Drug Treatment and In Vivo Imaging of Osteoblast-Osteoclast Interactions in a Medaka Fish Osteoporosis Model
概要
Small laboratory fish have become popular models for bone research on the mechanisms underlying human bone disorders and for the screening of bone-modulating drugs. In this report, we describe a protocol to assess the effect of alendronate on bone cells in medaka larvae with osteoporotic lesions.
Abstract
Bone-forming osteoblasts interact with bone-resorbing osteoclasts to coordinate the turnover of bone matrix and to control skeletal homeostasis. Medaka and zebrafish larvae are widely used to analyze the behavior of bone cells during bone formation, degeneration, and repair. Their optical clarity allows the visualization of fluorescently labeled bone cells and fluorescent dyes bound to the mineralized skeletal matrix. Our lab has generated transgenic medaka fish that express the osteoclast-inducing factor Receptor Activator of Nuclear-factor κB Ligand (RANKL) under the control of a heat shock-inducible promoter. Ectopic expression of RANKL results in the excess formation of activated osteoclasts, which can be visualized in reporter lines with nlGFP expression under the control of the cathepsin K (ctsk) promoter. RANKL induction and ectopic osteoclast formation leads to severe osteoporosis-like phenotypes. Compound transgenic medaka lines that express ctsk:nlGFP in osteoclasts, as well as mCherry under the control of the osterix (osx) promoter in premature osteoblasts, can be used to study the interaction of both cell types. This facilitates the in vivo observation of cellular behavior under conditions of bone degeneration and repair. Here, we describe the use of this system to test a drug commonly used in human osteoporosis therapy and describe a protocol for live imaging. The medaka model complements studies in cell culture and mice, and offers a novel system for the in vivo analysis of drug action in the skeletal system.
Introduction
The vertebrate skeleton provides structural support and protection for organs, allows mobility, and serves as a source of calcium. Throughout life, the extracellular bone matrix is continuously turned over to maintain bone stability and rigidity. This process requires the tightly coordinated activity and interplay of bone-forming osteoblasts and bone-resorbing osteoclasts. Osteoblasts are derived from multipotent mesenchymal progenitors and produce collagen to form the osteoid, the proteinaceous part of the bone matrix10. Osteoblasts interact with osteoclasts to achieve a balanced activity of both cell types, which is required to control bone homeostasis7. Because of these intricate regulatory interactions, responses to drug treatment and bone homeostasis cannot be fully examined using in vitro studies. Hence, there is a strong demand for animal models. Compared to the cell culture settings, in vivo models can provide valuable insight into the multicellular networks within the bone environment.
Numerous mouse models exist for a variety of human bone disorders including osteoporosis16. However, the size and accessibility of mouse embryos represent significant limitations for live imaging of skeletal processes. Small teleost fish, on the other hand, serve as an attractive alternative for in vivo imaging. Zebrafish (Danio rerio) and medaka (Oryzias latipes) have become popular animal models for skeletal research over the last two decades17, 19, 22, 24. Bone in teleost fish and in mammals is very similar, both on a structural and on a physiological level, and many of the key regulatory genes and signaling pathways are conserved3. As in mammals, teleost fish carefully regulate the activity of osteoblasts and osteoclasts to balance bone formation and resorption26. Most importantly, the optical clarity of fish larvae allows the use of fluorescent reporters to label bone cells and the calcified skeletal matrix8, 9, 12, 21, 23, which facilitates the observation of cellular processes in the living animal. In addition, a series of genetic tools has been generated to facilitate biomedically relevant research in fish. For medaka in particular, methods for targeted gene mutation by CrispR/Cas92, cell-lineage tracing6, and site-specific transgenesis14 have been recently established and are now widely in use15.
Small teleost larvae have been successfully used for chemical screens, which led to the discovery of several pharmacologically relevant drugs1, 18.
Fish larvae are tolerant to low concentrations of DMSO and are able to absorb compounds from their aquatic environment, either through the skin or through the gastrointestinal tract1, 5. Our lab previously reported transgenic medaka lines that express fluorescent reporters in bone cells under the control of various osteoblast- and osteoclast-specific promoters. These include premature osteoblasts (collagen 10a1, col10a1; osterix, osx)20, 21, mature osteoblasts (osteocalcin, osc)27, and osteoclasts (cathepsin K, ctsk)24. We also generated a transgenic line that expresses the osteoclast-inducing factor Receptor Activator of Nuclear-factor κB Ligand (RANKL) under the control of a heat shock-inducible promoter24.
Induction of RANKL in this system results in the ectopic formation of active osteoclasts. This leads to increased bone resorption and a severe osteoporosis-like phenotype, with drastically reduced mineralization in the vertebral bodies. We recently showed that osteoclast activity in this model can be blocked by the bisphosphonates etidronate and alendronate, two drugs commonly used in human osteoporosis therapy, thus validating medaka as a suitable model system for osteoporosis27.
Due to their large brood size, rapid development, and small size of embryos, transgenic medaka larvae are uniquely suited for the large-scale screening of osteoporosis drugs and for the in vivo analysis of bone cell behavior. Studies in medaka thus can efficiently complement experiments in cell cultures and in mice that are aimed at discovering new therapeutic targets and novel therapies for human bone disorders.
In the present study, we describe a protocol to treat medaka bone-reporter larvae with the common osteoporosis drug, alendronate. We also describe in detail how treated larvae are mounted and prepared for the live imaging of bone matrix and bone cells. These protocols can be easily adapted to other small chemical compounds that either work as bone anabolic or antiresorptive drugs.
Protocol
All experiments were performed in accordance with approved Institutional Animal Care and Use Committee (IACUC) protocols of the National University of Singapore (R14-293).
1. Fish Husbandry and the Collection of Embryos
- Raise WT, ctsk:nlGFP24, RANKL:HSE:CFP24, and osx:mCherry21 single- or compound-transgenic medaka fish at 26 °C under a controlled light cycle (14 h light, 10 h dark) to induce spawning.
- Daily spawning takes place during the first 30 min after the light is switched on. The eggs stick together through filaments and attach to the female's abdomen for several hours. Use a fine-meshed net to catch a female adult carrying an egg cluster. Let the fish briefly rest in the net and then gently massage the fish’s abdomen to carefully strip the fertilized egg cluster from the abdomen of the female.
NOTE: A healthy medaka female can produce 10 – 20 eggs each day for approximately 5 months. - Place the eggs into a 60 mm plastic Petri dish. Use a plastic pipette to rinse the embryos with 5 – 10 mL of 0.3x Danieau's solution (fish medium; 19.3 mM NaCl, 0.23 mM KCl, 0.13 mM MgSO4, 0.2 mM Ca(NO3)2, and 1.7 mM HEPES, pH 7.0). Add 1 mL of a 0.25% (w/v) methylene blue stock solution to 2.5 L of fish medium to prevent fungal growth.
- Gently roll egg cluster to form a knot of attachment filaments. Use forceps to carefully remove the attachment filaments from the fertilized egg clusters to obtain individual embryos (Figure 1A).
- Stage the embryos according to Iwamatsu 200413.
- Culture 20 – 30 embryos per 60 mm plastic Petri dish in a 28 °C incubator. Change the medium daily to ensure normal development of the embryos.
NOTE: The time around the hatching stage (8 – 9 d postfertilization, DPF) is especially critical for survival. Remove free-floating chorions to keep the medium clean and to ensure good larval survival rates.
2. Transgenic Embryo Screening
- Use a stereomicroscope equipped with a mercury lamp for fluorescence imaging and GFP, RFP, and CFP filters to screen transgenic embryos for fluorescent reporter expression using 40X magnification.
- Visually identify osx:mCherry embryos by mCherry reporter expression in the early-forming cranial bones, such as the cleithrum, on both sides of the posterior head (Figure 1B, arrow), and the parasphenoid, at a central position in the ventral cranium (Figure 1B, arrowhead).
NOTE: Reporter expression starts from 5 DPF onwards21. - Identify ctsk:nlGFP embryos by strong nlGFP expression in the head (Figure 1C, arrow) and tail (Figure 1C, arrowhead), starting from 6 DPF.
NOTE: Endogenous osteoclasts only form after 21 DPF. nlGFP-expressing cells at this early stage (6 DPF) are not osteoclasts but other, so far uncharacterized, ctsk-positive cells24. - Identify RANKL:HSE:CFP transgenic embryos by ubiquitous CFP expression after a short heat shock treatment for 20 min at 39 °C, conducted at 2 DPF or later for screening purposes.
NOTE: The RANKL and CFP transgenes are under the control of the same bidirectional Heat Shock Element (HSE). CFP expression indicates successful RANKL induction24. - Perform a 1.5 – 2 h heat shock treatment at 9 DPF or later to induce large numbers of ectopic osteoclasts in the trunk region, which consequently results in an osteoporosis-like phenotype24.
NOTE: Transgenic RANKL expression induced at 9 DPF results in an ectopic activation of dormant osteoclast progenitor cells, which endogenously is not triggered before 21 DPF. Use a water bath to obtain stable 39 °C conditions. Let the Petri dish containing medaka embryos float on the water surface. Make sure the lid of the petri dish is dry to prevent the sinking of the dish. - Screen embryos from compound lines, such as RANKL:HSE:CFP/ctsk:nlGFP double-transgenic and osx:mCherry/RANKL:HSE:CFP/ctsk:nlGFP triple-transgenic, according to the expression pattern of each individual transgene.
NOTE: Hemizygous and homozygous transgenic embryos were distinguished by different fluorescence levels of the reporter transgene. Homozygous embryos had a fluorescence intensity that was approximately doubled compared to that of hemizygous transgenics. Compound lines that were homozygous for both RANKL:HSE:CFP and ctsk:nlGFP were obtained by repeated incrossing over several generations. For triple-transgenic osx:mCherry/RANKL:HSE:CFP/ctsk:nlGFP fish, homozygous RANKL:HSE:CFP/ctsk:nlGFP fish were crossed with homozygous osx:mCherry carriers. The resulting heterozygous triple-transgenic progeny were raised and incrossed to obtain embryos homozygous for RANKL:HSE:CFP. The RANKL:HSE:CFP transgene must be homozygous in order to obtain the efficient induction of ectopic osteoclasts.
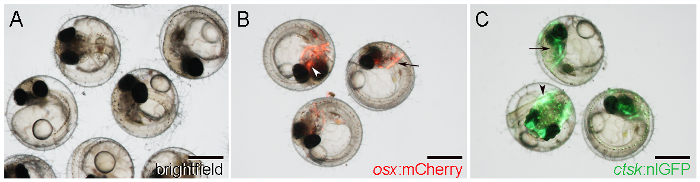
Figure 1: WT and Transgenic Medaka Embryos at 7 D Postfertilization (DPF). A. WT embryos observed with brightfield illumination. B. Transgenic embryos showing osx:mCherry expression around the cleithrum (arrow) and parasphenoid (arrowhead). C. Transgenic embryos showing ctsk:nlGFP expression in the head (arrow) and tail (arrowhead). Scale bars: 500 µm. Please click here to view a larger version of this figure.
3. Bisphosphonate Treatment of Medaka Larvae
- Prepare solutions containing different concentrations of bisphosphonates (BPs) for dose-response studies.
NOTE: The exemplary BP used in this protocol is alendronate.- Dissolve alendronate in fish medium at a concentration of 100 µg/mL to prepare a stock solution.
- Use a vortex mixer to ensure complete dissolution. Store the stock solution at 4 °C.
- Prepare different working solutions by diluting the stock solution with fish medium to a series of concentrations (i.e., 25, 37.5, 50, 62.5, and 75 µg/mL).
NOTE: Different drugs may have different Absorption, Distribution, Metabolism, and Excretion (ADME) parameters, which must be considered during testing using this medaka larvae system. Also, drug solubility and stability may vary when applied as an aqueous solution. Less water-soluble compounds might need to first be dissolved in organic solvents, such as DMSO. In this case, a stock solution is prepared in DMSO, which is then further diluted in fish medium. Note that the working solutions in water (fish medium) can be stored in the refrigerator for several wk. However, solutions containing DMSO must be stored at RT to prevent crystallization.
- Transfer medaka larvae into six-well plates (six larvae/well) for subsequent BP (alendronate) treatment.
- Remove the fish medium carefully using a clean plastic pipette and add a small volume (approx. 0.5 mL) of alendronate solution to each well.
- Avoid leftover fish medium, as the added BP solution might be diluted, which is especially critical for less-concentrated alendronate solutions.
- Remove a small volume of alendronate solution (up to 0.5 mL) from each well with a clean plastic pipette and replace it with a larger volume (4 mL) of alendronate solution.
- Change medium daily to ensure normal embryos development.
4. Live Staining of Mineralized-bone Matrix
- Dissolve 0.5 g of Alizarin complexone (ALC; alizarin-3-methyliminodiacetic acid) or 0.05 g of calcein in 50 mL of fish medium to prepare 1% and 0.1% stock solutions, respectively. Use a vortex mixer to ensure complete dissolution.
NOTE: Fish medium without the addition of methylene blue is used in this and the subsequent steps to reduce autofluorescence in the larvae. - Use a syringe and single-use filter (0.2 µm) to filter the staining solution. Store the filtered solution in the dark at RT.
NOTE: The color of the filtered, clear ALC staining solution is dark yellow to orange. The color of the filtered, clear calcein solution is bright yellow. Solutions can be used for several months. - Dilute the filtered ALC or calcein stock solution 1:10 in fish medium and incubate the medaka larvae for 1.5 – 2 h (0.1% ALC solution) or 2 – 2.5 h (0.01% calcein solution) in a 28 °C incubator if larvae between 9 and 17 DPF are used. Keep the samples in the dark.
- Transfer the larvae to fresh fish medium using a clean plastic pipette.
- Remove the fish medium with a clean plastic pipette and add fresh fish medium. Repeat this step for 3 – 4 times until no red- or yellow-stained solution (ALC or calcein, respectively) is left over. Leave the larvae in fish medium for 30 – 60 min before mounting them for imaging to avoid epifluorescence from the medium.
NOTE: 0.1% ALC staining solution is harmful to medaka larvae for extended exposure times. Incubation times of longer than 2 h affect the survival of the larvae. Concentration and staining time therefore need to be optimized for different stages in order to achieve optimal embryo survival and staining results.
5. Live Fluorescence Imaging
- Anesthetize the medaka larvae with 0.01% Tricaine (ethyl 3-aminobenzoate methanesulfonate) in fish medium.
NOTE: Anesthetized larvae become immobilized after 5 – 10 min in Tricaine solution and usually are lying either on their sides or their backs. - Use a plastic microloader to orientate the larvae according to the region of interest. The orientation of the larvae used in this protocol is lateral.
- Use a stereomicroscope with fluorescence illumination for imaging. Use high magnification when taking images, focusing on different parts of the larvae (head, anterior trunk, posterior trunk, and tail). Stitch individual images together at overlapping regions using a suitable image-processing software (insets in Figure 3G).
NOTE: This helps to improve the image quality of all relevant body parts in the correct focal plane. - Return the larvae to the fish medium for recovery after imaging.
6. Live Confocal Imaging
- Anesthetize the larvae with 0.01% Tricaine in fish medium for 5 – 10 min until they become immobilized.
- Dissolve low-melting agarose to 1.5% in fish medium by heating it in a microwave oven. Cool this solution to approximately 30 °C.
- Add 0.5 – 1 mL of liquid 1.5% low-melting agarose in fish medium to a glass-bottomed Petri dish. Transfer the anesthetized larvae into the solution using a clean plastic pipette.
NOTE: Take special precaution that the temperature of the liquid low-melting agarose is low enough to not harm the larvae. - Before the agarose solidifies, use a plastic microloader to push the larvae to the bottom of the Petri dish and orientate the larvae according to the region of interest. The orientation of the larvae used in this protocol is lateral.
NOTE: The samples are ready for confocal live imaging after the agarose completely solidifies. - Use a confocal microscope to acquire images.
- Use a 543 nm laser line for mCherry and ALC staining analyses. Use a 488 nm laser line for nlGFP and calcein staining analyses.
- After imaging, add fish medium to the Petri dish and use a pair of fine syringe needles (27 G x 1½") to carefully remove the larvae from the agarose. Transfer the larvae with residually attached agarose to a Petri dish with fish medium to recover.
- Process the images using an image analysis software27.
Representative Results
Abundant egg numbers, as well as the small size of the larvae, make medaka an excellent model for drug screening. A single six-well plate was used to culture up to 36 larvae, which was sufficient to provide statistically significant data. Another big advantage of using fish for skeletal analysis is the possibility of doing live imaging. The transparency of fish larvae allows the use of fluorescent proteins to label bone cells, as well as the use of dyes that bind to bone matrix in order to visualize mineralization. Fish larvae are easy to handle, and sample preparation for imaging is simple (Figure 2).
A RANKL:HSE:CFP/ctsk:nlGFP double-transgenic line was used to visualize the ectopic formation of RANKL-induced osteoclasts. Additionally, osx:mCherry/ RANKL:HSE:CFP/ctsk:nlGFP triple-transgenic larvae were used for the simultaneous detection of premature osteoblasts and differentiated osteoclasts (Figure 3). Overview images were taken with a stereomicroscope (Figure 3A – C and F – H), while confocal microscopy was used to visualize processes at the cellular level (Figure 3D, E, I, J arrowheads). ALC-stained bone matrix along the neural arches (na) and centra (c) (Figure 3D) was used as a reference to determine the position of fluorescently labeled bone cells (Figure 3D and I).
An advantage of simultaneously visualizing osteoclasts and osteoblasts in the same intact larvae is that the antiresorptive vs. bone-anabolic activities of a tested compound can be distinguished. For this, the distribution of osteoclasts and osteoblasts is determined along pre-existing and newly formed mineralized bone matrix. Successive staining of bone matrix with ALC (red) (Figure 4A, B) followed by calcein (green) (Figure 4C, D) reveals de novo mineralized bone matrix (green) (Figure 4E, F arrowheads). This assay allows for the quantification of the rate of bone formation. Increased de novo rates after drug treatment indicate a bone-anabolic effect of the tested compound. In contrast, persistence of pre-existing bone matrix points to an antiresorptive activity of the drug27. Both ALC and calcein labels in the larvae are stable for at least two weeks, allowing a continuous observation of new bone formation in medaka larvae in vivo.
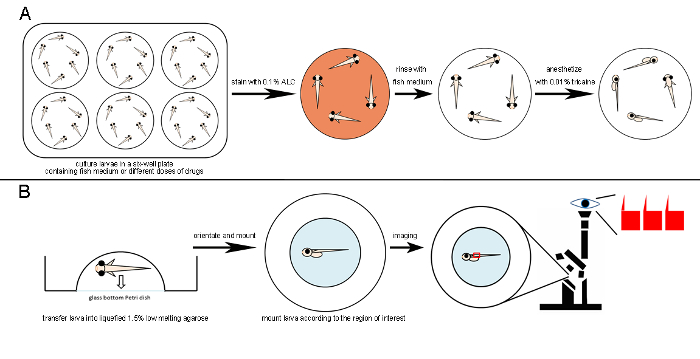
Figure 2: Schematic Diagram of Drug Treatment, Live ALC Staining, and Mounting for Confocal Live Imaging. A. A six-well plate is used for drug treatment to ensure sufficient space for the larvae. The mineralized-bone matrix is stained by incubating medaka larvae in 0.1% ALC for 1.5 – 2 h in the dark. Stained larvae are rinsed several times with fish medium. 0.01% Tricaine is used to anesthetize larvae. B. After the transfer of anesthetized larvae to lukewarm and liquid 1.5% low-melting agarose, their position is adjusted with a plastic microloader. The larvae are then mounted according to the region of interest (e.g., the vertebrate column is best imaged in the lateral view). After the agarose has solidified, the mounted sample is ready for confocal imaging. Please click here to view a larger version of this figure.
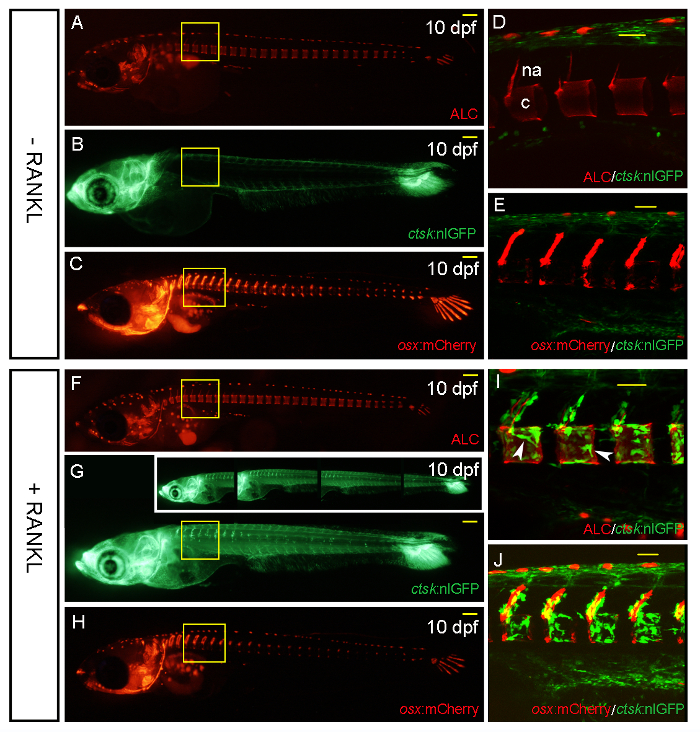
Figure 3: ctsk:nlGFP and osx:mCherry Expression in Transgenic Medaka Larvae at 10 DPF, without and after Heat Shock-induced RANKL Expression. A-C. Control larvae without RANKL induction. ALC-stained skeletal matrix (A); note the unspecific auto-fluorescence signal in the dorsally located row of pigment cells. ctsk:nlGFP expression in the head and tail (B) and the distribution of osx:mCherry-positive premature osteoblasts in the cranium, vertebral columns, and caudal fin (C). D. Confocal stack of the area boxed in A and B showing the absence of ectopic osteoclasts around the neural arches (na) and centra (c) at this developmental stage. E. Confocal stack of the area boxed in C showing osx:mCherry-expressing premature osteoblasts along the neural arches and at the edges of the centra. Osteoclasts are absent from the trunk without RANKL induction. F-J. Larvae after RANKL induction by heat shock at 9 DPF. F. ALC-stained skeletal matrix. G. ctsk:nlGFP-expressing osteoclasts forming in the vertebral column. Insets in G show individual images taken at higher magnification that are stitched together to result in the compound image in G. H. The distribution of osx:mCherry-expressing cells is not altered 1 d after RANKL induction. I. Confocal stack of the area boxed in F and G showing RANKL-induced osteoclasts forming around the neural arches, as well as the centra (arrowheads). J. Confocal stack of the area boxed in H showing ctsk:nlGFP-expressing osteoclasts next to osx:mCherry-labeled premature osteoblasts along the neural arches and the centra. Scale bars in A, B, C, F, G, and H: 100 µm. Scale bars in D, E, I, and J: 50 µm. Please click here to view a larger version of this figure.
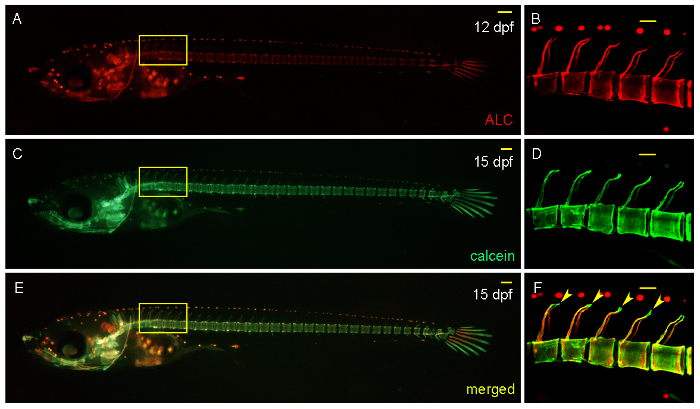
Figure 4: Analysis of De Novo Mineralization of Bone Matrix by the Successive Staining of Larvae with ALC and Calcein at 12 and 15 DPF, Respectively. A,B. ALC-stained skeletal matrix at 12 DPF. C,D. Calcein-stained skeletal matrix at 15 DPF in the same larvae. E,F. Merged image showing the newly mineralized bone matrix at the tips of the neural arches (green, arrows). B, D, and F. Confocal stack of the area boxed in A, C, and E, respectively. Scale bars in A, C, and E: 100 µm. Scale bars in B, D, and F: 50 µm. Please click here to view a larger version of this figure.
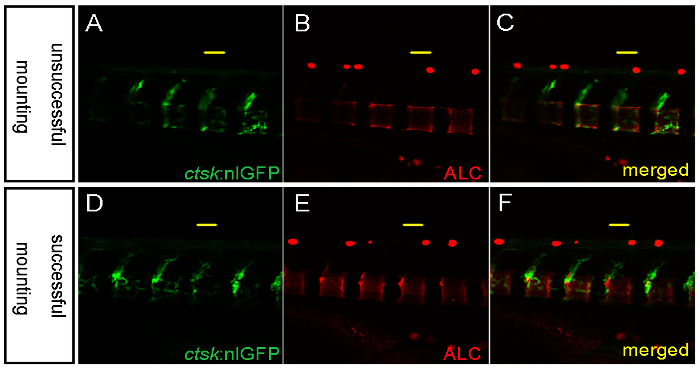
Figure 5: Representative Images Showing an Unsuccessful and a Successful Mounting for Confocal Imaging. A-C. Unsuccessful mounting in low-melting agarose results in part of the sample (left) lying outside of the focal plane. D-F. Successful mounting showing all regions of interest in the same focal plane. Scale bars: 50 µm. Please click here to view a larger version of this figure.
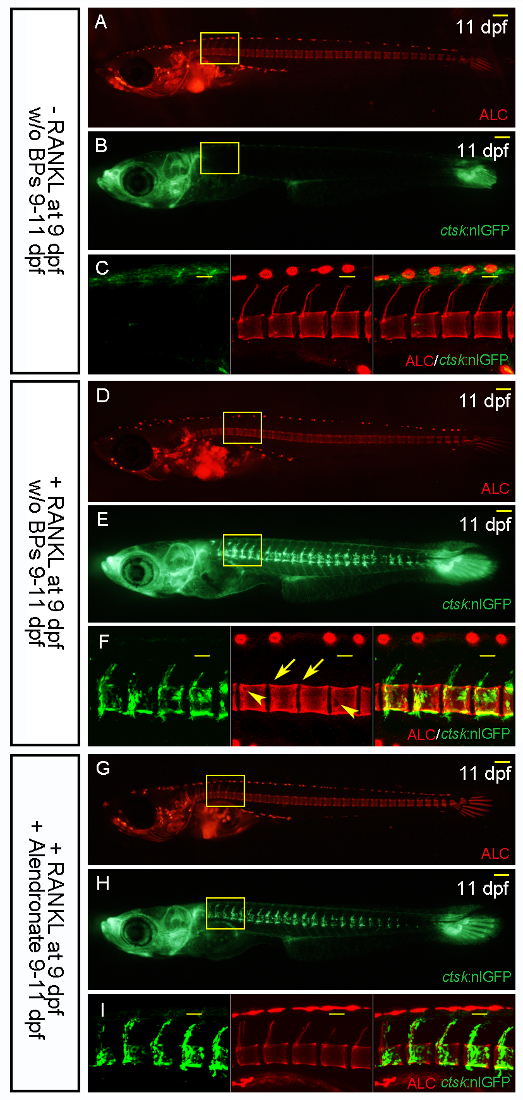
Figure 6: Alendronate Treatment Prevents Bone Resorption. ALC-stained skeletal matrix and ctsk:nlGFP-expressing osteoclasts in transgenic medaka larvae without RANKL induction (A-C), with heat shock-induced RANKL expression (D-F), and with the addition of alendronate on the same day as RANKL induction (G-I). C, F, and I. Confocal stacks of the areas boxed in A and B, D and E, G and H, respectively. C. ALC-stained intact vertebral columns without RANKL induction. F. RANKL-induced, ctsk-expressing osteoclasts form around the neural arches and the centra, resulting in the complete resorption of the mineralized matrix of the neural arches (arrows) and in defects to the centra (arrowheads). G. The addition of alendronate has no effect on the formation of ctsk:nlGFP-expressing osteoclasts, but it blocks bone resorption and leaves the neural arches and the centra intact. Scale bars in A, B, D, E, G, and H: 100 µm. Scale bars in C, F, and I: 50 µm. Please click here to view a larger version of this figure.
Discussion
Critical Steps within the Protocol
It is essential that the conditions for heat shock treatment are consistent and stable when comparing different samples. Stable temperature conditions guarantee similar levels of RANKL induction in the transgenic larvae and, consequently, comparable osteoclast formation, which can be confirmed by screening for ctsk:nlGFP expression. Ultimately, this leads to a similar degree of induced ectopic bone resorption and osteoporosis-like lesions, as validated by ALC staining. Such an experimental design then allows for the determination and comparison of the effects of various anti-resorptive drugs or the same drug applied at different concentrations.
Modifications and Troubleshooting
Fish larvae are very fragile and must be carefully handled during the mounting process for imaging. For optimal live confocal imaging, larvae are carefully positioned with a plastic microloader, rather than forceps, to avoid injuries (Figure 2B). The yolk extension is less sensitive to touch stimuli and can be used to orientate the larvae with a microloader, thereby avoiding larval movement during mounting. The region of interest should be mounted flat to ensure that the sample can be focused in one focal plane during imaging (Figure 5). Bone cell behavior is very dynamic and requires high temporal and spatial resolution during live imaging. Optimally, time-lapse imaging by confocal analysis is desired to follow osteoblast-osteoclast interactions over the course of several hours in the living larvae. However, this demands special conditions to keep the larvae alive and in a fixed position. For prolonged anesthesia, a solution of 0.001% Tricaine in fish medium is added to cover the solidified agarose during imaging. This prevents the agarose from drying out and prevents sudden twitching of the larvae during imaging over several hours.
Limitations of the Technique
Osteoporosis is a disease that mostly affects elderly humans. Therefore, an ideal in vivo model for osteoporosis drug screening should involve adult fish. So far, however, the technique of live imaging described in this report is limited to the embryonic and larval stages. Currently available confocal imaging does not allow live imaging of genetically labeled bone cells in adult fish due to limitations in penetration depth. The recent development of Light Sheet Fluorescence Microscopy (LSFM), which allows imaging deep into living tissue, together with the availability of less pigmented "see-through" medaka strains, makes it conceivable that this limitation will soon be overcome, and live imaging of bone cells in adult medaka fish after drug screening will be possible.
Significance of the Technique with Respect to Existing/Alternative Methods
A unique combination of live imaging and advanced genetic tools has made small teleost fish popular for biomedical research — bone research in particular — and as in vivo models for drug screening. With the possibility to analyze cell-cell interactions within an intact organism, transgenic medaka lines are uniquely suited for the live imaging of dynamic cellular processes during bone modeling and remodeling. Because they share many gene-regulatory networks for controlling bone homeostasis with mammals, in vivo studies in medaka can complement and even exceed in vitro studies using mammalian osteoblast and osteoclast cell-culture assays.
Future Applications or Directions after Mastering This Technique
Other than testing single compounds in a transgenic background, as described in this protocol, fish also offer simple approaches to examine combinations of various drugs for possible synergy or interference of compounds. Furthermore, the use of a combination of different reporter lines, for example, in the context of a particular mutant background, provides ample opportunities to study the behavior of bone cells during different stages of bone degeneration and repair, or in the presence of a bone-modulating drug. With its unique features that complement mouse assays, the medaka osteoporosis model provides an excellent in vivo approach to test and quantify the anabolic and antiresorptive effects of novel bone-modulating compounds.
開示
The authors have nothing to disclose.
Acknowledgements
This project was funded by grants from the Singapore Ministry of Education (MOE, grant number 2013-T2-2-126) and the National Institute of Health, USA (NIH, grant number 1R21AT008452-01A1). T.Y. received a graduate scholarship from the NUS Department of Biological Sciences. We thank the confocal unit of the NUS Centre for Bioimaging Sciences (CBIS) for their constant support.
Materials
| Alendronate | Sigma | A4978 | |
| alizarin-3-methyliminodiacetic acid, Alizarin Complexone | Sigma | A3882 | |
| Calcein | Sigma | C0875 | |
| ethyl 3-aminobenzoate methanesulfonate (Tricaine) | Sigma | A5040 | |
| ImageJ (1.4.3.67) | National Institute of Health (NIH) | https://imagej.nih.gov/ij/ | |
| LSM 510 Meta confocal | Zeiss | ||
| LSM Image Browser (4.2.0.121) | Zeiss | http://www.zeiss.com/microscopy/en_de/downloads/lsm-5-series.html | |
| Micro-loader | Eppendorf | 5242956003 | Eppendorf ep T.I.P.S 20 μl |
| NIS-Elements BR 3.0 software | Nikon | ||
| Photoshop CS6 (13.0.0.0) | Adobe | ||
| SMZ1000 stereomicroscope | Nikon |
参考文献
- Ablain, J., Zon, L. I. Of fish and men: using zebrafish to fight human diseases. Trends Cell Biol. 23 (12), 584-586 (2013).
- Ansai, S., Kinoshita, M. Targeted mutagenesis using CRISPR/Cas system in medaka. Biol Open. 3 (5), 362-371 (2014).
- Apschner, A., Schulte-Merker, S., Witten, P. E. Not all bones are created equal-using zebrafish and other teleost species in osteogenesis research. Methods Cell Biol. 105, 239-255 (2011).
- Bajoghli, B., Aghaallaei, N., Heimbucher, T., Czerny, T. An artificial promoter construct for heat-inducible misexpression during fish embryogenesis. Dev Biol. 271 (2), 416-430 (2004).
- Barrett, R., Chappell, C., Quick, M., Fleming, A. A rapid, high content, in vivo model of glucocorticoid-induced osteoporosis. Biotechnol J. 1 (6), 651-655 (2006).
- Centanin, L., Ander, J. J., Hoeckendorf, B., Lust, K., Kellner, T., Kraemer, I., Urbany, C., Hasel, E., Harris, W. A., Simons, B. D., et al. Exclusive multipotency and preferential asymmetric divisions in post-embryonic neural stem cells of the fish retina. Development. 141 (18), 3472-3482 (2014).
- Charles, J. F., Aliprantis, A. O. Osteoclasts: more than ‘bone eaters. Trends Mol Med. 20 (8), 449-459 (2014).
- DeLaurier, A., Eames, B. F., Blanco-Sanchez, B., Peng, G., He, X., Swartz, M. E., Ullmann, B., Westerfield, M., Kimmel, C. B. Zebrafish sp7:EGFP: a transgenic for studying otic vesicle formation, skeletogenesis, and bone regeneration. Genesis. 48 (8), 505-511 (2010).
- Du, S. J., Frenkel, V., Kindschi, G., Zohar, Y. Visualizing normal and defective bone development in zebrafish embryos using the fluorescent chromophore calcein. Dev Biol. 238 (2), 239-246 (2001).
- Eriksen, E. F. Cellular mechanisms of bone remodeling. Rev Endocr Metab Disord. 11 (4), 219-227 (2010).
- Hockendorf, B., Thumberger, T., Wittbrodt, J. Quantitative analysis of embryogenesis: a perspective for light sheet microscopy. Dev Cell. 23 (6), 1111-1120 (2012).
- Inohaya, K., Takano, Y., Kudo, A. The teleost intervertebral region acts as a growth center of the centrum: in vivo visualization of osteoblasts and their progenitors in transgenic fish. Dev Dyn. 236 (11), 3031-3046 (2007).
- Iwamatsu, T. Stages of normal development in the medaka Oryzias latipes. Mech Dev. 121 (7), 605-618 (2004).
- Kirchmaier, S., Hockendorf, B., Moller, E. K., Bornhorst, D., Spitz, F., Wittbrodt, J. Efficient site-specific transgenesis and enhancer activity tests in medaka using PhiC31 integrase. Development. 140 (20), 4287-4295 (2013).
- Kirchmaier, S., Naruse, K., Wittbrodt, J., Loosli, F. The genomic and genetic toolbox of the teleost medaka (Oryzias latipes). 遺伝学. 199 (4), 905-918 (2015).
- Komori, T. Animal models for osteoporosis. Eur J Pharmacol. 759, 287-294 (2015).
- Mackay, E. W., Apschner, A., Schulte-Merker, S. A bone to pick with zebrafish. Bonekey Rep. 2, 445 (2013).
- MacRae, C. A., Peterson, R. T. Zebrafish as tools for drug discovery. Nat Rev Drug Discov. 14 (10), 721-731 (2015).
- Mitchell, R. E., Huitema, L. F., Skinner, R. E., Brunt, L. H., Severn, C., Schulte-Merker, S., Hammond, C. L. New tools for studying osteoarthritis genetics in zebrafish. Osteoarthritis Cartilage. 21 (2), 269-278 (2013).
- Renn, J., Buttner, A., To, T. T., Chan, S. J., Winkler, C. A col10a1:nlGFP transgenic line displays putative osteoblast precursors at the medaka notochordal sheath prior to mineralization. Dev Biol. 381 (1), 134-143 (2013).
- Renn, J., Winkler, C. Osterix-mCherry transgenic medaka for in vivo imaging of bone formation. Dev Dyn. 238 (1), 241-248 (2009).
- Schilling, T. F., Kimmel, C. B. Segment and cell type lineage restrictions during pharyngeal arch development in the zebrafish embryo. Development. 120 (3), 483-494 (1994).
- Spoorendonk, K. M., Peterson-Maduro, J., Renn, J., Trowe, T., Kranenbarg, S., Winkler, C., Schulte-Merker, S. Retinoic acid and Cyp26b1 are critical regulators of osteogenesis in the axial skeleton. Development. 135 (22), 3765-3774 (2008).
- To, T. T., Witten, P. E., Renn, J., Bhattacharya, D., Huysseune, A., Winkler, C. Rankl-induced osteoclastogenesis leads to loss of mineralization in a medaka osteoporosis model. Development. 139 (1), 141-150 (2012).
- Wakamatsu, Y., Pristyazhnyuk, S., Kinoshita, M., Tanaka, M., Ozato, K. The see-through medaka: a fish model that is transparent throughout life. Proc Natl Acad Sci USA. 98 (18), 10046-10050 (2001).
- Witten, P. E., Huysseune, A. A comparative view on mechanisms and functions of skeletal remodelling in teleost fish, with special emphasis on osteoclasts and their function. Biol Rev Camb Philos Soc. 84 (2), 315-346 (2009).
- Yu, T., Witten, P. E., Huysseune, A., Buettner, A., To, T. T., Winkler, C. Live imaging of osteoclast inhibition by bisphosphonates in a medaka osteoporosis model. Dis Model Mech. 9 (2), 155-163 (2016).

