Multi-locus Variable-number Tandem-repeat Analysis of the Fish-pathogenic Bacterium Yersinia ruckeri by Multiplex PCR and Capillary Electrophoresis
概要
The Multi-Locus Variable-number tandem-repeat Analysis (MLVA) assay presented here enables inexpensive, robust and portable high-resolution genotyping of the fish-pathogenic bacterium Yersinia ruckeri. Starting from pure cultures, the assay employs multiplex PCR and capillary electrophoresis to produce ten-loci MLVA profiles for downstream applications.
Abstract
Yersinia ruckeri is an important pathogen of farmed salmonids worldwide, but simple tools suitable for epizootiological investigations (infection tracing, etc.) of this bacterium have been lacking. A Multi-Locus Variable-number tandem-repeat Analysis (MLVA) assay was therefore developed as an easily accessible and unambiguous tool for high-resolution genotyping of recovered isolates. For the MLVA assay presented here, DNA is extracted from cultured Y. ruckeri samples by boiling bacterial cells in water, followed by use of supernatant as template for PCR. Primer-pairs targeting ten Variable-number tandem-repeat (VNTR) loci, interspersed throughout the Y. ruckeri genome, are distributed equally amongst two five-plex PCR reactions running under identical cycling conditions. Forward primers are labelled with either of three fluorescent dyes. Following amplicon confirmation by gel electrophoresis, PCR products are diluted and subjected to capillary electrophoresis. From the resulting electropherogram profiles, peaks representing each of the VNTR loci are size-called and employed for calculating VNTR repeat counts in silico. Resulting ten-digit MLVA profiles are then used to generate Minimum spanning trees enabling epizootiological evaluation by cluster analysis. The highly portable output data, in the form of numerical MLVA profiles, can rapidly be compared across labs and placed in a spatiotemporal context. The entire procedure from cultured colony to epizootiological evaluation may be completed for up to 48 Y. ruckeri isolates within a single working day.
Introduction
Yersinia ruckeri, a Gram-negative bacterium and member of the Yersiniaceae family, causes yersiniosis in farmed salmonid fish worldwide1. It is readily diagnosed from infected fish by cultivation on many types of agar media, but until recently, little was known regarding the population structure and epizootiology of Y. ruckeri across the world and in different habitats (host species, etc.). Existing serotyping systems for Y. ruckeri are inconsistent, lack mutual compatibility and offer low epidemiological resolution. Some molecular studies on the bacterium have been conducted, employing techniques such as Multilocus sequence typing (MLST), Pulsed-field gel electrophoresis (PFGE) or whole-genome sequence (WGS) analysis2,3,4,5. However, MLST does not provide a sufficiently high resolution for routine infection tracing, while PFGE is labor demanding and produces results that are not readily portable across labs. While WGS analysis would provide a near ultimate resolution, the establishment and implementation of such analyses would prerequisite technical- and bioinformatics capabilities that yet remain restricted to a relatively small number of laboratories.
Multi-locus Variable-number tandem-repeat Analysis (MLVA) represents a simple and easily accessible molecular typing tool, which offers a genetic resolution in some cases almost matching that of WGS analysis6,7. The technique is based on repeat number variation in selected variable-number tandem-repeat (VNTR) loci, resulting in output data that is highly transportable, making comparison of profiled isolates towards online databases and across labs straightforward. Although MLST remains the gold standard for epidemiological typing of many bacterial pathogens, an increasing number of studies identify a significantly higher discriminatory power of MLVA8,9,10. Several protocols have also been published targeting fish-pathogenic bacteria, such as Francisella noatunensis, Edwardsiella piscicida and Renibacterium salmoninarum11,12,13.
The ten-loci MLVA protocol presented here, which recently formed the basis for an extensive Y. ruckeri population study14, involves extraction of DNA from agar-cultivated colonies, multiplex PCR and capillary electrophoresis (CE), followed by downstream in silico applications. For each examined isolate, two multiplex PCRs, both containing five fluorescently labelled primer pairs (6FAM, NED or VIC) each targeting individual VNTR regions, are run in parallel under identical conditions. Following verification of PCR amplicons by gel electrophoresis (GE), PCR products are diluted prior to CE analysis, and peaks representing the respective VNTR loci are size-called from the resulting electropherogram files. Together with locus-specific formulas accounting for minor, sequence-specific discrepancies in CE migratory patterns, VNTR CE size calls are then employed for calculating VNTR repeat counts which are concatenated into ten-digit MLVA profiles. These are used as input for epizootiological evaluations (e.g., by cluster analysis in Minimum spanning tree (MST) diagrams).
Protocol
CAUTION: For the entirety of the protocol, it is advisable to conduct all wet-lab procedures sterilely by use of lab coats, disposable gloves and sterile reagents and equipment. It is also advisable to prepare PCR reactions in a separate room (pre-PCR) not used for PCR amplification and/or handling of PCR products (post-PCR). Store all reagents as recommended by the manufacturer. See Table of Materials for further details on reagents, equipment and software used.
1. Bacterial Cultivation and Extraction of Genomic DNA
- Sow out Y. ruckeri pure cultures on any suitable agar type (the authors used 5% bovine blood agar) and incubate at 22 °C for 1-2 days, or 15 °C for 3-4 days.
- From each agar plate, pick a single representative colony with an inoculation loop and transfer to 1.5 mL centrifuge tubes containing 50 µL of ultrapurified water. Suspend, vortex briefly, and incubate for 7 min on a heating block at 100 °C.
- Centrifuge at 16 000 x g for 3 min and use a pipette to carefully transfer the supernatant into an empty 1.5 mL centrifuge tube. Proceed to next step using the supernatant as template DNA or store at -20 °C until such time.
2. Multiplex PCR setup and Cycling Conditions
NOTE: Each multiplex PCR reaction (two per Y. ruckeri isolate) should contain 12.5 µL of 2x Multiplex PCR Plus master mix, 0.1 to 0.2 µM of each appropriate primer pair (Table 1) and 3 µL of template DNA, adjusted to a final reaction volume of 25 µL by addition of RNase-free water. Aim to keep light exposure of the fluorescently labelled forward-primers at a minimum (e.g., by wrapping their storage tubes in aluminum foil).
- For each of the two multiplex PCR assays (Table 1), prepare master mixes as described above (without template DNA) according to the number of samples plus one positive and one negative control. Additionally, allow 10% surplus volume. Vortex the prepared master mixes gently at low speed.
- Distribute 22 µL of each master mix separately into individual wells on either PCR strips or plates, as appropriate for the number of samples, and add 3 µL of template to each well (for positive and negative controls, respectively, use DNA from a verified Y. ruckeri strain and ultrapurified water). Seal and centrifuge briefly.
- Run all samples on a PCR thermal cycler with the following programme: (i) 5 min at 95 °C (ii) 30 cycles of 0.5 min at 95 °C, 1.5 min at 60 °C, and 1 min at 72 °C, and (iii) 60 min at 68 °C, followed by cooling to 4 °C indefinitely. The program will complete in less than 3 h.
3. PCR Amplicon Confirmation by Gel Electrophoresis
- According to the manufacturer’s recommendations, prepare a volume of 1.5% (w/v) agarose gel in 1x tris-borate-EDTA (TBE) buffer appropriate for the number of PCR reactions to be tested. Prior to casting, add 5 µL of fluorescent nucleic acid dye per 50 µL of gel solution and mix. Use trays and combs as appropriate for casting, leaving an appropriate number of wells free for DNA reference ladders.
- After setting, submerge the gel in 1x TBE-buffer in a GE system. Mix 5 µL of PCR product together with 2 µL of loading dye and transfer to gel wells. Add 5 µL of DNA ladder in empty wells for reference.
- Run the gel at 110 V per 15 cm for approximately 1 h and use a UV-based gel imaging/visualisation system to verify the presence of multiple (up to five) bands representing PCR amplicons (see example in Figure 1). Discard the gel. Proceed to the next step or store remaining PCR products at 4 °C until further processing.
4. Capillary Electrophoresis Setup and Run Conditions
- Following confirmation of PCR amplicons, dilute PCR products 1:10 (v/v) in purified water. Seal, mix and centrifuge briefly.
- Working in a fume cupboard, prepare a volume of master mix consisting of 9 µL of formamide and 0.5 µL of size standard per PCR product (allow 10% surplus volume). Vortex briefly and distribute 9.5 µL into wells on a plate appropriate for the available CE system, before adding 0.5 µL of diluted PCR product. Seal, mix and centrifuge briefly.
CAUTION: Handle with care. Mixing formamide with water generates formic acid, which is toxic. - Using a PCR thermal cycler, denature the samples at 95 °C for 3 min before cooling to 4 °C indefinitely. Centrifuge briefly and load the plate onto a calibrated CE system according to the manufacturer’s instructions.
- Run fragment analysis CE using reagents as appropriate for the apparatus of choice and the following settings: 60 °C; 5 s injections at 1.6 kV (32 V per cm); 32 min run time at 15 kV (300 V per cm). CE fragment analysis of 24 wells on a 24-capillary (50 cm) will typically take approximately 50 min.
5. VNTR Size Calling, Repeat Count Calculation and MLVA Profiling
NOTE: Step 5.1 describes Y. ruckeri VNTR CE size calling from electropherogram files, using the specific software listed in Table of Materials. Consult the software manual for additional details and troubleshooting. For use of other software, consult appropriate manuals.
- Import CE result files (two per Y. ruckeri isolate). Set Analysis Method to Microsatellite Default and select the appropriate product choice under Size Standard, prior to pressing the Analyze button. Verify the correct identification of size standard fragments through the Size Match Editor and rectify any visibly erroneous allocations.
- Having selected the sample(s) to be read, hit the Display Plots button and press Ctrl+A to enable view of the Sizing Table. While in the top panel, hold down Ctrl while clicking on the five peaks representing the VNTR amplicons (use zooming tool as needed).
NOTE: For each of the multiplex PCR products, the electropherogram will show five peaks distributed amongst the three dyes employed (see 5' dye labelling of forward primers in Table 1 and the two examples in Figure 2). - Press Ctrl+G to filter the Sizing Table, showing only characteristics of the five highlighted peaks, and record CE size calls for each VNTR locus (with reference to Table 1) for downstream application.
- Having selected the sample(s) to be read, hit the Display Plots button and press Ctrl+A to enable view of the Sizing Table. While in the top panel, hold down Ctrl while clicking on the five peaks representing the VNTR amplicons (use zooming tool as needed).
- In order to account for biased amplicon mobility patterns during CE, calculate accurate VNTR repeat counts according to the formula provided below, employing VNTR CE size calls together with locus-specific variables (see Table 1). For efficiency, it is advisable to automate this process (e.g., by using a spreadsheet template).
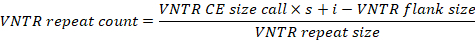
- Round calculated VNTR repeat counts off to the nearest integer and concatenate into ten-digit strings, each representing the MLVA profile of a single Y. ruckeri isolate.
6. Minimum Spanning Tree Cluster Analysis of MLVA Data
NOTE: Step 6 describes the creation of MST diagrams from Y. ruckeri MLVA data, using the specific software listed in Table of Materials. Consult the software manual for additional details and troubleshooting. For use of other software, consult appropriate manuals.
- Create a new database and opt to activate the MLVA plugin.
- Import Y. ruckeri MLVA profiles and metadata by selecting Character type data followed by Import fields and characters (further sub-selection depending on storage format). When prompted, specify import rules according to the content of the import file: In the Destination type column, classify VNTR repeat counts as Character value: VNTR, and the miscellaneous metadata as Entry information: Entry info field.
NOTE: For comparison and context, it is also possible to import the entire dataset published (open access) together with the original paper employing the present MLVA protocol14. MLVA profiles and metadata on the diverse collection of Y. ruckeri isolates (n = 484) scrutinised in that study is available from its supplemental material (Tables S1 and S2) through the following link: https://aem.asm.org/content/84/16/e00730-18/figures-only#fig-data-additional-files - In the Experiment type panel, open the VNTR entry and set minimum and maximum values for each VNTR locus to 0 and 100, respectively. Under General settings, set the number of decimal digits to 0 and select Numbers under Data type. Check to consider absent values as zero.
- Select imported samples destined for MST cluster analysis and click the Create new comparison button (in Comparison panel).
- If desired for the visual presentation of the MST, allocate the samples to colored groups (e.g., according to a particular metadata trait) by employing the various options available in the Groups panel.
NOTE: Groups can also be created/altered retrospectively, subsequent to the following steps. - Select Advanced cluster analysis… and MST for categorical data to generate an MST diagram based on the chosen samples.
- Further modify the visual presentation of the MST as preferred (e.g., by adding partitioning parameters, node/branch labelling, crosslinks, legends, etc). See example in Figure 3.
NOTE: A cluster (clonal complex) partitioning threshold of ≤4/10 non-identical VNTR loci, in addition to hiding of branch connections representing >5/10 non-identical VNTR loci, has previously been employed for MST cluster analysis based on MLVA data generated using this protocol14. Provided the aforementioned dataset of 484 Y. ruckeri MLVA profiles was imported, those samples can also be included for MST cluster analysis (as described above) to provide a global and historical context. This will e.g. facilitate identification of any samples affiliated with previously described clonal complexes, as well as those representing yet undescribed lineages. Depending on available metadata, the resulting MST diagram can be scrutinised in different ways, e.g. to discover eventual clustering patterns linked to particular traits (geography, host, time etc.). - If needed, export the finalized MST in a desired format using the Export image selection.
- If desired for the visual presentation of the MST, allocate the samples to colored groups (e.g., according to a particular metadata trait) by employing the various options available in the Groups panel.
Representative Results
Following multiplex PCR as described here, a typical GE image verifying the presence of multiple amplicons from each PCR reaction is shown in Figure 1. Downstream CE fragment analysis performed on verified PCR products will, for each Y. ruckeri isolate examined, result in two electropherogram files used for size calling of the respective VNTR loci (Figure 2). From analysis of 484 diverse Y. ruckeri isolates, no overlap in amplicon size range was observed between VNTR loci labelled with the same dye in the same multiplex reaction (Table 1)14. Each of the electrophoretic peaks can, therefore, be unambiguously identified by color.
Following import of MLVA profiles and relevant metadata into the preferred software, MST diagrams can be constructed as described for scrutiny of any epidemiological patterns of interest in the material. Consult appropriate manuals for additional options available in the respective software. As an example, Figure 3 shows comparison by MST of MLVA profiles for Y. ruckeri isolates recovered from fish associated with five different salmon farms in Norway.
The consistent repeat sizes of the ten VNTR loci, as well as their in vitro and in vivo stability, have previously been verified in the original study based upon this protocol14. Briefly, this was done using Sanger sequencing (repeat size), and by MLVA typing of multiple isolates following serial passages (in vitro) and from within individual disease outbreaks (in vivo). Moreover, the environmental stability of the loci over time was examined by typing multiple 'house strain' isolates recovered over several years from persistently infected freshwater production sites for Atlantic salmon.
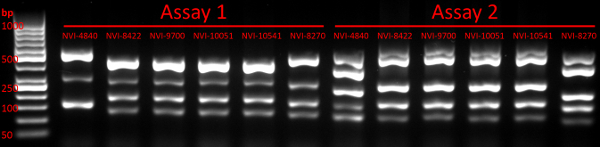
Figure 1: Gel electrophoresis verifying the presence of multiple PCR products. The image confirms the presence of multiple PCR amplicons in all 12 lanes containing samples, with the first lane representing the DNA ladder used. The sizes of selected ladder fragments have been indicated, as have the PCR assay and strain (see Table S1 in Gulla et al. 201814) affiliation of each lane. Please click here to view a larger version of this figure.
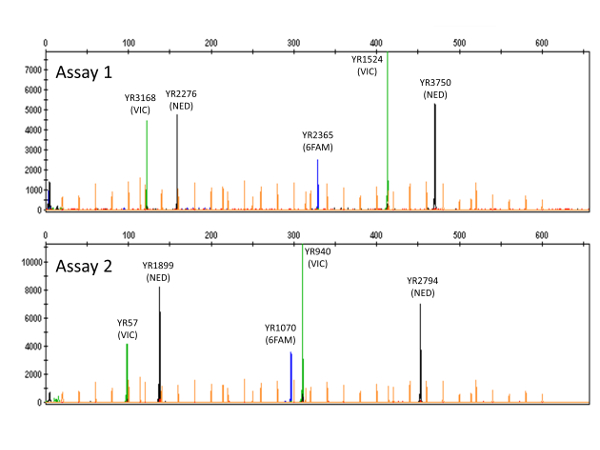
Figure 2: Electropherograms showing peaks corresponding to VNTR amplicons. Names of the different VNTR loci are indicated, with dye labels (VIC = green; NED = black; 6FAM = blue) in parentheses. The two electropherograms (PCR assay 1 top; PCR assay 2 bottom) originate from typing of a single Y. ruckeri isolate. Orange peaks (dye LIZ) represent the size standard employed. Please click here to view a larger version of this figure.
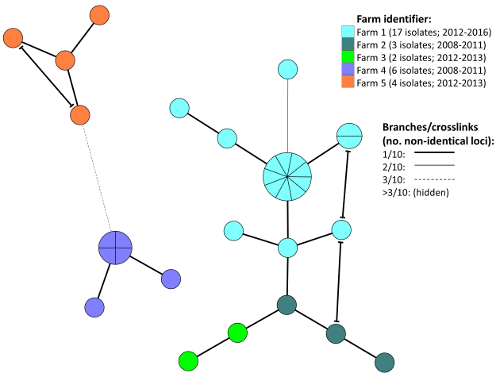
Figure 3: Example Minimum spanning tree for epidemiological evaluation. The diagram is based on MLVA profiles from Y. ruckeri isolates recovered from Atlantic salmon in five different Norwegian farms (1-5; see legend) experiencing recurrent yerisniosis outbreaks. A clear clustering tendency linked to farm origin can be observed. Crosslinks show all possible connections involving ≤1/10 non-identical VNTR loci (see legend). Please click here to view a larger version of this figure.
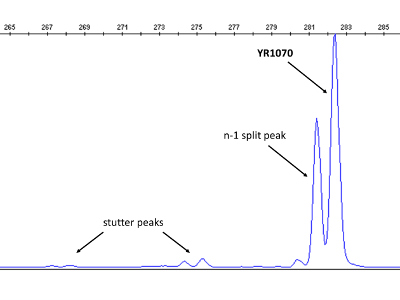
Figure 4: Electropherogram visualizing stutter and split peaks. In this case, both occur simultaneously, which is not always the case. The longer and taller peak, representing the YR1070 VNTR locus, can be readily distinguished. The display is magnified and shows only blue dye peaks. Please click here to view a larger version of this figure.
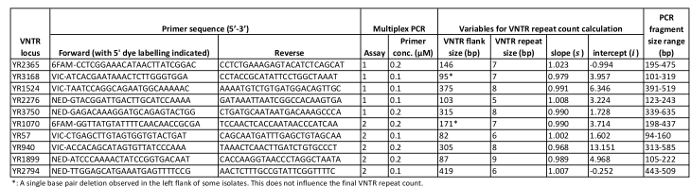
Table 1: VNTR locus characteristics. Relevant characteristics of the ten Y. ruckeri VNTR regions targeted in the present MLVA protocol.
Discussion
Both multiplex PCRs presented here have appeared relatively robust in the face of poor template DNA quality, but lack of PCR amplification was nevertheless occasionally observed when using templates with extremely high DNA concentrations. These issues were readily resolved by diluting the templates prior to PCR. Other methods for DNA extraction than the one employed here may also be used (e.g., commercial kits).
Although five amplicons are expected from each multiplex PCR reaction, five visually distinguishable bands should not always be expected from GE, as some (differently labelled) VNTR loci within the same reaction have overlapping size ranges. The final PCR extension time of 60 min may be shortened if required, but will likely result in the increased occurrence of split peaks in subsequent CE electropherograms (see below). Notably, as the purpose of the GE step is purely for qualitative verification of PCR amplicons, the run time, voltage and/or gel recipe may be adjusted as preferred. If particularly weak bands are observed by GE, it may be advisable to reduce the dilution factor of those samples prior to CE.
While the CE protocol described here was run on a specific commercial capillary electrophoresis apparatus (see Table of Materials), different CE systems may have different sample requirements, which may in turn prompt some modifications to the protocol. Refer to the manual of the respective CE system manufacturer for instructions on appropriate reagents/equipment, calibration etc. for fragment analysis. There is also a possibility that the biased amplicon mobility patterns observed during CE may differ, relatively, across CE systems and/or machines, as has previously been documented for other MLVA protocols15,16. If occurring to an extent where final (rounded) VNTR repeat counts become affected, this means the locus-specific variables s and i (Table 1), used to determine VNTR repeat counts, must be re-calibrated. This involves linear regression on plots comparing accurate sequence sizes versus CE size calls, as described by Gulla et al. 201814.
Split peaks and stutter peaks, both well-known artefacts in CE based MLVA typing17, may be observed in electropherograms during size calling (Figure 4). While stutter peaks should be disregarded, the longer peak should consistently be selected for downstream applications in the case of split peaks separated by a single base pair. Moreover, absent peaks indicating lack of particular VNTR loci are rare, but may occur, in which case a repeat count of '0' should be assigned. If the starting culture from which DNA is extracted is not pure (i.e., contains more than one Y. ruckeri sub-type), multiple tall peaks corresponding to different alleles of the same locus/loci may be observed following CE. Secondary cultivations must then be performed from single colonies prior to new DNA extraction for re-typing.
As stated in the protocol, template DNA for PCR should by default be extracted from pure cultures of Y. ruckeri. In a few cases, however, egg-fluid samples testing positive for Y. ruckeri by qPCR (Ct-values < 27) were successfully MLVA typed directly, without prior culturing, using an increased amount of genomic DNA (extracted with commercial kit) as template. Although this approach has not been extensively tested nor verified, it does indicate the potential of this MLVA assay for examination of complex biological matrices containing DNA from a range of different organisms in addition to Y. ruckeri.
The entire MLVA typing procedure presented here, from DNA extraction to epizootiological evaluation, may be completed in a single working day. However, the number of samples examined is in a sublinear relationship with the time required for DNA extraction, PCR and CE, and the method is therefore much more time efficient when running multiple samples simultaneously. This is nevertheless the case for most lab-based methods, and as a tool for epidemiological subtyping of Y. ruckeri, the combination of high resolution, simplicity and portability makes this MLVA assay superior to previously published protocols4,5. It has also been used to verify the limited epidemiological relevance of Y. ruckeri serotyping14.
Through a comprehensive MLVA based population study involving 484 Y. ruckeri isolates recovered from a range of spatiotemporal origins and habitats (host fish, environment, etc.), our understanding regarding the epizootiology and population structure of this important fish pathogen was substantially increased14. MLVA typing enabled the tracing of clones disseminated anthropogenically over decades, presumably through transport of fish, as well as identification of locally confined strains. Moreover, while some clonal complexes of the bacterium could clearly be associated with disease in particular fish hosts (rainbow trout and Atlantic salmon, respectively), others were only recovered from environmental sources and/or clinically unaffected fish specimens. The applicability of the method is thus not only limited to infection tracing, as it may also provide information of potential relevance e.g. for vaccine development, risk assessment, and maintenance of national biosecurity. It is currently in active use at the Norwegian Veterinary Institute as a tool for investigating Y. ruckeri diagnoses in Norwegian aquaculture.
開示
The authors have nothing to disclose.
Acknowledgements
The present study was funded by The Norwegian Seafood Research Fund, FHF (project nos. 901119 and 901505). We wish to thank all contributors of bacterial isolates and samples used during method development.
Materials
| 22°C/15°C incubator | As preferred. | NA | |
| 5' Labeled Primer, 10K PMOL, Desalted, Dry | Thermo Fisher Scientific | 450007 | Sequences and labelling in Table 1. Prepare working aliquouts in TE-buffer. |
| Agarose, universal, peqGOLD | VWR | 732-2789P/732-2788 | Used during gel electrophoresis. |
| Avant 3500xL Genetic Analyzer | Thermo Fisher Scientific | A30469 | Used for capillary electrophoresis fragment analysis. |
| BioNumerics7 modules | Applied Maths | NA | Used for generating Minimum spanning trees from MLVA data. |
| Centrifuge(s) | As preferred. | NA | |
| Custom DNA Oligo, 25N, Desalted, Dry | Thermo Fisher Scientific | A15612 | Sequences in Table 1. Prepare working aliquouts in TE-buffer. |
| DNA Gel Loading Dye (6X) | Thermo Fisher Scientific | R0611 | Loading dye used during gel electrophoresis. |
| Eppendorf Safe-Lock Tubes, 1.5 mL | Eppendorf | 30120086 | Centrifuge tubes used during DNA extraction. |
| Freezer | As preferred. | NA | |
| Fume cupboard | As preferred. | NA | |
| Gel electrophoresis system | As preferred. | NA | |
| GelRed Nucleic Acid Stain, 10,000X in water | Biotium | 41003 | Fluorescent nucleic acid dye used during gel electrophoresis. |
| GeneMapper Software 5 | Thermo Fisher Scientific | 4475073 | Used for reading electropherograms from capillary electrophoresis. |
| GeneRuler 50 bp DNA Ladder, ready-to-use | Thermo Fisher Scientific | SM0373 | DNA ladder used during gel electrophoresis. |
| GeneScan 600 LIZ dye Size Standard v2.0 | Thermo Fisher Scientific | 4408399 | Size standard used during capillary electrophoresis. |
| Heating block | As preferred. | NA | |
| Hi-Di Formamide | Thermo Fisher Scientific | 4311320/4440753 | Deionized formamide used during capillary electrophoresis. Prepare working aliquouts. |
| Milli-Q water | NA | NA | Purified water used during PCR and capillary electrophoresis. Standard recipe; produced in-house. |
| Multiplex PCR Plus Kit | Qiagen | 206151/206152 | |
| PCR thermal cycler | As preferred. | NA | |
| POP-7 Polymer for 3500 Dx/3500xL Dx Genetic Analyzers | Thermo Fisher Scientific | A26077/4393713/4393709 | Separation matrix used during capillary electrophoresis. |
| Pure culture Yersinia ruckeri | NA | NA | E.g. cryopreserved or fresh. |
| RNase-free water | Qiagen | NA | In: Multiplex PCR Plus Kit |
| Tris-borate-EDTA (TBE) buffer | NA | NA | Standard recipe; produced in-house. |
| Trypsine soy agar/bovine blood agar | NA | NA | Standard recipe; produced in-house. |
| UV-based gel imaging/visualisation system | As preferred. | NA | |
| Vortexer | As preferred. | NA |
参考文献
- Barnes, A. C. Enteric redmouth disease (ERM) (Yersinia ruckeri). Fish Diseases and Disorders, Vol 3: Viral, Bacterial and Fungal Infections. 3, 484-511 (2011).
- Barnes, A. C., et al. Whole genome analysis of Yersinia ruckeri isolated over 27 years in Australia and New Zealand reveals geographical endemism over multiple lineages and recent evolution under host selection. Microbial Genomics. 2 (11), 000095 (2016).
- Calvez, S., Mangion, C., Douet, D. G., Daniel, P. Pulsed-field gel electrophoresis and multi locus sequence typing for characterizing genotype variability of Yersinia ruckeri isolated from farmed fish in France. Veterinary Research. 46, 73 (2015).
- Bastardo, A., Ravelo, C., Romalde, J. L. Multilocus sequence typing reveals high genetic diversity and epidemic population structure for the fish pathogen Yersinia ruckeri. Environmental Microbiology. 14 (8), 1888-1897 (2012).
- Wheeler, R. W., et al. Yersinia ruckeri biotype 2 isolates from mainland Europe and the UK likely represent different clonal groups. Diseases of Aquatic Organisms. 84 (1), 25-33 (2009).
- Eyre, D. W., et al. Comparison of multilocus variable-number tandem-repeat analysis and whole-genome sequencing for investigation of Clostridium difficile transmission. Journal of Clinical Microbiology. 51 (12), 4141-4149 (2013).
- Sun, M., et al. Multiple Locus Variable-Number Tandem-Repeat and Single-Nucleotide Polymorphism-Based Brucella Typing Reveals Multiple Lineages in Brucella melitensis Currently Endemic in China. Frontiers in Veterinary Science. 4, (2017).
- Bouchouicha, R., et al. Comparison of the performances of MLVA vs. the main other typing techniques for Bartonella henselae. Clinical Microbiology and Infection. 15, 104-105 (2009).
- Dahyot, S., et al. Multiple-Locus Variable Number Tandem Repeat Analysis (MLVA) and Tandem Repeat Sequence Typing (TRST), helpful tools for subtyping Staphylococcus lugdunensis. Scientific Reports. 8 (1), (2018).
- Elberse, K. E. M., Nunes, S., Sá-Leão, R., van der Heide, H. G. J., Schouls, L. M. Multiple-locus variable number tandem repeat analysis for Streptococcus pneumoniae: Comparison with PFGE and MLST. PLoS ONE. 6 (5), (2011).
- Matejusova, I., et al. Multilocus variable-number tandem-repeat genotyping of Renibacterium salmoninarum, a bacterium causing bacterial kidney disease in salmonid fish. BMC microbiology. 13, 285 (2013).
- Duodu, S., et al. An improved multiple-locus variable-number of tandem repeat analysis (MLVA) for the fish pathogen Francisella noatunensis using capillary electrophoresis. BMC Veterinary Research. 9, 252 (2013).
- Abayneh, T., Colquhoun, D. J., Austin, D., Sørum, H. Multilocus variable number tandem repeat analysis of Edwardsiella piscicida isolates pathogenic to fish. Journal of Fish Diseases. 37 (11), 941-948 (2014).
- Gulla, S., et al. Multilocus variable-number tandem-repeat analysis of Yersinia ruckeri confirms the existence of host specificity, geographic endemism, and anthropogenic dissemination of virulent clones. Applied and Environmental Microbiology. 84 (16), (2018).
- Pasqualotto, A. C., Denning, D. W., Anderson, M. J. A cautionary tale: Lack of consistency in allele sizes between two laboratories for a published multilocus microsatellite typing system. Journal of Clinical Microbiology. 45 (2), 522-528 (2007).
- Lista, F., et al. Genotyping of Bacillus anthracis strains based on automated capillary 25-loci multiple locus variable-number tandem repeats analysis. BMC Microbiology. 6, 33 (2006).
- . Thermo Fisher Scientific DNA Fragment Analysis by Capillary Electrophoresis Available from: https://www.thermofisher.com/content/dam/LifeTech/global/Forms/PDF/fragment-analysis-chemistry-guide.pdf (2014)

