Executing Complexity-Increasing Queries in Relational (MySQL) and NoSQL (MongoDB and EXist) Size-Growing ISO/EN 13606 Standardized EHR Databases
概要
This study compares relational and non-relational (NoSQL) standardized medical information systems. The computational complexity of the response times of querying such database management systems (DBMS) is computed using doubling-sized databases. These results help the discussion of the appropriateness of each database approach to different scenarios and problems.
Abstract
This research shows a protocol to assess the computational complexity of querying relational and non-relational (NoSQL (not only Structured Query Language)) standardized electronic health record (EHR) medical information database systems (DBMS). It uses a set of three doubling-sized databases, i.e. databases storing 5000, 10,000 and 20,000 realistic standardized EHR extracts, in three different database management systems (DBMS): relational MySQL object-relational mapping (ORM), document-based NoSQL MongoDB, and native extensible markup language (XML) NoSQL eXist.
The average response times to six complexity-increasing queries were computed, and the results showed a linear behavior in the NoSQL cases. In the NoSQL field, MongoDB presents a much flatter linear slope than eXist.
NoSQL systems may also be more appropriate to maintain standardized medical information systems due to the special nature of the updating policies of medical information, which should not affect the consistency and efficiency of the data stored in NoSQL databases.
One limitation of this protocol is the lack of direct results of improved relational systems such as archetype relational mapping (ARM) with the same data. However, the interpolation of doubling-size database results to those presented in the literature and other published results suggests that NoSQL systems might be more appropriate in many specific scenarios and problems to be solved. For example, NoSQL may be appropriate for document-based tasks such as EHR extracts used in clinical practice, or edition and visualization, or situations where the aim is not only to query medical information, but also to restore the EHR in exactly its original form.
Introduction
NoSQL (Not only SQL) DBMS have recently emerged as an alternative to traditional Relational DBMS (RDMBS). RDBMS have dominated the way data were stored in database systems for decades. Well-studied and understood relational algebra and calculus have guaranteed the efficiency and consistency of RDBMS1. NoSQL systems will not become substitutes for relational systems, but they could behave advantageously in certain scenarios and under several conditions.
Some of these particular scenarios and conditions would occur when designing the database persistence of Electronic Health Record (EHR) systems used to manage and store medical information. In order to be interoperable and sustainable in practice, several international standards such as ISO/EN 13606, openEHR, and HL72,3,4,5 have been used to standardize EHR extracts.
Several standards such as ISO/EN 13606 and openEHR have separated information and knowledge into two different levels of abstraction, represented by the Reference Model (RM) and special data structures called archetypes. This separation is often called the dual model and it permits EHR systems to be semantically interoperable and medical knowledge to evolve without reprogramming the whole EHR system and, consequently, to be maintainable and sustainable in practice6. However, the dual model implemented in standardized EHR systems requires that the organization of information follows a specific structure, and this has profound consequences in the way the database persistence level of the system is designed7.
Object Relational Mapping (ORM)8 is one methodology to implement an EHR system using the relational database paradigm. ORM exhaustively maps the structure of standardized EHR extracts XML (eXtensible Markup Language) files used by the system for a relational database. ORM constructs many relational tables exhaustively following the structure of the standardized EHR extracts XML files. These relational tables are related through many foreign keys and the resulting system may not be very efficient.
Several relational improvements to ORM have been proposed. openEHR's Node+Path9 reduces the number of relational tables by serializing subtrees of the whole extract XML file into BLOBs (binary large objects). However, this simplification causes complex retrieval logic, damaging complex queries. Archetype Relational Mapping (ARM)10 generates a database model driven by archetypes, building a new relational schema based on mappings between archetypes and relational tables. Consequently, some of the non-medical information of the EHR extract is lost.
Many document-based NoSQL databases store whole documents as entire BLOBs respecting an original XML or JSON (JavaScript Object Notation) format. This means that no relational tables are constructed. These NoSQL databases have no schema and they do not support either joins or (ACID) properties11, i.e.,atomicity, consistency, isolation, or durability. As a result, they may be very inefficient if an element of a document references elements of the same or other such documents utilizing an indirection link. This happens because, in order to maintain consistency, the entirety of the referenced documents have to be processed sequentially. However, a non-relational database may be still appropriate if the main task performed by the DBMS is a document-based task. This is because data may remain in a form more closely approximating its true representation using a document-based NoSQL database, though this is also due to the special persistence policies accomplished by EHR medical documents (see discussion section).
The purpose of these methods is to showcase several experiments to compare the implementation of the persistence layer of a standardized EHR system using three different DBMSs: one relational (MySQL) and two NoSQL (document-based MongoDB and native XML eXist). Their computational complexity has been computed and compared using three different increasing size databases and six different complexity-increasing queries. The three database servers have been installed and configured locally in the same computer where the queries have been executed. See the Table of Materials for technical details.
Concurrency experiments have also been conducted in order to compare the performance of relational MySQL and NoSQL MongoDB DBMSs. The described ORM improvements (Node+Path and ARM) have also been compared using relevant appropriate results from the literature10.
Database management systems are evolving continuously at an accelerating rate. No one would think about this exponential development when the only existing paradigm was the relational model. To take an example, see for instance12, where a model was proposed to implement response-time improved relational databases retaining the ACID properties.
Protocol
1. Build a Relational MySQL DBMS to Store Three Double Sized Standardized EHR Extracts Databases
- Import the W3C (World Wide Web Consortium) XML Schema corresponding to the ISO/EN13606 RM and the ISO21090 Data types into a 'Java IDE' (Integrated Development Environment).
NOTE: ISO stands for International Standards Organization. EN stands for European Norm. - Execute the JAXB (Java XML Binding) plug-in in the IDE; this produces Java classes corresponding to the structure of the elements of the EHR extracts XML files.
- Tag manually the Java classes produced with JPA labels. These labels refer to the cardinalities and other relations between the relational tables of the MySQL database.
- Import the libraries of the JPA (Java Persistence API) into the IDE and execute the method that builds a MySQL database out of the tagged Java classes.
- Create three directories with 5,000, 10,000 and 20,000 realistic EHR extracts XML files.
- Execute the JPA method to load an XML extract into the MySQL DBMS on all the extracts of the 5,000 extracts directory.
- Repeat step 1.6 twice, once with the 10,000 extracts directory and once with the 20,000 extracts directory.
2. Build a NoSQL MongoDB DBMS to Store Three Double Sized Standardized EHR Extracts Databases
- Process each of the three directories containing 5,000, 10,000 and 20,000 realistic EHR extracts XML files with a standard program to convert XML files to JSON files, such as json.org.XML. Three directories with 5,000, 10,000 and 20,000 JSON files should be produced.
- Launch a MongoDB GUI (Graphic User Interface, see Table of Materials).
- Launch the MongoDB 2.6 server executing the mongod program from a DOS (Disk Operating System) system window.
- Connect the MongoDB GUI to the localhost server using port 27017.
- Select the "Connect" menu.
- Write a name for the connection (for instance 'first').
- Write localhost:27017 in the DB Server textbox.
- Press the "Connect" button ; a tree with the current databases should appear on the left.
- Build a database containing 5,000 standardized EHR extracts.
- Click on the name of the connection at the top of the tree on the left.
- Select the "File" menu.
- Choose "Add Database".
- Enter the name of the database in the dialog that appears.
- Click OK.
- Build a collection containing 5,000 standardized EHR extracts.
- Click on the name of the database in the tree on the left.
- Select menu "Database".
- Choose "AddCollection".
- Enter the name of the collection in the dialog that appears.
- Click " create".
- Click on the name of the collection.
- Select the "Import" menu.
- Choose radio button ''JSON – mongo shell / / mongoexport".
- Click "next".
- Press the "Add Source Files" button.
- Navigate on the computer using the dialog.
- Open the directory containing 5,000 JSON extract files.
- Select all the files in the directory.
- Press "Open". The list of JSON files should appear in the Import dialog.
- Press "Next"; a preview of the new collection in the database appears on the left.
- Press "Next".
- Press "Start Import". The progress of the import appears down on the left, indicating the number of files imported and the elapsed time.
- Repeat steps 5 and 6 to build a collection of 10,000 standardized EHR extracts.
- Repeat steps 5 and 6 to build a collection of 20,000 standardized EHR extracts.
3. Build a NoSQL eXist DBMS to Store Three Double Sized Standardized EHR Extracts Databases
- Launch the eXist database.
- Using the database's icon, open the Java Admin Client.
- Enter the admin password.
- Press the "Connect" button.
- Build a collection containing 5,000 standardized EHR extracts.
- In the toolbar, select the menu "Create a new Collection".
- In the dialog that appears, type the name of the new collection.
- Click "accept"; the new collection will appear.
- Select the name of the collection.
- In the toolbar, select the menu "Store files in the database".
- Navigate on the computer using the dialog box.
- Select the directory containing 5,000 standardized XML extract files.
- Click the button "Select the files or directories to store". Note that a dialog box appears showing the progress, the files being stored, and the percentage of the database created.
- Repeat step 5 to build a collection containing 10,000 standardized EHR extracts.
- Repeat step 5 to build a collection containing 20,000 standardized EHR extracts.
4. Design and Execute in the 3 Relational MySQL Databases 6 Complexity-Increasing Queries
- Design six complexity-increasing queries according to the archetypes used by the EHR extracts.
- Program an SQL script with the first query on the MySQL database. The SQL must adapt to the special structure of the MySQL database due to extracts standardization (archetypes). The database maps the whole structure of the extracts. As a result, the SQL query is rather complex.
- Identify the attributes of the databases that would speed up the response time of the queries if an index was built on them, then construct such indexes, though most indexes are built automatically by the DBMS.
- If a query needs a non-automatically built index, build it manually.
- Connect to the MySQL server (Supplementary Figure 1).
- Select and click on the database name on the left.
- Select and click on the relational table where the indexed field resides.
- Click on the tab "Structure".
- Select and click on the column where the index will be built.
- Click on "index". Note that the SQL sentence building the index appears, and a message stating that the sentence has been built successfully appears.
- Execute the first query.
- Select and click on the database name on the left.
- Click on the tab "SQL".
- Write or paste the SQL code of the first query (see Supplementary Figure 2).
- Press "continue". Note that the first screen of the list of results appears, along with a message with the execution time of the query.
- Repeat the execution 5 times and compute the average response time.
- Repeat step 5 with queries 2 through 6.
- Do the whole process three times, with the 5,000, 10,000 and 20,000 extracts databases.
5. Design and Execute in the 3 NoSQL MongoDB Databases 6 Complexity-Increasing Queries
- Launch the MongoDB GUI (see Table of Materials).
- Launch the MongoDB 2.6 server executing the mongod program from a DOS system window (see Supplementary Figure 3).
- Follow step 2.4 to connect the MongoDB GUI to the localhost server using port 27017.
- Select and expand the MongoDB database on the left side.
- Select the collection.
- Click on the "Collection" menu in the toolbar.
- Execute the first MongoDB query.
- Double-click the "Query Builder" button.
- Double-click on the "Query field" of the Query Builder at the right.
- Write the field of the MongoDB query in the field textbox of the query panel (See Supplementary Figure 4).
- Write the value of the MongoDB query in the value textbox of the query panel.
NOTE: This query should be something like {"ns3:EHRExtract.allCompositions.content.items.parts.parts.name.ns2:originalText. value": "Descripcion"}. The field and the value are quoted and separated by semicolon. - Double-click on the Projection field of the Query Builder
- Write the first projection in the projection textbox (see Supplementary Figure 5).
- Double-click on the projection field to add a new projection textbox.
- Write the second projection in the projection textbox.
NOTE: A projection selects a part of the document retrieved by the query. These should be something like {"ns3:EHRExtract. allCompositions.content.items.parts.parts.value.value": 1} and {"ns3: EHRExtract.all Compositions.content.items.parts.parts.value.nullFlavor" : 1} - Click on the blue play button to execute the query.
- Visualize the query code in the Query Code tab.
- View the details of the result in the Explain tab: number of results, execution time in milliseconds.
- View, expand, and examine the results in the Result tab.
- If further processing of the query is required, write a Java program with the MongoDB Java driver with the query and a method to process the results.
- Repeat the execution 5 times and compute the average response time.
- Do step 5.7 for the remaining 2 through 6 queries.
- Repeat the whole process in the 5,000, 10,000 and 20,000 extracts MongoDB databases.
6. Design and Execute in the 3 NoSQL eXist Databases 6 Increasing-Complexity Queries
- Launch the eXist DBMS.
- Open the Java Admin Client.
- Press the button "connect to the database".
- Select the database and click on it.
- Click on the menu "Consult database using XPath"; the consult dialog box appears.
- Execute the first XPath query (see Supplementary Figure 6).
- Write or paste the XPath code of the first query in the upper part of the dialog box.
- Click on the menu "Execute" in the toolbar of the dialog box.
- View XML results using the "XML" tab in the lower part of the dialog box.
- View number of results and compilation and execution time at the bottom of the dialog box.
- Repeat the execution 5 times and compute the average response time.
- Repeat step 6 for queries 2 through 6.
- Do the whole process three times, for the 5,000, 10,000 and 20,000 extracts eXist databases.
7. Design and Execute a Concurrency Experiment using the MySQL and MongoDB 5,000 Extracts Databases
NOTE: The eXist database has been removed from the experiment at this juncture due to worse performance in the previous experiments.
- Select the queries with the three shortest time responses in the previous experiments using the 5,000 extracts databases (typically under several seconds).
- Identify and manually build appropriate attribute indexes for those queries, if necessary.
- Program two Java multithread applications, one for MySQL and the other for MongoDB; each application will have three different priority threads, one for each query selected in step 1.
- Execute and compute the CPU (Central Processing Unit) use distribution for each thread (query).
- Execute each multithread application, clicking on the execute button five times during each 10-min span, and compute the most executed (highest priority) query average throughput and the average time response of the three queries.
- View the queries in execution, with priorities and execution time.
- Compute average throughput and average response time of each of the three queries.
Representative Results
Six different queries performed on realistic standardized EHR extracts containing information about the problems of patients, including their names, initial and final dates and severity, are shown in Table 1.
Average response times of the six queries in the three doubling-size databases in each DBMS are shown in Tables 2-4. Figures 1-6 show the same results graphically (notice that the vertical axes use very different scales throughout these figures).
The strong linear behavior of computational complexity is evident throughout all queries of the NoSQL databases, although with appropriate caution due to the relatively small size of the 3 datasets used. However, the relational ORM database shows an unclear linear behavior. The MongoDB database has a much flatter slope than the eXist database.
Results by the improved relational systems discussed in the introduction published in the literature may be found in Table 5. Interpolating MongoDB results from Table 3 with similar queries and database sizes of ARM results from Table 5 equals both database systems in Q1, but favors MongoDB in Q3.
The results of the concurrency experiments may be found in Table 5 and Table6. MongoDB beats MySQL both in throughput and response time. In fact, MongoDB behaves better in concurrency than in isolation, and stands as an impressive database in concurrent execution.
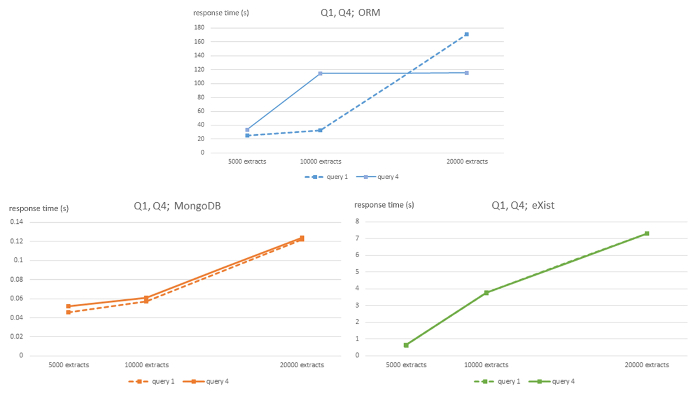
Figure 1: Algorithmic complexity of ORM MySQL, MongoDB, and eXist DBMS for queries Q1 and Q4. This figure has been modified from7 using Creative Commons license (http://creativecommons.org/ licenses/by/4.0/) and shows response times in seconds for 5,000, 10,000 and 20,000-sized EHR extracts databases for each DBMS and queries Q1 and Q4. Please click here to view a larger version of this figure.
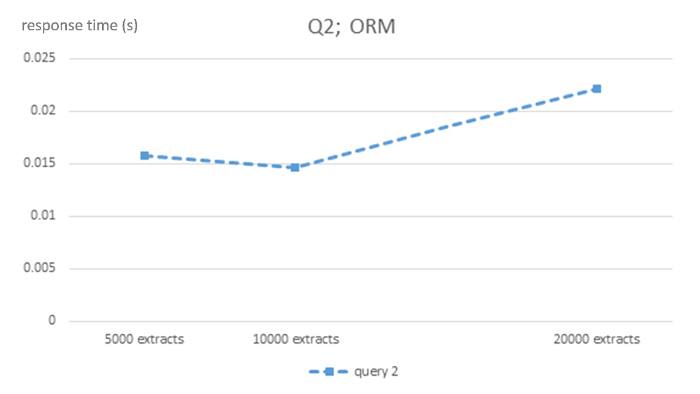
Figure 2: Algorithmic complexity of ORM MySQL DBMS for query Q2. This figure shows response times in seconds for 5,000, 10,000 and 20,000-sized EHR extracts ORM MySQL database for query Q2. Please click here to view a larger version of this figure.
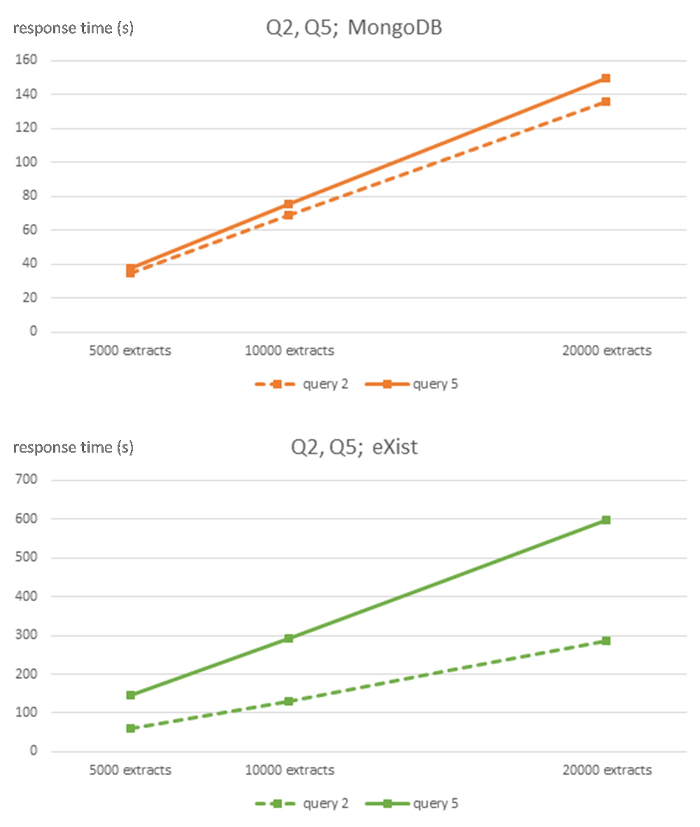
Figure 3: Algorithmic complexity of MongoDB and eXist DBMS for queries Q2 and Q5. This figure has been modified from7 using Creative Commons license (http://creativecommons.org/licenses/ by/4.0) and Shows response times in seconds for 5,000, 10,000, and 20,000-sized EHR extracts databases for each DBMS and queries Q2 and Q5. Please click here to view a larger version of this figure.
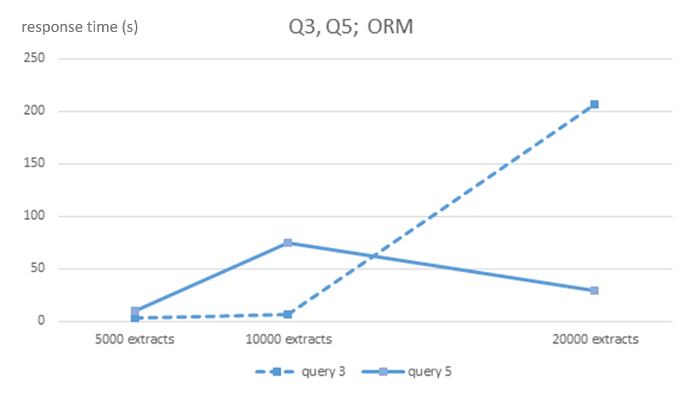
Figure 4: Algorithmic complexity of ORM MySQL DBMS for queries Q3 and Q5. Shows response times in seconds for 5,000, 10,000 and 20,000-sized EHR extracts databases for each DBMS and queries Q3 and Q5. Please click here to view a larger version of this figure.
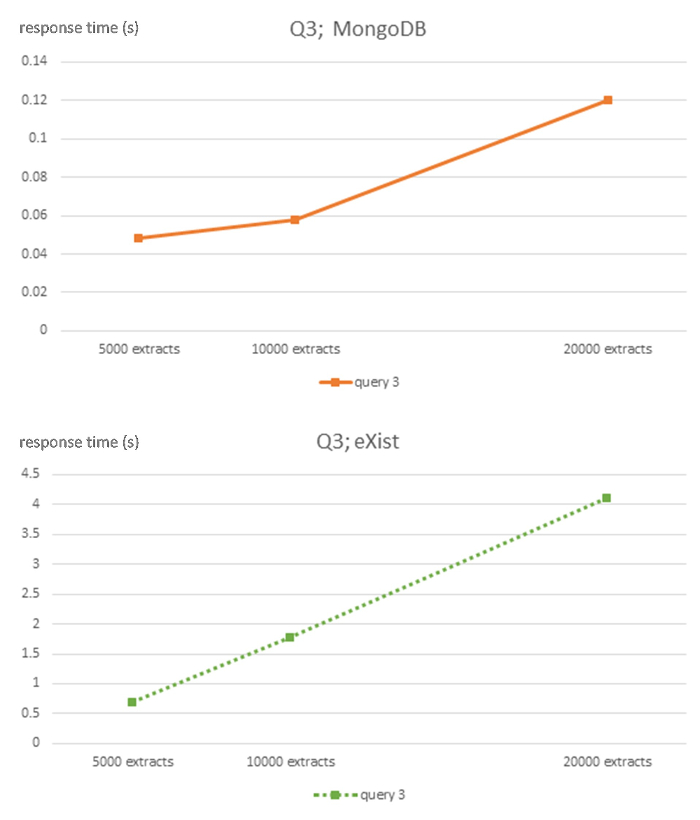
Figure 5: Algorithmic complexity of eXist and MongoDB DBMS for query Q3. This figure has been modified from7 using Creative Commons license (http://creativecommons.org/licenses/ by/4.0/ ) and shows response times in seconds for 5,000, 10,000 and 20,000-sized EHR extracts databases for each DBMS and query Q3. Please click here to view a larger version of this figure.
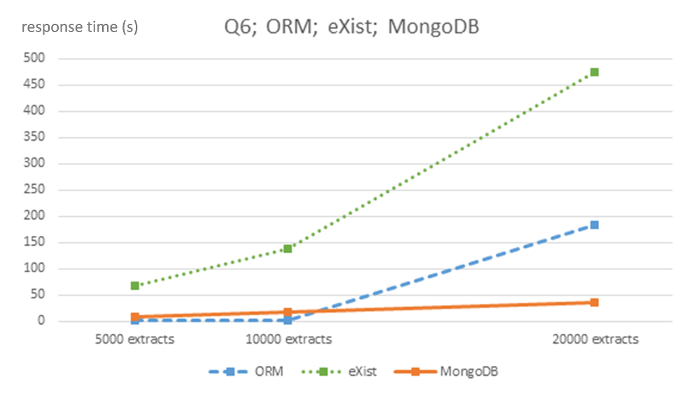
Figure 6: Algorithmic complexity of ORM MySQL, eXist and MongoDB DBMS for query Q6. This figure has been modified from7 using Creative Commons license (http://creativecommons.org/licenses/ by/4.0/) and shows response times in seconds for 5,000, 10,000 and 20,000-sized EHR extracts databases for each DBMS and query Q6. Please click here to view a larger version of this figure.
| Query | |
| Q1 | Find all problems of a single patient |
| Q2 | Find all problems of all patients |
| Q3 | Find initial date, resolution date and severity |
| of a single problem of a single patient | |
| Q4 | Find initial date, resolution date and severity |
| of all problems problem of a single patient | |
| Q5 | Find initial date, resolution date and severity |
| of all problems problem of all patients | |
| Q6 | Find all patients with problem 'pharyngitis', |
| initial date >= '16/10/2007', resolution date | |
| <= '06/05/2008' and severity 'high' |
Table 1: The six queries performed on the relational and NoSQL databases containing standardized EHR extracts about problems of patients. This table has been modified from7 using Creative Commons license (http://creativecommons.org/ licenses/by/4.0/) and shows the six complexity-growing queries performed on the three size-growing databases for each DBMS expressed in natural language.
| ORM/MySQL | 5000 docs | 10,000 docs | 20,000 docs |
| Q1 (s) | 25.0474 | 32.6868 | 170.7342 |
| Q2 (s) | 0.0158 | 0.0147 | 0.0222 |
| Q3 (s) | 3.3849 | 6.4225 | 207.2348 |
| Q4 (s) | 33.5457 | 114.6607 | 115.4169 |
| Q5 (s) | 9.6393 | 74.3767 | 29.0993 |
| Q6 (s) | 1.4382 | 2.4844 | 183.4979 |
| Database size | 4.6GB | 9.4GB | 19.4GB |
| Total extracts | 5000 | 10,000 | 20,000 |
Table 2: Average response times in seconds of the six queries on doubling-size databases of the ORM MySQL relational DBMS. This table shows six response times for each query for the three doubling-sized databases using the ORM MySQL relational DBMS and the in-memory size of the three databases.
| MongoDB | 5000 docs | 10,000 docs | 20,000 docs | slope (*10exp(-6)) |
| Q1 (s) | 0.046 | 0.057 | 0.1221 | 5.07 |
| Q2 (s) | 34.5181 | 68.6945 | 136.2329 | 6,780.99 |
| Q3 (s) | 0.048 | 0.058 | 0.1201 | 4.81 |
| Q4 (s) | 0.052 | 0.061 | o.1241 | 4.81 |
| Q5 (s) | 38.0202 | 75.4376 | 149.933 | 7460.85 |
| Q6 (s) | 9.5153 | 18.5566 | 36.7805 | 1,817.68 |
| Database size | 1.95GB | 3.95GB | 7.95GB | |
| Total extracts | 5000 | 10,000 | 20,000 |
Table 3: Average response times in seconds of the six queries on doubling-size databases of the MongoDB NoSQL DBMS. This table has been modified from7 using Creative Commons license (http://creativecommons.org/ licenses/by/4.0/) and shows the six response times of each query for the three doubling-sized databases using the NoSQL MongoDB DBMS and the in-memory size of the three databases. The linear slope of each query is also shown.
| eXist | 5000 docs | 10,000 docs | 20,000 docs | slope (*10exp(-6)) |
| Q1 (s) | 0.6608 | 3.7834 | 7.3022 | 442.76 |
| Q2 (s) | 60.7761 | 129.3645 | 287.362 | 15,105.73 |
| Q3 (s) | 0.6976 | 1.771 | 4.1172 | 227.96 |
| Q4 (s) | 0.6445 | 3.7604 | 7.3216 | 445.17 |
| Q5 (s) | 145.3373 | 291.2502 | 597.7216 | 30,158.93 |
| Q6 (s) | 68.3798 | 138.9987 | 475.2663 | 27,125.82 |
| Database size | 1.25GB | 2.54GB | 5.12GB | |
| Total extracts | 5000 | 10,000 | 20,000 |
Table 4: Average response times in seconds of the six queries on doubling-size databases of the eXist NoSQL DBMS. This table has been modified from7 using Creative Commons license (http://creativecommons.org/ licenses/by/4.0/) and shows the six response times of each query for the three doubling-sized databases using the NoSQL eXist DBMS and the in-memory size of the three databases. The linear slope of each query is also shown.
| ARM paper | ARM (s) | Node+Path (s) | |
| Q1 | Query 2.1 | 0.191 | 24.866 |
| Q3 | Query 3.1 | 0.27 | 294.774 |
| Database size | 2.90GB | 43.87GB | |
| Total extracts | 29,743 | 29,743 |
Table 5: Average response times in seconds of queries similar to Q1 and Q3 of the improved relational systems presented in10. This table has been modified from7 using Creative Commons license (http://creativecommons.org/ licenses/by/4.0/) and shows the two most-similar queries to Q1 and Q3 presented in10 corresponding to two improved relational database systems and their response times. The two database sizes are also shown.
| ORM/MySQL | Throughput | Response time |
| Q1 (s) | 4,711.60 | 0.0793 |
| Q3 (s) | 4,711.60 | 0.1558 |
| Q4 (s) | 4,711.60 | 0.9674 |
Table 6: Average throughput and response time in seconds of queries Q1, Q3 and Q4 of ORM MySQL relational DBMS in concurrent execution. This table has been modified from7 using Creative Commons license (http://creativecommons.org/ licenses/by/4.0/) and shows the highest average throughput of the three single-patient queries and their average response times in the concurrent execution experiment using the ORM MySQL relational system.
| MongoDB | Throughput | Response time |
| Q1 (s) | 178,672.60 | 0.003 |
| Q3 (s) | 178,672.60 | 0.0026 |
| Q4 (s) | 178,672.60 | 0.0034 |
Table 7: Average throughput and response time in seconds of queries Q1, Q3 and Q4 of MongoDB NoSQL DBMS in concurrent execution. This table has been modified from7 using Creative Commons license (http://creativecommons.org/ licenses/by/4.0/) and shows the highest average throughput of the three single-patient queries and their average response times in the concurrent execution experiment using the MongoDB NoSQL system.
Supplementary Figure 1: The screenshot shows the software screen to connect to the MySQL server. Please click here to download this figure.
Supplementary Figure 2: The screenshot shows the SQL interface to the MySQL server where the first SQL query has been written. Please click here to download this figure.
Supplementary Figure 3: The MongoDB 2.6 localhost server is launched using a DOS system window by executing the server mongod. Please click here to download this figure.
Supplementary Figure 4: The screenshot shows the query written in the textboxes of the Query Builder as shown in steps 5.7.1 through 5.7.4. The screenshot illustrates step 5.7.3. Please click here to download this figure.
Supplementary Figure 5:The screenshot shows the step 5.7.6. Please click here to download this figure.
Supplementary Figure 6: The screenshot illustrates the writing of the XPath query in theupper part of the dialog. Please click here to download this figure.
Discussion
This protocol shows that pure relational ORM systems do not seem practical for single-patient queries (Q1, Q3, and Q4) since response times are slower, probably due to a high number of relational tables performing many expensive join operations, and due to the storage system used by the specific kind of database. NoSQL databases store data in a document-based fashion, while relational systems use a table-based fashion that spreads each document throughout the whole database. NoSQL systems show a linear slope, with MongoDB performing considerably faster than eXist DBMS. In concurrency, MongoDB also behaves much better than relational MySQL ORM7.
This protocol presents a troubleshooting protocol for the results presented in7 regarding the ORM MySQL DBMS. The MySQL system has been updated to the latest version and the results have been slightly modified. In addition, a critical point in document-based NoSQL systems such as MongoDB is that they may preserve consistency when storing medical information7 because when an EHR extract is updated, it is not overwritten, but a whole new extract with the new data is generated and stored in the system, and the original extract is maintained. This is a strict requirement of medical information, because some medical practitioners may have made important medical decisions based on the original data.
The improved relational ARM system drastically diminishes the number of relational tables and improves relational performance. However, since it modifies the relational schema, medical information held by the extracts may be queried, but extracts cannot be recovered in their exact original forms.
For very big databases in secondary use (research), it is not clear which database system is more appropriate, since the all-patient queries (Q2 and Q5) behave better in ORM than in NoSQL systems, but these systems perform better than the simplified relational systems in 12. We consider Q6 a special query in between clinical practice and secondary use whose behavior cannot be determined by the results yielded by these experiments.
However, one limitation of the method is the inavailability of direct experiments comparing the improved relational ARM system with NoSQL MongoDB regarding single-patient, medical practice queries with exactly the same data used in the protocol. We maintained the results interpolating Table 3 and Table 5 regarding single-patient queries until the experiment including optimized ARM in the protocol was performed. We leave these experiments for future applications. One critical step within the protocol is the selection of free database, similar software versions from recent years, so that we may compare the exact state-of-the-art of the three technologies.
This is one of the first attempts to directly compare relational and NoSQL systems using actual, realistic, standardized medical information. However, the specific system to be used depends much on the actual scenario and problem to be solved8.
開示
The authors have nothing to disclose.
Acknowledgements
The authors would like to thank Dr Dipak Kalra, leader of the EHRCom task force that defined the ISO/EN 13606 standard and his team from University College London for their kind permission to use the ISO/EN 13606 W3C XML Schema.
This work was supported by Instituto de Salud Carlos III [grant numbers PI15/00321, PI15/00003, PI1500831, PI15CIII/00010 and RD16CIII].
Materials
| MySQL 5.7.20 | MySQL experiments | ||
| Red Hat Enterprise Linux Server release 7.4 (Maipo), 2.60GHz, RAM 16GB | |||
| MongoDB 2.6 | MongoDB experiments | ||
| Windows 7, 2.66GHz, RAM 12GB | |||
| eXist 3.0RC1 | eXist experiments | ||
| Windows 7, 2.66GHz, RAM 12GB | |||
| Studio 3T 5.5.1 | 3T Software Labs Gmbh | MongoDB GUI |
参考文献
- Codd, E. F. A relational model for large shared data banks. Comm ACM. 13 (6), 377-387 (1970).
- Kalra, D., Lloyd, D. . ISO 13606 electronic health record communication part 1: reference model. , (2008).
- Kalra, D., et al. . Electronic health record communication part 2: archetype interchange specification. , (2008).
- Kalra, D., Beale, T., Heard, S. The openEHR foundation. Stud. Health Technol. Inform. 115, 153-173 (2005).
- Beale, T. Archetypes: constraint-based domain models for future proof information systems. OOPSLA, Workshop Behav Semant. , (2002).
- Sánchez-de-Madariaga, R., et al. Examining database persistence of ISO/EN 13606 standardized electronic health record extracts: relational vs. NoSQL approaches. BMC Medical Informatics and Decision Making. 32 (2), 493-503 (2017).
- Ireland, C., Bowers, D., Newton, M., Waugh, K. Understanding object-relational mapping: a framework based approach. Int. J. Adv. Softw. 2, 202-216 (2009).
- . Node+Path Persistence Available from: https://openehr.atlassian.net/wiki/spaces/dev/pages/6553626/Node+Path+Persistence (2017)
- Wang, L., Min, L., Wang, R., et al. Archetype relational mapping – a practical openEHR persistence solution. BMC Medical Informatics and Decision Making. 15, 88 (2015).
- Kaur, K., Rani, R. Managing data in healthcare information systems: many models, one solution. Computer. March. , 52-59 (2015).
- Sabo, C., Pop, P. C., Valean, H., Danciulescu, D. An innovative approach to manage heterogeneous information using relational database systems. Advances in Intelligent Systems and Computing. 557, (2017).

