the laser scanning confocal microscope with the 25x, 1.05 NA, water immersion objective for fluorescent imaging.
- Select the proper channel with the respective excitation laser (405 nm, 488 nm, 561 nm, and 640 nm) and PMT pair according to the targeted antibodies in the dropdown menu on the microscope software. Press the LIVE button for real-time image acquisition.
- Adjust the focus position by a manual knob based on the real-time fluorescence signal. Optimize the laser power, pixel-dwell time, and PMT gain in the microscope software according to the real-time fluorescence signal to avoid a dim signal or oversaturation.
NOTE: Evaluate the appearance and features based on the reported structures of different antibodies to eliminate potential antibody cross-reactivities and to avoid crosstalk between different fluorescence channels. - Hit the LSM button to start acquiring correlative SRS and fluorescent images. First, collect volumetric fluorescence images on immuno-labeled samples. Once fluorescence imaging is finished, switch the microscope light path to the infrared (IR) transparent condition.
- Change the detection channel from fluorescence PMT to SRS on the microscope software. Open the shutter for the SRS laser and perform SRS volumetric imaging on the field of view of the same sample with the same range of z. The normal range is between 100-200 µm and can go up to 700 µm.
NOTE: There is slight chromatic aberration that causes a shift in z-position between the fluorescent images and the SRS images because of the wavelength difference in the excitation lasers. Manual side-by-side comparison between the SRS and fluorescence z-stack images is needed. Look for the exact matching features from both the SRS and the fluorescence channel to match the z-position precisely.
7. Construction, training, and validation of U-Net architecture
NOTE: Installation on Linux is recommended. A graphics card with >10 GB of RAM is required.
- Setting up the environment
- Install Anaconda or Miniconda on a 3-5.3.0-linux-x86_64. Clone or download the following files: https://github.com/Li-En-Good/VISTA. Create a Conda environment for the platform using the following command line:
conda env create -f environment.yml
- Install Anaconda or Miniconda on a 3-5.3.0-linux-x86_64. Clone or download the following files: https://github.com/Li-En-Good/VISTA. Create a Conda environment for the platform using the following command line:
- Training the prediction model
- Pair the directories of corresponding SRS images with ground truth images in a csv file. Place the directories of SRS images under path_signal and ground truth images under path_target columns.
- Put the csv file in the folder data/csvs. Modify the configuration in scripts/train_model_2d.sh if needed. Activate the environment using the command line:
conda activate fnet - Initiate model training using the command line:
./scripts/train_model_2d.sh <file name of the csv file> 0
The training will then start. The losses for each iteration will be shown in the command line and saved with the model in the folder save_models/<file name for the csv file>.
- Validate the images in the training set and test set using the command line:
./scripts/train_model_2d.sh <file name of the csv file> 0
The prediction results will be saved in the folder results/<file name of the csv file>.
8. VISTA combined with U-Net predictions for protein-specific multiplexity in label-free images
- Modify the csv file in data/csvs/<file name of the csv file>/test.csv. Replace the directories of both path_signal and path_target with the directory of new SRS images.
- Remove the prediction results from the training, which is the folder results/<file name of the csv file>. Run predictions using the command line:
./scripts/train_model_2d.sh <file name of the csv file> 0
The prediction results will be saved in the folder results/<file name of the csv file>/test.
Representative Results
After establishing the working principle of the imaging and analysis method, image registration was done to evaluate the expansion ratio and to ensure isotropic expansion during sample processing (Figure 1A,B). Both untreated and VISTA samples were imaged while targeting the bond vibration at 2940 cm−1, which originates from CH3 of endogenous proteins. In untreated samples, the protein-rich structures like nuclei were dark due to the overwhelming lipid content from surrounding tissues22 (Figure 1A). After sample processing that includes the delipidation treatment, the resulting image showed the same feature with an inversed contrast (Figure 1B). The shapes and the relative positions of nuclei and vessels were completely unaltered (Figure 1A,B; numbered structures), confirming that the treatment is an isotropic process. By comparing the sizes of the corresponding nuclei, it was concluded that the method achieves 3.4 times expansion in brain tissue samples as compared to untreated samples22,23.
Knowing the expansion ratio in brain tissue, VISTA can now resolve new features in the label-free SRS images that were previously unresolvable. Although actin and tubulin structures have been the gold standard for super-resolution demonstrations, the resolution improvements on actin and tubulin structures have been well-characterized by fluorescence-based sample-expansion strategies using similar hybridization chemistry28. Moreover, imaging specific actin/tubulin structures is less feasible with this technique because the signal comes from the total ensemble of endogenous proteins, where cytoskeleton structures like tubulins would not have sufficient contrast (signal-to-background ratio) to be clearly distinguished. Hence, we decided to pursue imaging other nanoscale structures. We showed that features can be captured from mouse cortexes down to 150 nm (Figure 1C,D). Based on the dispersive patterns around neuronal dendrites, the observed small structures are likely dendritic spine heads7, which have a size of 146 nm (Figure 1D). In addition, the method was used to image the fibrillar structures in Aβ plaques, which are believed to have a thickness around 100 nm29,30. Indeed, it was demonstrated that ~130 nm fibrillar structures can be resolved in a representative diffusive Aβ plaque using this method (Figure 1E,F).
As VISTA enables effective protein retention and protein imaging22, one can clearly distinguish protein-rich nucleoli in nuclei and the ribbon-like cytoskeleton structures in the cytosols of cultured HeLa cells (Figure 2A, arrowhead). The method was further applied to study poly-glutamine (polyQ) aggregates that are transiently expressed in mammalian cells (Figure 2B,C). The results confirmed that the aggregates, as an expectedly densely packed structure, were expanded isotropically by comparing the same aggregate structures before and after expansion across multiple replicate samples23. High-resolution structures that are absent/blurred in the normal-resolution SRS images were obtained using this method. The VISTA-aggregate images revealed fibril-like protrusions on the periphery of the polyQ aggregates and a hollow structure in the center (Figure 2B, arrowhead). The observation that protrusions seamlessly attach to cytosolic contents might suggest that aggregates engage with functional proteins in cytosol. In hindsight, the capacity to expand dense aggregates also becomes plausible because the fixation reagent formaldehyde and the hydrogel monomers acryl amide and sodium acrylates are all small molecules that can diffuse in and out of protein aggregates. Once the aggregate co-polymerizes with monomers into hydrogel, the expansion process should proceed as normal.
We then applied this method to mouse brain tissue to further extend its scope. Although tissue samples pose challenges like reduced permeability, increased thickness, and heterogeneous mechanical strength, the mouse brain samples were successfully imaged using this method (Figure 2D). Similar to cell samples, protein-rich structures including cell nuclei, blood vessels, and neuronal processes were observed (Figure 2D, arrowhead). The limitation of brain tissues is that only 3.4 times expansion was achieved, which makes the effective resolution in brain samples 99 nm22. We validated the structural origin of SRS signals by correlative dye and antibody staining, in which DAPI stains for nuclei and lectin stains for blood vessels (Figure 2E,F). Neuronal cell bodies and processes were also delineated by immunofluorescence from NeuN and MAP222. With the trained convolutional neural network (CNN) algorithm utilizing the correlative fluorescence images as ground truth, the single-channel images were then segmented into specific protein-structure channels for multiplex images22.
Finally, we aimed to interrogate pathologic Aβ plaques in the brains of 5xFAD mice, a well-known animal model for Alzheimer's disease31. After following the procedures, a 3-dimensional SRS image of amyloid plaques deposition in brain tissues was acquired (Figure 2G). Puncta with high protein concentrations were observed (Figure 2G, orange arrow), representing the core of the Aβ plaque. Such image also revealed the peripheral Aβ plaques (Figure 2G, magenta arrowhead), which are often neglected by conventional Congo red staining that only targets the Aβ core. When combined with the trained segmentation algorithm, the label-free image could be transformed into a target-specific multiplex image (Figure 2F) and can be performed jointly with immunofluorescence23 to study plaque-astrocyte and plaque-microglia microenvironment interactions32 in a comprehensive and high-throughput manner.
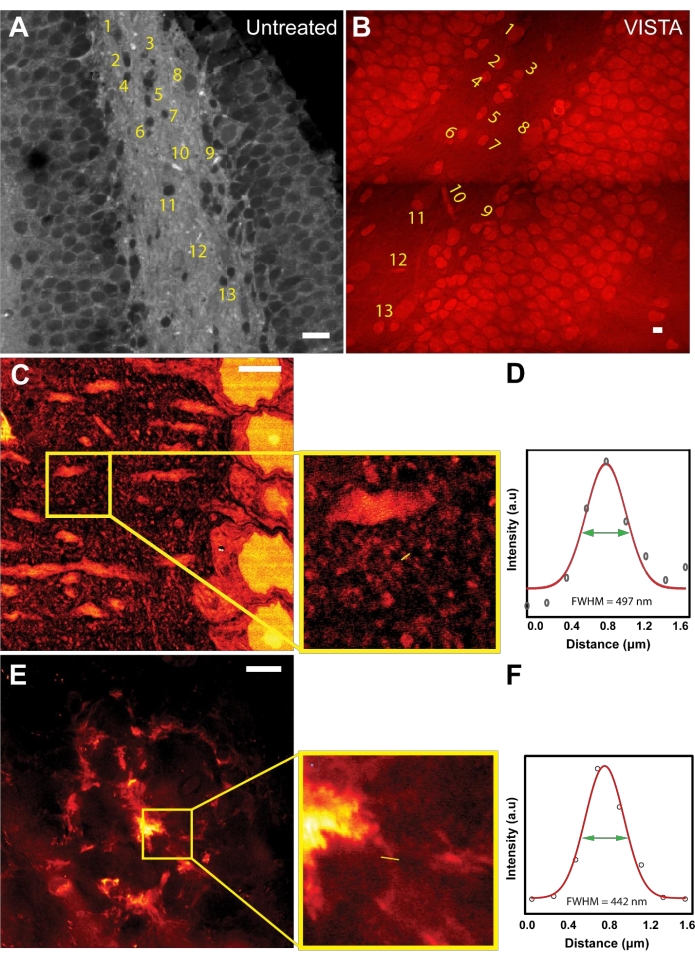
Figure 1: Sample expansion strategy enables super-resolution label-free imaging in mouse brain tissues. (A) An SRS image at CH3 frequency of mouse hippocampus. (B) A VISTA image in the same field of view of mouse hippocampus. Labeled area shows the corresponding features before and after treatments. (C) A VISTA image of a normal mouse cortex that shows finer features. Inset shows the region of interest. (D) Resolution quantification for the fine structure observed in the expanded sample. FWHM of 497 nm corresponds to an effective resolution of 146 nm with 3.4 times expansion. (E) A VISTA image of an amyloid-beta (Aβ) plaque in mouse brain tissues. Inset shows the enlarged region of interest. (F) Resolution quantification for the extrusion fiber structure of the expanded amyloid-beta plaque. FWHM of 442 nm corresponds to an effective resolution of 130 nm with 3.4 times expansion. Scale bars = 20 µm. Please click here to view a larger version of this figure.
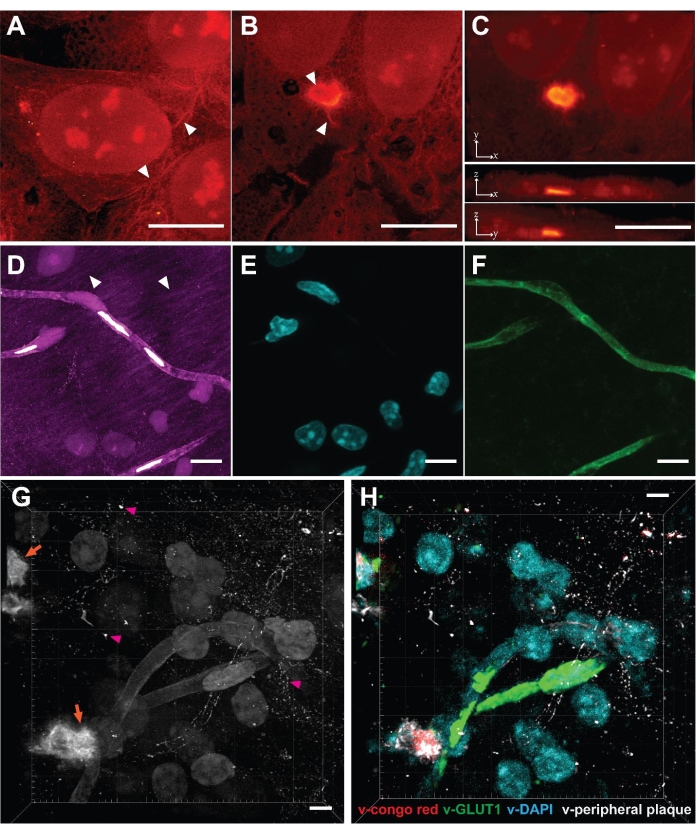
Figure 2: Label-free super-resolution volumetric imaging in cells and tissues enabled by VISTA. (A) Volumetric image of a normal HeLa cell. Arrowhead: cytoskeleton-like structure. (B) Single z-slice image of a polyQ aggregate expressed in HeLa cells. Arrowhead: hollow structure and fibril extrusions. (C) Maximum intensity projections of x-y, x-z, and y-z directions show the volumetric view of the polyQ aggregate-containing cell. (D) Volumetric image of a coronal section of mouse brain. Arrowhead: neuronal processes. (E) Fluorescence image of nuclei (DAPI staining) at the same sample region shows 1 to 1 correlation with nuclei in the VISTA image. (F) Fluorescence image of blood vessels (anti-lectin) at the same sample region shows a 1 to 1 correlation with vessel structures in the VISTA image. (G) Volumetric image of Aβ (orange arrow) containing brain tissue. Pink arrowhead: peripheral Aβ plaque. (H) Multiplex image from (G), predicted by the trained image segmentation algorithm. v-congo red represents the core of the Aβ plaque; v-GLUT1 represents blood vessels; v-DAPI represents nuclei; v-peripheral plaque represents the Aβ plaque not stained by the Congo red dye. Scale bars = 10 µm. The length scale is in terms of distance before expansion (adjusted for different expansion ratios). Please click here to view a larger version of this figure.
Discussion
In summary, we present the protocol for VISTA, which is a label-free modality to image protein-rich cellular and subcellular structures of cells and tissues. By targeting endogenous CH3 from proteins in hydrogel-embedded cell and tissues, the method achieves an effective imaging resolution down to 78 nm in biological samples and resolves minor extrusion in Huntingtin aggregates and fibrils in Aβ plaques. This technique is the first instance to report sub-100 nm resolution for label-free imaging modalities22. Compared to existing expansion methods6,7,8,28,33,34, the technique inherits the merit of label-free SRS imaging and, hence, is free from photobleaching, inactivation, or quenching caused by laser illuminations. In addition, as a label-free method, it circumvents the demanding, inefficient, and potentially artifact-causing antibody-labeling that is always involved in methods such as DISCO12,35 and ExM33,34 and, thus, offers high-throughput sample preparation and uniform imaging throughout tissues. To address the lack of multiplexity in the label-free approach, VISTA, implemented with a CNN-based image segmentation algorithm25, provides protein-specific multi-component images without any labels in brain tissues22. The method was further applied on 5xFAD mouse brains and enabled a holistic volumetric view of aggregates core and periphery fibrils, nuclei, and blood vessels23. We envision that VISTA would scale up well for larger samples such as primate or human brain slices and could, ultimately, be useful for clinical investigations.
There are three essential steps that ensure the successful implementation of this method. First, maximum protein retention in the hydrogel sample hybrid is crucial and required22. To achieve this goal, the fixation condition was modified to contain a high concentration of acrylamide28 and to replace the protein digestion procedure with high-concentration detergent delipidation that saves significant protein loss from protein digestion. The addition of AA quenches the intermolecular crosslinking of proteins and enables the isotropic expansion without protein digestions28. In a previous study, deuterated monomers were used to prove that the aliphatic CH bonds in acrylamide hydrogels only give rise to constant background22. Second, proper correlations between SRS and immuno-labeling and distinctions between different protein targets need to be established. As the method relies on image-segmentation algorithms to add multiplexity to monochromatic SRS images, crosstalk between different protein targets in immunofluorescence will significantly compromise the quality of images. We meticulously selected protein-rich structures that are obvious in SRS images and validated their corresponding immunofluorescence features. Third, before using the model to predict fluorescence patterns from new SRS data sets, the validity and reliability of the trained machine-learning model should be testified. Distinct features that are not included in the training sets will likely cause issues in prediction. If the prediction results are not satisfying, the user should try to include more data for training and avoid predicting patterns that are not included in the training sets. Pearson's correlations of the testing sets and validation sets should also be monitored to ensure the quality of the prediction22,23. It is suggested to have at least 100 corresponding image sets for training.
While the method has immense potential for biological studies, there are certain limitations awaiting creative solutions. First and foremost, the sensitivity needs further improvement. The detection limit of label-free simulated Raman scattering is in the low millimolar range, and the isotropic expansion of samples in three dimensions significantly dilutes chemical bonds and weakens the signal. We, hence, are limited to imaging the total ensemble of endogenous proteins, which lacks specificity and multiplexity. Combining VISTA with ultrasensitive SRS36 could possibly extend this to image low-abundance proteins and study aggregate structures and compositions at super-resolution level by targeting orthogonal chemical bonds37. Second, the current 3.4 times expansion ratio in brain tissues only gives moderate resolution improvement. Although we have already resolved minor extrusions in Aβ plaques that were previously indistinguishable, higher resolution is always desirable. In this case, innovations in protein-anchoring and hydrogel chemistry would greatly benefit. For example, different gel formulation could enable larger expansion ratios for even higher image resolution38,39,40. New procedures in sample processing would allow it to be applied with widely available FFPE histology samples38,41, making it even better suited for large-scale clinical studies.
開示
The authors have nothing to disclose.
Acknowledgements
We acknowledge the Caltech Biological Imaging Facility for software support. L.W. acknowledges the support of the National Institutes of Health (NIH Director's New Innovator Award, DP2 GM140919-01), Amgen (Amgen Early Innovation Award), and the start-up funds from the California Institute of Technology.
Materials
| 1.0 M Tris pH 8 | Sigma-Aldrich | 648314 | |
| 16% Paraformaldehyde | Electron microscopy science | 15710 | diluted to 4% in PBS |
| 25x water immersion objective | Olympus | XLPLN25XWMP2 | NA 1.05 |
| 5XFAD Mice | Mutant Mouse Resource and Research Centers and the Jackson Laboratory | B6SJL-Tg (APPSwFlLon, PSEN1*M146L*L286 V) 6799Vas/Mmjax | Alzheimer brain |
| 60x water immersion objective | Olympus | UPLSAPO60XWIR | NA 1.2 |
| Acrylamide | Sigma-Aldrich | A9099 | |
| ammonium persulfate | Sigma-Aldrich | A3678 | |
| anti-MAP2 | Cell Signaling Technology | 8707 | |
| anti-NeuN | Cell Signaling Technology | 24307 | |
| borosilicate coverslip #1.5 | Fisher Scientific | 1254581 | |
| C57BL/6J Mice | Jackson Laboratory (JAX) | 664 | Normal mice |
| D2O | Sigma-Aldrich | 151882 | for SRS calibration |
| DAPI | Thermo Fisher | D1306 | |
| DMEM | GIBCO | 10566-016 | |
| FBS | GIBCO | A4766 | |
| glass slide 3" x 1" x 1 mm | VWR | 16004-430 | |
| goat anti-chicken IgY, Alexa Fluor 647 | Invitrogen | A-21449 | |
| goat anti-mouse IgG, Alexa Fluor 647 | Invitrogen | A-21236 | |
| goat anti-rabbit IgG, Alexa Fluor 488 | Invitrogen | A-11034 | |
| goat anti-rat IgG, Alexa Fluor 568 | Invitrogen | A-11077 | |
| Grace Bio-Labs Press-To-Seal silicone isolators | Sigma-Aldrich | GBL664108 | microscope spacer |
| Htt-97Q-GFP Plasmid | Gift from Prof. R. Kopito and Prof. F.-U.Hartl. | ||
| Laser scanning microscope | Olympus | FV3000 | laser scanning confocal microscope |
| lipofectamine 3000 | Thermo Fisher | L3000001 | transfection agent |
| Lycopersicon Esculentum Lectin DyLight®594 (lectin) | Vector Laboratories | DL-1177-1 | |
| Microscope spacer | Grace Bio-Labs | 621502 | |
| N,N′-methylenebisacrylamide (BIS) | Sigma-Aldrich | M1533 | bought as 2% solution in water |
| Nuclease free water | Thermo Fisher | 10977-015 | |
| Penicillin-Streptomycin | GIBCO | 15140-122 | |
| poly-strene beads | Sigma-Aldrich | 43302 | for resolution characterization |
| Sodium Acrylate | Sigma-Aldrich | 408220 | |
| sodium dodecyl sulfate | Sigma-Aldrich | 71725 | |
| soft-wool paint brush #3 | TANIS | 000333 | |
| SRS Laser | A.P.E | picoEmerald | 2ps pulse width |
| tetramethylethylenediamine | Sigma-Aldrich | T9281 | |
| Tissue culture flask 25 cm2 | Corning | 430639 | |
| Triton X-100 | Sigma-Aldrich | T8787 | |
| Tween-20 | Sigma-Aldrich | P9416 | |
| tweezer | Fine Science Tool | 11295-51 | |
| Vibrotome | Leica | VT1200S | the vibratome |
参考文献
- Ntziachristos, V. Going deeper than microscopy: The optical imaging frontier in biology. Nature Methods. 7 (8), 603-614 (2010).
- Lavis, L. D., Bright Raines, R. T. ideas for chemical biology. ACS Chemical Biology. 3 (3), 142-155 (2008).
- Tsien, R. Y. Constructing and exploiting the fluorescent protein paintbox (Nobel Lecture). Angewandte Chemie (International Edition in English). 48 (31), 5612-5626 (2009).
- Huang, B., Bates, M., Zhuang, X. Super-resolution fluorescence microscopy. Annual Review of Biochemistry. 78, 993-1016 (2009).
- Sahl, S. J., Hell, S. W., Jakobs, S. Fluorescence nanoscopy in cell biology. Nature Reviews Molecular Cell Biology. 18 (11), 685-701 (2017).
- Wassie, A. T., Zhao, Y., Boyden, E. S. Expansion microscopy: Principles and uses in biological research. Nature Methods. 16 (1), 33-41 (2019).
- Chen, F., Tillberg, P. W., Boyden, E. S. Expansion microscopy. Science. 347 (6221), 543-548 (2015).
- Gambarotto, D., et al. Imaging cellular ultrastructures using expansion microscopy (U-ExM). Nature Methods. 16 (1), 71-74 (2019).
- Demchenko, A. P. Photobleaching of organic fluorophores: Quantitative characterization, mechanisms, protection. Methods and Applications in Fluorescence. 8 (2), 022001 (2020).
- Murray, E., et al. scalable proteomic imaging for high-dimensional profiling of intact systems. Cell. 163 (6), 1500-1514 (2015).
- Kim, S. -. Y., et al. Stochastic electrotransport selectively enhances the transport of highly electromobile molecules. Proceedings of the National Academy of Sciences. 112 (46), 6274-6283 (2015).
- Liebmann, T., et al. Three-dimensional study of Alzheimer’s disease hallmarks using the IDISCO clearing method. Cell Report. 16 (4), 1138-1152 (2016).
- Min, W., Freudiger, C. W., Lu, S., Xie, X. S. Coherent nonlinear optical imaging: Beyond fluorescence microscopy. Annual Review of Physical Chemistry. 62 (1), 507-530 (2011).
- Saar, B. G., et al. Video-rate molecular imaging in vivo with stimulated Raman scattering. Science. 330 (6009), 1368-1370 (2010).
- Cheng, J. -. X., Xie, X. S. Vibrational spectroscopic imaging of living systems: An emerging platform for biology and medicine. Science. 350 (6264), 1054-1063 (2015).
- Ji, M., et al. Rapid, label-free detection of brain tumors with stimulated Raman scattering microscopy. Science Translational Medicine. 5 (201), (2013).
- Wei, M., et al. Volumetric chemical imaging by clearing-enhanced stimulated Raman scattering microscopy. Proceedings of the National Academy of Sciences. 116 (14), 6608-6617 (2019).
- Ji, M., et al. Label-free imaging of amyloid plaques in Alzheimer’s disease with stimulated Raman scattering microscopy. Science Advances. 4 (11), 39 (2018).
- Silva, W. R., Graefe, C. T., Frontiera, R. R. Toward label-free super-resolution microscopy. ACS Photonics. 3 (1), 79-86 (2016).
- Gong, L., Zheng, W., Ma, Y., Huang, Z. Higher-order coherent anti-stokes Raman scattering microscopy realizes label-free super-resolution vibrational imaging. Nature Photonics. 14 (2), 115-122 (2020).
- Watanabe, K., et al. Structured line illumination Raman microscopy. Nature Communication. 6 (1), 10095 (2015).
- Qian, C., et al. Super-resolution label-free volumetric vibrational imaging. Nature Communications. 12 (1), 3648 (2021).
- Lin, L. -. E., Miao, K., Qian, C., Wei, L. High spatial-resolution imaging of label-free in vivo protein aggregates by VISTA. Analyst. 146 (13), 4135-4145 (2021).
- Mutlu, A. S., Chen, T., Deng, D., Wang, M. C. Label-free imaging of lipid storage dynamics in Caenorhabditis elegans using stimulated Raman scattering microscopy. Journal of Visualized Experiments. (171), e61870 (2021).
- Ounkomol, C., Seshamani, S., Maleckar, M. M., Collman, F., Johnson, G. R. Label-free prediction of three-dimensional fluorescence images from transmitted-light microscopy. Nature Methods. 15 (11), 917-920 (2018).
- Falk, T., et al. U-Net: Deep learning for cell counting, detection, and morphometry. Nature Methods. 16 (1), 67-70 (2019).
- Yang, B., et al. Single-cell phenotyping within transparent intact tissue through whole-body clearing. Cell. 158 (4), 945-958 (2014).
- Ku, T., et al. Multiplexed and scalable super-resolution imaging of three-dimensional protein localization in size-adjustable tissues. Nature Biotechnology. 34 (9), 973-981 (2016).
- Mlodzianoski, M. J., et al. Active PSF shaping and adaptive optics enable volumetric localization microscopy through brain sections. Nature Methods. 15 (8), 583-586 (2018).
- Querol-Vilaseca, M., et al. Nanoscale structure of amyloid-β plaques in Alzheimer’s disease. Scientific Reports. 9 (1), 5181 (2019).
- Oakley, H., et al. Intraneuronal beta-amyloid aggregates, neurodegeneration, and neuron loss in transgenic mice with five familial Alzheimer’s disease mutations: Potential factors in amyloid plaque formation. Journal of Neuroscience. 26 (40), 10129-10140 (2006).
- Bartels, T., De Schepper, S., Hong, S. Microglia modulate neurodegeneration in Alzheimer’s and Parkinson’s diseases. Science. 370, 66-69 (2020).
- Chen, F., et al. Nanoscale imaging of RNA with expansion microscopy. Nature Methods. 13 (8), 679-684 (2016).
- Tillberg, P. W., et al. Protein-retention expansion microscopy of cells and tissues labeled using standard fluorescent proteins and antibodies. Nature Biotechnology. 34 (9), 987-992 (2016).
- Renier, N., et al. IDISCO: A simple, rapid method to immunolabel large tissue samples for volume imaging. Cell. 159 (4), 896-910 (2014).
- Zhuge, M., et al. Ultrasensitive vibrational imaging of retinoids by visible preresonance stimulated Raman scattering microscopy. Advanced Science. 8 (9), 2003136 (2021).
- Miao, K., Wei, L. Live-cell imaging and quantification of PolyQ aggregates by stimulated Raman scattering of selective deuterium labeling. ACS Central Science. 6 (4), 478-486 (2020).
- Klimas, A., et al. Nanoscale imaging of biomolecules using molecule anchorable gel-enabled nanoscale in-situ fluorescence microscopy. Research Square. , (2021).
- Shi, L., et al. Super-resolution vibrational imaging using expansion stimulated Raman scattering microscopy. Advanced Science. , (2022).
- M’Saad, O., et al. All-optical visualization of specific molecules in the ultrastructural context of brain tissue. bioRxiv. , (2022).
- Zhao, Y., et al. Nanoscale imaging of clinical specimens using pathology-optimized expansion microscopy. Nature Biotechnology. 35 (8), 757-764 (2017).

.
