Differentiation, Maintenance, and Analysis of Human Retinal Pigment Epithelium Cells: A Disease-in-a-dish Model for BEST1 Mutations
概要
Here we present a protocol to differentiate retinal pigment epithelium (RPE) cells from human pluripotent stem cells bearing patient-derived mutations. The mutant cell lines may be used for functional analyses including immunoblotting, immunofluorescence, and patch clamp. This disease-in-a-dish approach circumvents the difficulty of obtaining native human RPE cells.
Abstract
Although over 200 genetic mutations in the human BEST1 gene have been identified and linked to retinal degenerative diseases, the pathological mechanisms remain elusive mainly due to the lack of a good in vivo model for studying BEST1 and its mutations under physiological conditions. BEST1 encodes an ion channel, namely BESTROPHIN1 (BEST1), which functions in retinal pigment epithelium (RPE); however, the extremely limited accessibility to native human RPE cells represents a major challenge for scientific research. This protocol describes how to generate human RPEs bearing BEST1 disease-causing mutations by induced differentiation from human pluripotent stem cells (hPSCs). As hPSCs are self-renewable, this approach allows researchers to have a steady source of hPSC-RPEs for various experimental analyses, such as immunoblotting, immunofluorescence, and patch clamp, and thus provides a very powerful disease-in-a-dish model for BEST1-associated retinal conditions. Notably, this strategy can be applied to study RPE (patho)physiology and other genes of interest natively expressed in RPE.
Introduction
It has been documented that at least five retinal degenerative diseases are caused by genetic mutations in the BEST1 gene1,2,3,4,5,6,7,8, with the number of reported mutations already over 200 and still increasing. These BEST1-associated diseases, also known as bestrophinopathies, cause progressive vision loss and even blindness, and there are currently no effective treatments. The protein product of BEST1, namely BESTROPHIN1 (BEST1), is a Ca2+-activated Cl– channel (CaCC) specifically expressed in the retinal pigment epithelium (RPE) of the eyes5,6,8,9. Importantly, a clinical phenotype of BEST1-associated diseases is the reduced visual response to light stimuli, called light peak (LP) measured in the electrooculogram10,11; LP is believed to be mediated by a CaCC in RPE12,13,14. In order to better comprehend the pathological mechanisms of BEST1 mutations and to work towards potential therapies, it is essential to study mutant BEST1 channels endogenously expressed in human RPE cells.
However, obtaining RPE cells directly from live patients is highly impractical. Although native RPE cells can be harvested from biopsies of human cadavers and fetuses, the difficult accessibility to these sources significantly limits scientific research. Therefore, it is critical to have alternative RPE sources other than human eyes. This call has been answered by recent advancements in stem cell techniques, as functional RPE cells can now be differentiated from human pluripotent stem cells (hPSCs), including embryonic stem cells (hESCs) and induced pluripotent stem cells (hiPSCs), the latter being generated by reprogramming primary skin fibroblasts from donors16,17,18. Importantly, the self-renewal and pluripotency of hPSCs ensure a reliable source to generate RPEs, while the patient-specificity of hiPSCs and genomic modification potential of hESCs (e.g., by CRISPR) offer a versatile disease-in-a-dish model for desired BEST1 mutations.
hPSC-RPE has several advantages over mice RPE models: 1) BEST1 knockout mice do not display any retinal abnormality19, raising the possibility of different genetic requirement of BEST1 in RPE between mice and humans; 2) only 3% of human RPE cells are binucleate, in contrast to 35% in mice20; 3) hiPSC-RPE potentiates autologous transplantation in clinical treatment of retinal disorders21. Nevertheless, animal models are still indispensable for studying RPE physiology and pathology in a live system, and the oncogenic potential of hiPSC cannot be overlooked.
The procedure here describes a useful and moderately simple hPSC to RPE differentiation protocol that can be used for research and clinical purposes. This protocol uses nicotinamide (vitamin B3) to augment differentiation of hPSCs to neural tissue, which is further induced to differentiate into RPE by treatment with activin-A. Nicotinamide treatment has been shown to increase the number of pigmented cells (a sign of differentiation into RPE), possibly by attenuating the apoptotic activity of differentiating cells22. The resulting hPSC-RPE cells display the same key markers, cobblestone morphology, and cellular functionality as native human RPE cells22. Thus, in a research setting, the resulting hPSC-RPE cells are suitable for downstream functional analyses including immunoblotting, immunostaining, and whole-cell patch clamp, for which detailed experimental procedures are also provided. Clinically, RPE cells derived from stem cells have shown great potential for transplantation treatment of macular degeneration in both animal studies and human trials23.
Protocol
1. Differentiation of hPSC to RPE
- Maintain and passage hPSCs as previously described18.
NOTE: All cells (including hPSCs and hPSC-RPEs) are grown in 37 °C at 5% CO2 throughout the duration of the growth and differentiation protocols. - Prior to differentiation, split confluent hPSCs to pre-coated 6-well plates.
- To coat plates, thaw basement membrane matrix on ice for about 1 h, suspend liquidized matrix in 4 °C DMEM medium at a 1:50 dilution, add 800 µL coating mixture to each well, and incubate for at least 1 h at 37 °C.
- When coating is complete, aspirate the mixture and immediately add 1.5–2 mL medium into the wells.
- To lift hPSCs from the old culture plates, incubate cells with PBS plus 0.5 mM EDTA for 5 min at room temperature, aspirate the solution, and resuspend cells in small clumps with culture medium.
- Seed cells at ~20% confluency in the new plates.
NOTE: Store basement membrane matrix in 100 µL aliquots at -20 °C and use one aliquot to coat a 6-well plate. The time for coating can be extended up to 4 h at 37 °C, or overnight at room temperature. It is important to suspend and seed hPSCs as small clumps of 3–5 cells rather than single cells.
- When cells are grown to full confluency, replace the culture medium with differentiation medium (4 mL/well) (Table 1, without activin-A). Culture for 14 days, changing the medium (4 mL/well) 3x/week.
NOTE: The hPSC plate density doubles approximately every 24 h. Significant cell death is expected after the start of the differentiation procedure. Aggregates of dead cells and debris in the wells can be seen by the naked eye during medium changes. - From day 15 to day 28, supplement differentiation medium with 100 ng/mL human activin-A (Table 1, with human activin-A). Change the medium (4 mL/well) 3x/week.
- Stop activin-A supplementation starting on day 29 (Table 1, without human activin-A). Continue cell culturing in differentiation medium for 8–10 weeks until pigmented hPSC-RPE clusters appear (Figure 1A-B).
NOTE: Clusters of pigmented cells can be seen as small dark dots on a cell culture plate by the naked eye (Figure 1A-B, top). Individual pigmented cells with the signature cobblestone shape can be seen under a 20X microscope (Figure 1A-B, bottom).
2. Isolation and Culture of Differentiated RPE Cells
- Remove the differentiation medium, wash once with PBS, add 1 mL of 0.05% trypsin plus 1 U/µL of collagenase into each well, and incubate at 37 °C for 20–30 min. Remove the trypsin/collagenase and gently add 2 mL of pre-warmed RPE medium into each well of the 6-well plate (Table 2).
NOTE: Use an inverted microscope to monitor the morphology of the hPSC-RPE cells, which are polygonal before trypsin/collagenase treatment, and become round after sufficient digestion. - Use a Bunsen burner to pull glass Pasteur pipettes.
- With both hands, hold a long (9 inch) Pasteur pipette at both ends so it is horizontal, and above the Bunsen burner about 2/3 down the length of the thin part of the pipette.
- Once the glass becomes soft from the flame, quickly pull the two ends apart, such that a micro scraper-like tip is formed at the new end of the pipette (Figure 2).
- Repeat several times to create multiple "cell scrapers". Spray the pulled pipettes with 70% ethanol for sterilization and air-dry them in the cell culture hood.
- Under a microscope, use a micro "cell scraper" to gently dissociate pigmented hPSC-RPE clusters from the plate.
- Gently tap (do not scrape) directly on the pigmented cells, which will float up into the culture medium once they dissociate from the bottom of the well.
- Immediately use a 20 µL micropipette to gently suction the dissociated hPSC-RPE cells and transfer them to a pre-coated 6-well plate with RPE medium24.
- Collect ~5 x 103 cells (~10% confluency as observed under a 20X microscope) in each pre-coated well of a 12-well plate by repeating steps 2.3.1–2.3.2 multiple times, as needed. Bring the final volume of medium to 2 mL.
NOTE: To prevent dissociated hPSC-RPE cells from floating away, avoid movement of the plates during the dissociation process such that the dissociated cells are floating in the medium but still remain locally concentrated.
- Culture the newly isolated hPSC-RPE cells (P0) on 12-well plates in RPE medium for another 6-8 weeks to allow them to form a pigmented monolayer (Figure 1C).
- Change the medium 3x/week. When changing the medium, leave approximately 1/3 volume of the old medium in each well, and add 2/3 volume of fresh, pre-warmed RPE medium. At this stage, use 2 mL of RPE medium for each well in a 12-well plate (thus, exchange 1.3 mL of medium each time).
NOTE: The monolayer of P0 hPSC-RPE cells may form domes, suggesting that the cells are actively transporting fluids, and are anchored by tight junctions25. Therefore, dome formation is an indicator of mature RPE status.
3. RPE Fate Validation by Immunoblotting
- Make the hPSC-RPE cell lysate using mammalian protein extraction buffer supplemented with proteinase inhibitor cocktail. Incubate the cell lysate on ice for 30 min before centrifugation at 13,000 x g for 15 min at 4 °C to discard undissolved cell debris. Measure the total protein concentration of lysate.
- Denature 40 µg of total protein in Laemmli SDS sample buffer at 75 °C for 10 min. Separate the protein samples on a 10% Tris-glycine SDS-PAGE gel at a constant voltage of 90 V for 15 min and then 150 V until the dye front reaches the bottom of the gel.
- Transfer the proteins onto a nitrocellulose membrane in pre-cooled Tris-glycine buffer supplemented with 10% methanol at 100 V for 1 h.
- Block the membrane in Tris buffered saline with 0.1% non-ionic detergent (TBST) and 5% non-fat dry milk for 1 h at room temperature.
- Incubate the membrane with primary antibodies diluted in the blocking buffer at 4 °C overnight. Dilute the antibodies targeting RPE specific marker proteins as follows: RPE65, 1:1,000; CRALBP, 1:1,000; BEST1, 1:1,000. Dilute the antibody against β-actin at 1:10,000 to detect β-actin as a loading control.
- Wash the membrane with TBST 3 times, with 5 min for each wash.
- Incubate the membrane with fluorophore conjugated goat anti-mouse IgG secondary antibody diluted 1:10,000 in blocking buffer for 1 h at room temperature.
- Wash the membrane with TBST 4 times, with 10 min for each wash.
- Detect the proteins using an infrared imaging system (Figure 3).
4. Checking BEST1 Subcellular Localization by Immunostaining
- Wash hPSC-RPE cells with 2 mL of PBS once and fix in 2 mL of 4% paraformaldehyde for 45 min at room temperature.
- Wash with 2 mL of PBS twice, and incubate the cells in 2 mL of PBS with 0.1% non-ionic surfactant and 2% donkey serum for 45 min to block the non-specific binding sites.
- Incubate the cells with BEST1 antibody (1:200 dilution) for 2 h at room temperature.
- Wash the cells with 2 mL of PBS 3 times. Incubate with fluorophore conjugated secondary IgG (1:1,000) for 1 h at room temperature.
- Wash with 2 mL of PBS 3 times to remove unbound secondary antibody.
- Observe stained cells by confocal microscopy (Figure 4).
5. Recording Ca2+-dependent Cl– Current in hPSC-RPE by Whole-cell Patch Clamp
- 24–72 h before the patch clamp, split a fully confluent well (on a 12-well plate) of mature P0 hPSC-RPE cells (cobblestone-shaped and pigmented). Aspirate the medium, wash once with PBS, and add 1 mL of 0.05% trypsin plus 1 U/µL collagenase into the well.
NOTE: Pre-warm the trypsin/collagenase to 37 °C right before use. - Incubate at 37 °C for 8 min.
NOTE: After this step, the hPSC-RPE cells are still adherent and connected. - Use a 1 mL micropipette to gently wash the cells off the plate, and transfer cells in the digestion mixture to a 15 mL conical tube. Wash the well with 1 mL of pre-warmed trypsin/collagenase to collect the residual cells and combine them into the 15 mL conical tube.
NOTE: The hPSC-RPE cells are still in large clumps at this point. Do not try to break the cell clumps by vigorous pipetting. Some clumps may stick to the inner wall of the 1 mL tip. - Incubate the conical tube in a 37 °C dry bath for 8 min.
- Use a 1 mL micropipette to gently pipette up-and-down 10–15 times the digestion mix.
NOTE: Cell clumps become much smaller but still visible. Avoid vigorous pipetting. - Repeat the previous two steps.
NOTE: Most of the cells should be in single-cell suspension after pipetting. There may still be some tiny cell clumps, which are acceptable. Optional: Incubate the conical tube at 37 °C for an additional 5 min followed by gentle pipetting. The total time in the trypsin/collagenase at 37 °C should not exceed 30 min (8 + 8 + 8 + 5 = 29). - Add 5 mL of pre-warmed RPE medium to the 15 mL conical tube containing the digested cells. Spin at 200 x g for 5 min to collect the cells. Re-suspend and seed cells on pre-coated 35 mm dishes at 10–20% confluency (e.g., a 100% confluent well on a 12-well plate to five 35 mm dishes for 10% confluency).
NOTE: At all times, avoid vigorous pipetting, which may cause significant cell death as represented by apparent dark cell debris in the supernatant after centrifugation. Well-separated single-cell seeding is essential for patch clamp recording. - Perform the whole-cell patch clamp as described previously18,26,27,28,29,30,31. Acquire current traces from a family of step potentials (-100 to +100 mV from a holding potential of 0 mV) (Figure 5).
NOTE: The recipes of internal and external solutions are described in Table 3 and Table 4, respectively.
Representative Results
The most technically challenging step is the manual isolation, which aims to achieve a high purity of a differentiated P0 hPSC-RPE population. After a successful isolation, >90% cells in the P0 population will grow and mature to display signature RPE morphologies (Figure 1C). The existence of a minor portion of non-RPE or immature RPE cells in the P0 population is almost inevitable, but will not interfere with the downstream experiments as long as the number of pigmented and cobblestone-shaped hPSC-RPE cells is the overwhelming majority.
With the hPSC-RPE based disease-in-a-dish approach, each patient-specific BEST1 mutation can be comprehensively characterized in vivo for its protein expression (immunoblotting, Figure 3), membrane trafficking (immunostaining, Figure 4), and ion channel function (whole-cell patch clamp, Figure 5). These results will provide critical information for elucidating the pathological mechanisms of the channel mutations, and for developing personalized medicine.
As BEST1 is predominantly expressed in RPE cells, it can be used as a cellular marker for the validation of a mature RPE status. It should be noted that some BEST1 mutations might affect its protein expression, so other well-established RPE markers such as RPE65 and CRALBP still need to be evaluated (Figure 3).
Matured P0 hPSC-RPE cells can be maintained for 2-3 months before splitting (to P1) for whole-cell patch clamp. P0 cells older than 3 months are not recommended for patch clamp but can still be used for immunoblotting and immunostaining experiments.
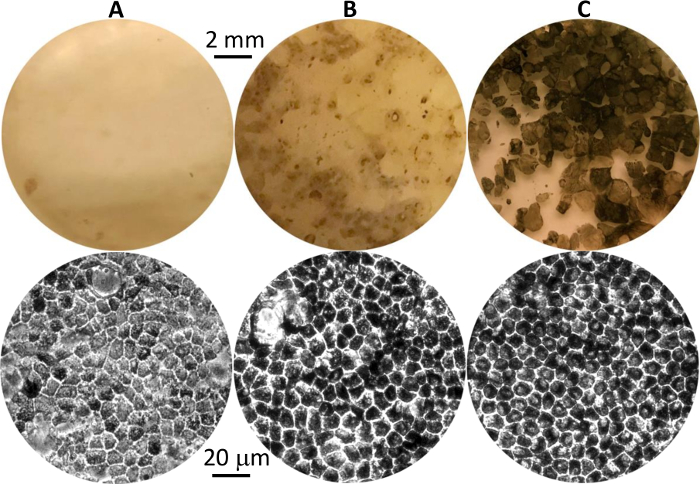
Figure 1: Differentiated hPSC-RPE cells at different stages. Representative images of pigmented hPSC-RPE clusters in culture plates at first appearance (A), before isolation (B), and after growth to maturity post isolation (C). Eye-view and 20X phase contrast images are shown in the top and bottom panels, respectively. Please click here to view a larger version of this figure.

Figure 2: Representative image of the micro cell scraper pulled from a Pasteur pipette. Please click here to view a larger version of this figure.
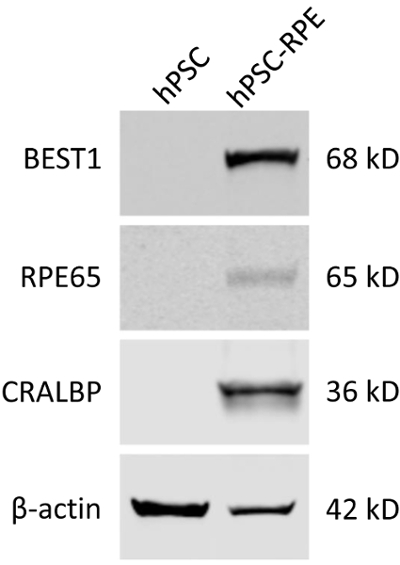
Figure 3: Validation of the mature RPE status of differentiated hPSC-RPE cells by immunoblotting. Blots showing the expression of RPE-specific protein markers BEST1, RPE65, and CRALBP in WT hPSC and hPSC-RPE cells. Please click here to view a larger version of this figure.
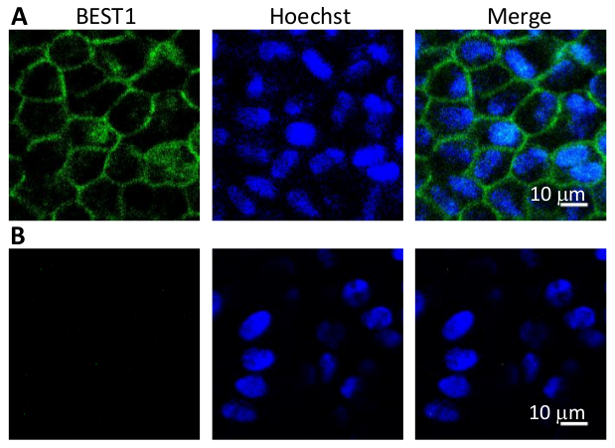
Figure 4: Immunostaining of BEST1 in WT hPSC-RPEs. (A) Representative immunofluorescent images showing the membrane localization of WT BEST1 in cobblestone-shaped hPSC-RPE cells. (B) Immunostaining negative control omitting the BEST1 primary antibody.
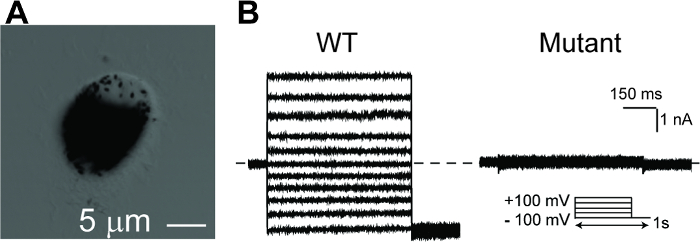
Figure 5: Whole-cell patch clamp recordings of hPSC-RPEs. (A) A single hPSC-RPE cell for whole-cell patch clamp. (B) Representative current traces recorded from a BEST1 WT hPSC-RPE and a BEST1 mutated hPSC-RPE at peak Ca2+. The voltage protocol used to elicit currents is shown in the Insert. Scale bar = 1 nA, 150 ms. See Tables 3 and Table 4 for patch clamp preparation details. Please click here to view a larger version of this figure.
| Reagent | Amount |
| Knock-Out (KO) DMEM | 500 mL |
| KO serum replacement | 15% (75 mL) |
| Nonessential amino acids | 1% (5 mL) |
| Glutamine | 1% (5 mL) |
| Penicillin-streptomycin | 1% (5 mL) |
| Nicotinamide | 10 mM |
| Human activin-A* | 100 ng/mL |
| *Human activin-A is supplemented during days 15–28 of the differentiation protocol. | |
Table 1: RPE Differentiation Medium.
| Reagent | Amount |
| MEM (α modification) | 500 mL |
| Fetal Bovine Serum | 5% (25 mL) |
| N1 supplement | 1% (5 mL) |
| Glutamine-penicillin-streptomycin | 1% (5 mL) |
| Nonessential amino acids | 1% (5 mL) |
| Taurine | 125 mg |
| Hydrocortisone | 10 µg |
| Triiodo-thyronin | 0.0065 µg |
Table 2: RPE Culture Medium.
| Reagent | Concentration |
| CsCl | 130 mM |
| MgCl2 | 1 mM |
| EGTA | 10 mM |
| Magnesium ATP | 2 mM |
| HEPES (pH 7.4) | 10 mM |
| CaCl2* | Varies |
| Glucose** | ~ 5 mM |
| *Add CaCl2 to obtain desired Ca2+ concentration | |
| ** Use glucose to adjust osmolarity to 290–295 mOsm/L | |
Table 3: Patch Clamp Internal Solution.
| Reagent | Concentration |
| NaCl | 140 mM |
| KCl | 5 mM |
| MgCl2 | 1 mM |
| CaCl2 | 2 mM |
| HEPES (pH 7.4) | 10 mM |
| Glucose* | ~ 5 mM |
| *Use glucose to adjust osmolarity to 300–305 mOsm/L. | |
Table 4: Patch Clamp External Solution.
Discussion
The most important procedure for the disease-in-a-dish approach is to differentiate hPSCs with a disease-causing mutation to the correct cell lineage, which is RPE for BEST1. Thus, after each differentiation experiment, the resulting hPSC-RPE cells should be carefully validated for their mature status by RPE-specific morphologies and protein markers16,17,18. To minimize clonal artifact, multiple hiPSC clones from the same patient or multiple hESC clones with the same mutation should be used whenever possible.
Both hiPSC and hESC lines can be differentiated to RPE with the same protocol. hiPSCs with or without a mutation in the BEST1 gene can be reprogrammed from the skin fibroblasts of BEST1 patients or healthy donors, respectively. hESCs bearing a mutation in BEST1 can be genetically engineered from the parental hESC line, which has the wild-type (WT) BEST1. The hiPSC-RPE and hESC-RPE routes have their own advantages and disadvantages. The former is more patient-specific and clinically relevant, particularly to personalized medicine. However, it is worth noting that there are several limitations for the hiPSC-RPE approach: 1) it is logistically difficult to access a full spectrum of BEST1 patient samples, as donations from patients cannot always be granted, and some mutations are very rare in the first place; 2) non-specific phenotypes may result from different genetic backgrounds and physical conditions of the donors; 3) the hiPSC to RPE differentiation efficiency varies for different donors, such that some cases can be technically challenging to obtain hiPSC-RPEs. On the other hand, the hESC-RPE route, although more artificial, offers a versatile platform to generate isogenic hESC-RPEs with desired mutations on demand for non-biased functional investigations.
Mature RPE cells are pigmented, polygonal, and connected by tight junctions in a monolayer. After splitting into a single cell population for patch clamp, the freshly reseeded hPSC-RPE cells will lose the polygonal shape, but still retain pigmentation for several days (Figure 5A). Patch clamp is performed 24–72 h after cell splitting, during which time the pigmentation can still be used as a visible marker to select cells with good RPE status. A gentle cell split resulting in a majority of well-separated single cells without significant cell death is key to the success of the patch clamp.
Several other differentiation protocols using different cell culture media and growth factors have been documented in the literature21,32. The time required for differentiation of hPSC to RPE is similar in all these protocols including ours, while it is hard to tell if there is any significant difference in the differentiation efficiency without a side-by-side comparison. It should be noted that in the current protocol, hPSC-RPE cells are grown in regular flat bottom plates but not on plates with membrane inserts, so the hPSC-RPE generated by this procedure may not best recapitulate the polarity of RPE cells in vivo. Therefore, the techniques described above are mostly suited in a research setting33, although the generated hPSC-RPE cells can also be cultured in Transwell plates to form monolayer RPE sheets for clinical purposes17.
開示
The authors have nothing to disclose.
Acknowledgements
This project was funded by NIH grants EY025290, GM127652, and University of Rochester start-up funding.
Materials
| Knock-Out (KO) DMEM | ThermoFisher | 10829018 | |
| KO serum replacement | ThermoFisher | 10829028 | |
| Nonessential amino acids | ThermoFisher | 11140050 | |
| Glutamine | ThermoFisher | 35050061 | |
| Penicillin-streptomycin | ThermoFisher | 10378016 | |
| Nicotinamide | Sigma-Aldrich | N0636 | |
| Human activin-A | PeproTech | 120-14 | |
| MEM (a modification) | Sigma-Aldrich | M4526 | |
| Fetal Bovine Serum | VWR | 97068-085 | |
| N1 supplement | Sigma-Aldrich | N6530 | |
| Glutamine-penicillin-streptomycin | Sigma-Aldrich | G1146 | |
| Nonessential amino acids | Sigma-Aldrich | M7156 | |
| Taurine | Sigma-Aldrich | T0625 | |
| Hydrocortisone | Sigma-Aldrich | H0386 | |
| Triiodo-thyronin | Sigma-Aldrich | T5516 | |
| mTeSR-1 medium | Stemcell Technologies | 5850 | |
| Matrigel | Corning | 356230 | |
| Collagenase | Gibco | 17104019 | |
| Trypsin | VWR | 45000-664 | |
| M-PER mammalian protein extraction reagent | Pierce | 78501 | |
| proteinase inhibitor cocktail | Sigma-Aldrich | 4693159001 | |
| RPE65 antibody | Novus Biologicals | NB100-355 | |
| CRALBP antibody | Abcam | ab15051 | |
| BEST1 antibody | Novus Biologicals | NB300-164 | |
| Beta Actin antibody | ThermoFisher | MA5-15739 | |
| Alexa Fluor 488-conjugated donkey anti-mouse IgG | ThermoFisher | A-21202 | |
| Goat anti-mouse IgG | ThermoFisher | SA5-35521 | |
| Goat anti-Rabbit IgG | LI-COR Biosciences | 926-68071 | |
| Hoechst 33342 | ThermoFisher | 62249 | |
| HEKA EPC10 patch clamp amplifier | Warner Instruments | 895000 | |
| Patchmaster | Warner Instruments | 895040 |
参考文献
- Allikmets, R., et al. Evaluation of the Best disease gene in patients with age-related macular degeneration and other maculopathies. Hum Genet. 104 (6), 449-453 (1999).
- Burgess, R., et al. Biallelic mutation of BEST1 causes a distinct retinopathy in humans. Am J Hum Genet. 82 (1), 19-31 (2008).
- Davidson, A. E., et al. Missense mutations in a retinal pigment epithelium protein, bestrophin-1, cause retinitis pigmentosa. Am J Hum Genet. 85 (5), 581-592 (2009).
- Kramer, F., et al. Mutations in the VMD2 gene are associated with juvenile-onset vitelliform macular dystrophy (Best disease) and adult vitelliform macular dystrophy but not age-related macular degeneration. Eur J Hum Genet. 8 (4), 286-292 (2000).
- Marquardt, A., et al. Mutations in a novel gene, VMD2, encoding a protein of unknown properties cause juvenile-onset vitelliform macular dystrophy (Best’s disease). Hum Mol Genet. 7 (9), 1517-1525 (1998).
- Petrukhin, K., et al. Identification of the gene responsible for Best macular dystrophy. Nat Genet. 19 (3), 241-247 (1998).
- Yardley, J., et al. Mutations of VMD2 splicing regulators cause nanophthalmos and autosomal dominant vitreoretinochoroidopathy (ADVIRC). Invest Ophthalmol Vis Sci. 45 (10), 3683-3689 (2004).
- Yang, T., Justus, S., Li, Y., Tsang, S. H. BEST1: the Best Target for Gene and Cell Therapies. Mol Ther. 23 (12), 1805-1809 (2015).
- Marmorstein, A. D., et al. Bestrophin, the product of the Best vitelliform macular dystrophy gene (VMD2), localizes to the basolateral plasma membrane of the retinal pigment epithelium. Proc Natl Acad Sci U S A. 97 (23), 12758-12763 (2000).
- Boon, C. J., et al. The spectrum of ocular phenotypes caused by mutations in the BEST1 gene. Prog Retin Eye Res. 28 (3), 187-205 (2009).
- Marmorstein, A. D., Cross, H. E., Peachey, N. S. Functional roles of bestrophins in ocular epithelia. Prog Retin Eye Res. 28 (3), 206-226 (2009).
- Fujii, S., Gallemore, R. P., Hughes, B. A., Steinberg, R. H. Direct evidence for a basolateral membrane Cl- conductance in toad retinal pigment epithelium. Am J Physiol. 262, C374-C383 (1992).
- Gallemore, R. P., Steinberg, R. H. Effects of DIDS on the chick retinal pigment epithelium. II. Mechanism of the light peak and other responses originating at the basal membrane. J Neurosci. 9 (6), 1977-1984 (1989).
- Gallemore, R. P., Steinberg, R. H. Light-evoked modulation of basolateral membrane Cl- conductance in chick retinal pigment epithelium: the light peak and fast oscillation. J Neurophysiol. 70 (4), 1669-1680 (1993).
- Li, Y., Nguyen, H. V., Tsang, S. H. Skin Biopsy and Patient-Specific Stem Cell Lines. Methods Mol Biol. 1353, 77-88 (2016).
- Milenkovic, A., et al. Bestrophin 1 is indispensable for volume regulation in human retinal pigment epithelium cells. Proc Natl Acad Sci U S A. 112 (20), E2630-E2639 (2015).
- Moshfegh, Y., et al. BESTROPHIN1 mutations cause defective chloride conductance in patient stem cell-derived RPE. Hum Mol Genet. 25 (13), 2672-2680 (2016).
- Li, Y., et al. Patient-specific mutations impair BESTROPHIN1’s essential role in mediating Ca2+-dependent Cl- currents in human RPE. Elife. 6, (2017).
- Marmorstein, L. Y., et al. The light peak of the electroretinogram is dependent on voltage-gated calcium channels and antagonized by bestrophin (best-1). J Gen Physiol. 127 (5), 577-589 (2006).
- Volland, S., Esteve-Rudd, J., Hoo, J., Yee, C., Williams, D. S. A comparison of some organizational characteristics of the mouse central retina and the human macula. PLoS One. 10 (4), e0125631 (2015).
- Kamao, H., et al. Characterization of human induced pluripotent stem cell-derived retinal pigment epithelium cell sheets aiming for clinical application. Stem Cell Reports. 2 (2), 205-218 (2014).
- Idelson, M., et al. Directed differentiation of human embryonic stem cells into functional retinal pigment epithelium cells. Cell Stem Cell. 5 (4), 396-408 (2009).
- Mandai, M., et al. Autologous Induced Stem-Cell-Derived Retinal Cells for Macular Degeneration. N Engl J Med. 376 (11), 1038-1046 (2017).
- Maminishkis, A., et al. Confluent monolayers of cultured human fetal retinal pigment epithelium exhibit morphology and physiology of native tissue. Invest Ophthalmol Vis Sci. 47 (8), 3612-3624 (2006).
- Maruotti, J., et al. A simple and scalable process for the differentiation of retinal pigment epithelium from human pluripotent stem cells. Stem Cells Transl Med. 2 (5), 341-354 (2013).
- Yang, T., He, L. L., Chen, M., Fang, K., Colecraft, H. M. Bio-inspired voltage-dependent calcium channel blockers. Nat Commun. 4, 2540 (2013).
- Yang, T., Hendrickson, W. A., Colecraft, H. M. Preassociated apocalmodulin mediates Ca2+-dependent sensitization of activation and inactivation of TMEM16A/16B Ca2+-gated Cl- channels. Proc Natl Acad Sci U S A. 111 (51), 18213-18218 (2014).
- Yang, T., et al. Structure and selectivity in bestrophin ion channels. Science. 346 (6207), 355-359 (2014).
- Yang, T., Puckerin, A., Colecraft, H. M. Distinct RGK GTPases differentially use alpha1- and auxiliary beta-binding-dependent mechanisms to inhibit CaV1.2/CaV2.2 channels. PLoS One. 7 (5), e37079 (2012).
- Yang, T., Suhail, Y., Dalton, S., Kernan, T., Colecraft, H. M. Genetically encoded molecules for inducibly inactivating CaV channels. Nat Chem Biol. 3 (12), 795-804 (2007).
- Yang, T., Xu, X., Kernan, T., Wu, V., Colecraft, H. M. Rem, a member of the RGK GTPases, inhibits recombinant CaV1.2 channels using multiple mechanisms that require distinct conformations of the GTPase. J Physiol. 588 (Pt 10), 1665-1681 (2010).
- Gong, J., et al. Differentiation of Human Protein-Induced Pluripotent Stem Cells toward a Retinal Pigment Epithelial Cell Fate. PLoS One. 10 (11), e0143272 (2015).
- Zhang, Y., et al. ATP activates bestrophin ion channels through direct interaction. Nat Commun. 9 (1), 3126 (2018).

