Affinity Precipitation of Active Rho-GEFs Using a GST-tagged Mutant Rho Protein (GST-RhoA(G17A)) from Epithelial Cell Lysates
概要
The method presented here describes an assay to follow activation of RhoA specific GDP/GTP Exchange Factors (GEFs) in cultured cells by making use of a mutant RhoA GST fusion protein that has high affinity for activated GEFs. GEFs are precipitated from cell lysates, detected by Western blotting and quantified by densitometry.
Abstract
Proteins of the Rho family of small GTPases are central regulators of the cytoskeleton, and control a large variety of cellular processes, including cell migration, gene expression, cell cycle progression and cell adhesion 1. Rho proteins are molecular switches that are active in GTP-bound and inactive in GDP-bound state. Their activation is mediated by a family of Guanine-nucleotide Exchange Factor (GEF) proteins. Rho-GEFs constitute a large family, with overlapping specificities 2. Although a lot of progress has been made in identifying the GEFs activated by specific signals, there are still many questions remaining regarding the pathway-specific regulation of these proteins. The number of Rho-GEFs exceeds 70, and each cell expresses more than one GEF protein. In addition, many of these proteins activate not only Rho, but other members of the family, contributing further to the complexity of the regulatory networks. Importantly, exploring how GEFs are regulated requires a method to follow the active pool of individual GEFs in cells activated by different stimuli. Here we provide a step-by-step protocol for a method used to assess and quantify the available active Rho-specific GEFs using an affinity precipitation assay. This assay was developed a few years ago in the Burridge lab 3,4 and we have used it in kidney tubular cell lines 5,6,7. The assay takes advantage of a “nucleotide free” mutant RhoA, with a high affinity for active GEFs. The mutation (G17A) renders the protein unable to bind GDP or GTP and this state mimics the intermediate state that is bound to the GEF. A GST-tagged version of this mutant protein is expressed and purified from E. coli, bound to glutathione sepharose beads and used to precipitate active GEFs from lysates of untreated and stimulated cells. As most GEFs are activated via posttranslational modifications or release from inhibitory bindings, their active state is preserved in cell lysates, and they can be detected by this assay8. Captured proteins can be probed for known GEFs by detection with specific antibodies using Western blotting, or analyzed by Mass Spectrometry to identify unknown GEFs activated by certain stimuli.
Protocol
1. Transformation of E. coli with the pGEX-RhoA(G17A) Construct
- Prepare LB-Agar by dissolving 2.5 g LB and 1.5 g Agar in 100 ml dH2O. Autoclave and cool to an estimated 50-55 °C, which as a rule of thumb, is when the flask can be held comfortably.
- Prepare Ampicillin (Amp) stock by dissolving 50 mg/ ml in dH2O. Syringe filter and freeze unused aliquots. Add 100 μl of Amp stock (final concentration 50 μg/ml) to the LB-Agar from 1.1. Swirl to mix and pour into 10 cm bacterial dishes (15-20 ml/dish). Allow it to solidify (15-30 min.) and store unused plates inverted at 4 °C for 2-3 weeks.
- To transform E. Coli, quickly thaw an aliquot of DH5α competent cells in an ice bath. Add 1 μl of pGEXRhoA(G17A) DNA diluted to 25-50 ng/μl. Flick the tube to mix and incubate on ice for 30 minutes. Heat shock at 42 °C for 45 seconds and place back on ice for 2 minutes. Add 900 μl SOC medium and grow for one hour at 37 °C with shaking.
- Spread 50-100 μl of the transformed bacteria on an LB-Agar-Amp plate using a bent sterile Pasteur pipette. Incubate the plate right side up in a 37 °C incubator for 5 minutes and then invert and grow overnight.
- A single colony will be picked from the plate for preparation of the GST-tagged protein (step 2.1). For future use, wrap and store plates inverted at 4 °C for about 3 weeks. In addition, bacterial stocks can be prepared for more prolonged storage by growing individual colonies in 2 ml sterile LB-Amp overnight at 37 °C with shaking. Mix an aliquot with sterile 80% glycerol in a 1:1 ratio and freeze at -80 °C.
2. Preparation of GST-RhoA(G17A) Beads
- Prepare LB by adding 25 g LB to 1 L dH20 and autoclaving. When cool, add 50 μl Amp from stock to 50 ml LB (50 μg/ml final concentration). Inoculate with a well isolated colony of transformed bacteria and grow overnight at 37 °C with agitation. When at full density (OD600 > 1.0) dilute with 450 ml LB-Amp and grow for an additional 30 minutes at 37 °C.
- Prepare a 100 mM stock solution of Isopropyl B-D-thiogalactopyranoside (IPTG) by dissolving 0.238 g in 10 ml dH2O. Store in aliquots at -20 °C. Induce bacteria to produce Rho protein by adding 500 μl 100 mM IPTG to 500 ml culture (a final concentration of 100 μM). Reduce temperature to 22-24 °C and grow for ~16 h hours.
- Spin culture at 3600 g for 10 minutes at 4 °C. If needed, the 500 ml culture can be divided into 50 ml tubes for centrifugation. Freeze pellet(s) for at least 1 hour (or preferably overnight) at -80 °C.
- Prepare 200 ml lysis buffer containing 20 mM HEPES (0.95 g)/ pH 7.5; 150 mM NaCl (1.75 g); 5 mM MgCl2 (0.203 g); 1% TX-100 (2 ml). Prepare stock solutions of 1M DTT (1.542 g in 10 ml dH2O) and 100 mM PMSF (0.174 g/10 ml EtOH). To prepare lysis buffer +, supplement 10 ml with 1mM DTT (10 μl of stock) and 1 mM PMSF (100 μl of stock) and one Complete Mini Protease Inhibitor tablet.
- Working on ice, add 10 ml lysis buffer+ to the pellets from step 2.3. Resuspend thoroughly by gentle vortexing and pipetting. Avoid foaming. Sonicate on ice for 1 minute at setting 4 with 50% pulse. Spin the sonicated lysate at 15,000-20,000 g for 15 minutes at 4 °C, and remove the clarified sonicate (supernatant) to a sterile capped 15 ml tube.
- Prepare the Glutathione Sepharose by gently mixing the original tube containing a 75% slurry and transfer 335 μl into a 15 ml tube. Use a wide bore tip to pipette beads. Add 10 ml cold PBS, and spin 500 g for 5 minutes at 4 °C. Discard the supernatant, add 1 ml lysis buffer+ to the beads and spin as for previous wash. Discard the supernatant and add lysis buffer+ to make a 50% slurry.
- Add 250 μl of equilibrated bead slurry to the supernatant from step 2.5. Rotate at 4 °C for 45 minutes.
- Prepare 500 ml HBS containing 20 mM HEPES (2.38 g)/pH 7.5 and 150 mM NaCl (4.38 g) in dH2O. Prepare stock solutions of 1M MgCl2 (0.952 g in 10 ml dH2O) and 1M DTT (1.542 g in 10 ml dH2O). To prepare HBS+, supplement 100 ml just before use with 5 mM MgCl2 (50 μl from stock) and 1 mM DTT (100 μl from stock).
- Spin the beads from step 2.7 at 500 g for 5 minutes at 4 °C. Discard the supernatant and wash beads 2x with 10 ml lysis buffer+, and 2x with 10 ml HBS+. After the final wash, make a 50% slurry by resuspending the beads in HBS+ supplemented with BD BaculoGold protease inhibitor (20 μl of 50x BD BaculoGold/ml).
- Dilute 10 μl of the final beads preparation with 2x Laemmli sample buffer containing β-mercaptoethanol. Make Bovine Serum Albumin (BSA) standards. Use a 2 mg/ml stock (0.02 g of BSA in 10 ml dH2O). Mix 10 μl of stock with 10 μl Laemmli (20 μg final); 5 μl of stock with 5 μl of dH2O and 10 μl Laemmli (10 μg final); and 2.5 μl stock with 7.5 μl dH2O and 10 μl Laemmli (5 μg final). Boil all samples (5 min). Spin the bead sample and run supernatant with BSA standards and molecular weight markers on a 10% SDS-polyacrylamide gel.
- Prepare the Comassie Blue stain (0.1 g in 10 ml Acetic Acid, 40 ml Methanol and 50 ml dH2O) and the destain solution (500 ml dH2O, 400 ml methanol and 100 ml acetic acid). Store at room temperature. Stain the gel for 20-30 minutes, remove the dye (it can be reused multiple times) and rinse with destain solution twice. Continue to destain with gentle shaking for several hours until bands are clearly visible.
- Estimate the concentration of GST-RhoA(G17A) coupled to the beads using the BSA standards as a reference (Fig 2). Aliquot an equal volume of beads containing ~10-15 μg protein into 1.5 ml micro centrifuge tubes. Store beads at 4 °C to use within a day. Freeze at -80 °C in HBS+/glycerol in a 3:1 ratio to use within a few days.
3. GEF Pulldown Assay with Nucleotide-free RhoA(G17A) Beads
- Grow cells in 10 cm dishes to confluence. Serum deprive for at least 3 hours and treat as required.
- Prepare lysis buffer+ as in step 2.4. Prepare enough lysis buffer for 700 μl/dish plus some extra amount to allow for pipetting errors. Add the protease inhibitors just before use.
- Working on ice, remove culture medium from the dishes and wash with ice-cold HBS. Remove all the HBS and add 700 μl lysis buffer+ to each dish. Swirl plates to cover all areas, scrape and collect lysates into numbered 1.5 ml tubes. Spin at 15,000 g for 1 min at 4 °C. The supernatant will be used for the assay.
- If your cell number is equivalent in all dishes being tested, you can omit doing a protein assay, and move to step 3.5. Otherwise measure the protein concentration of each supernatant using Bio-Rad quick protein assay and equalize the supernatant for volume and concentration. The amount of total protein depends on the cell types used (typically 1-1.5 mg protein for LLC-PK1 cells).
- Remove 30 μl of each supernatant and mix with 30 μl 2x reducing Laemmli sample buffer, boil and set aside for step 3.7. Add remaining supernatants to aliquots of the GST-RhoA(G17A) beads from step 2.12. Rotate for 45 minutes at 4 °C.
- Spin beads at 6800 g for 10 seconds at 4 °C. Discard the supernatant and wash the beads 3x with lysis buffer, spinning in the same way between washes. Completely remove the final wash using a 1 cc syringe fitted with a 30 G needle and add 20 μl 2x reducing Laemmli sample buffer. Boil for 5 min. Spin to pellet beads and either run the supernatant immediately (preferable) or store it at -80 °C for later analysis.
- Run 20 μl total cell lysates and all of the precipitated protein samples on the appropriate percentage SDS-polyacrylamide gel for the size of GEF you are studying. Detect your GEF of choice by Western blotting using a specific antibody.
4. Representative Results
Part 1 and 2 of the protocol describes preparation of GST-RhoA(G17A) coupled to GSH-sepharose beads and its testing by SDS-PAGE (see outline of protocol on Fig 1). A typical Coomassie stained gel is shown on Fig 2. The sample with the eluted protein should contain a single band at approximately 50 kDa (Fig 2, lane 6). The concentration of the protein can be estimated using the BSA reference samples. In the example on Fig 2, the concentration of RhoA(G17A) is estimated to be 15 μg/10 μl. Thus, aliquots of 10 μl/tube were prepared. The typical yield in our hand is 15-20 μg protein from 10 ml bacterial lysis. Part 3 of the protocol describes the affinity precipitation assay (see overview on Fig 1). A successful GEF assay detecting activation of the exchange factor GEF-H1 is shown on Fig 3. The RhoA(G17A) protein captured some GEF-H1 from the control (untreated) cell lysates, suggesting that GEF-H1 has basal activity. The amount precipitated however increases in cells treated with the inflammatory cytokine Tumor Necrosis Factor -α (TNF- α), consistent with the notion that TNF-α activates GEF-H1 5,7. Importantly, the total cell lysates show similar amounts of GEF-H1 in the control and the treated sample, suggesting that the treatment did not alter GEF-H1 levels and the input used in the assay is equal.
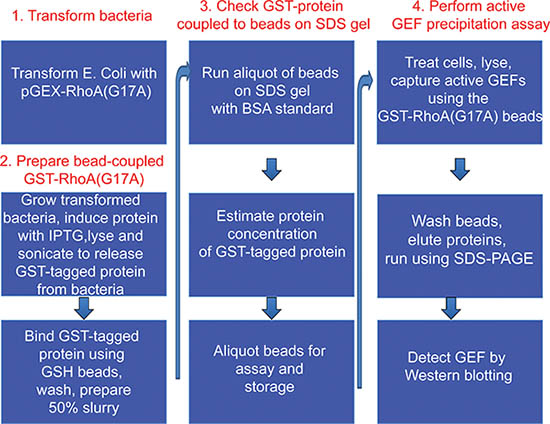
Figure 1. Overview of the protocol.
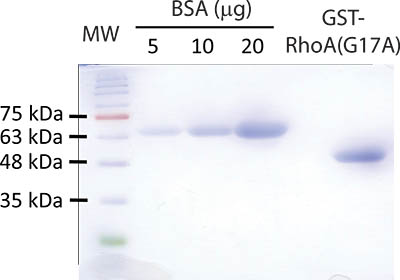
Figure 2. Representative result of the bead preparation protocol. A Coomassie stained gel with successful GST-RhoA(G17A) bead preparation is shown. Bead sample and BSA protein standards were separated by SDS-PAGE using a 10% acrylamide gel. To test the beads, 10 μl of the final bead slurry containing GST-RhoA(G17A) is diluted 1:1 with reducing Laemmli sample buffer and boiled for 5 minutes. The beads were spun briefly and the supernatant loaded on the gel. The following samples were loaded: Lane 1: molecular weight marker (MW) (FroggaBio BLUeye prestained protein ladder); Lanes 2-4: 5, 10 and 20 μg Bovine Serum Albumin (BSA); Lane 5 is empty; Lane 6: 10 μl of the freshly prepared RhoA(G17A) beads. After separation is completed, the gel is stained using Coomassie Blue and subsequently destained to reveal proteins. The molecular weight of the GST-RhoA(G17A) protein is roughly 50 kDa and runs around the level of the 48 kDa marker. The concentration of the GST-Rho protein in this particular sample is estimated to be around 15 μg/10 μl slurry.
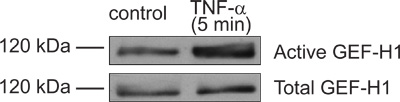
Figure 3. Representative GEF activation assay showing TNF-α-induced activation of GEF-H1. Confluent LLC-PK1 cells were treated with 10 ng/ml TNF-α for 5 minutes. Following treatment the cells were lysed and active GEFs were captured using GST-RhoA(G17A) bound beads. The presence of GEF-H1 in the precipitated proteins (top blot) and total cell lysate samples (bottom blot) was detected using Western blotting with an anti-GEF-H1 antibody (Cell Signaling). Please note the increased amount of precipitated GEF-H1 in TNF-α-treated versus non-treated cells relative to the equivalent inputs, indicating a successful result.
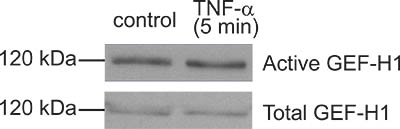
Figure 4. A “bad result”. Although the GEF pulldown assay shown here resulted in some GEF-H1 captured by the beads, the amounts precipitated from control and TNF-α-treated cells are the same. Thus TNF-α, a know activator of GEF-H1 in this case did not induce activation. In this particular experiment subsequent troubleshooting suggested that the TNF-α used was not fresh enough and probably degraded.
Discussion
The method presented here is the only available non-radioactive activation assay for GEFs that can follow the active pool of GEFs in cells. The assay is similar to the precipitation assays used for following activation of small GTPases as well as GEFs against Rac and Cdc42. Those assays use different GST-tagged proteins and have slight differences from the one described here, however the basic steps are the same. Thus, this protocol can easily be adapted for other small GTPase and GEF activation assays.
The presented GEF assay was recently modified for application for nuclear fractions 9,10. With further modifications, testing of GEF activation in other subcellular compartments might also be possible.
We use the presented method to study activation of GEFs in epithelial cell lines5,6,7. With some optimization, this assay should be adequate to detect GEFs from any cell line. When adapting to a specific cell type, find the optimal cell number, lysis buffer volume, and detection method for the GEF to be tested (a good antibody for Western blotting is important). For initial setup of the assay it is advisable to use a stimulus that is known to activate the GEF of interest. When using an unknown stimulus, always use a positive control to verify that your assay is working. This assay can be used to detect activation of known GEFs by Western blotting. However, it is also adequate to identify unknown GEFs. For this, captured GEFs from control and stimulated samples should be analyzed on a Coomassie-stained gel. Bands that appear only in stimulated samples might contain activated GEFs and can be sent for identification by Mass Spectrometry (e.g. 5).
Finally, a cautionary note. The assay is based on the assumption that the posttranslational modifications rendering GEFs active are preserved after cell lysis. Indeed, this is clearly the case for a number of GEFs, and since its establishments, this assay has been used by various groups to detect activation of different GEFs, including GEF-H1, p115RhoGEF and XPLN (e.g. 5,11,12,13). Preservation of the active state however might not happen in the case of all GEFs, and therefore it is conceivable that this assay will not work for all GEFs. It also has to be noted, that many GEFs exert activity towards more than one small GTPases. Thus, when a specific GEF is studied, it is recommended to complement this assay with GEF precipitation assays for other small GTPases, as well as functional studies examining RhoA, Rac1 and Cdc42.
Critical steps in the protocol:
Colonies of transformed bacteria should be picked from fresh, properly prepared plates to ensure adequate selection by Amp, good outgrowth and yield. Transformation conditions for competent cells obtained from other sources may vary and should be consulted.
All steps of the protein preparation protocol (from step 2.3) and the assay (from step 3.2) should be performed at 4°C with cooled solutions and centrifuges.
Bacterial lysis (step 2.5) should be thorough and complete in order to obtain a homogeneous suspension. When lysing the bacteria, vortex and pipette the lysate alternately, while maintaining it at 4°C and ensure sonication is done on ice to prevent denaturing the protein. If using a different model of sonicator, conditions may need to be adjusted. Incubation of sonicate with the beads should always be done at 4°C on a rotator to ensure sufficient binding, and care should be taken to keep the timing consistent.
GST-Rho mutants are somewhat unstable when expressed in bacteria, so it is best to use prepared beads right away or within a few days.
The precipitation assay (Part 3) is time and temperature sensitive, as active GEFs can be easily lost from the cell lysate, so steps should be performed as quickly as possible.
The following section contains some trouble-shooting tips.
No or very low amount of mutant Rho protein in the final bead preparation: This may be caused by inefficient induction, insufficient lysis of the bacteria, or a loss of the protein during the preparation process or storage. To help troubleshoot some of these possibilities, samples of bacteria can be analyzed before and after induction. If there is poor induction of the protein repeat the process using a colony from a freshly streaked plate or from re-transformed competent cells. Different IPTG concentrations and induction times should also be tested. If the lysis is insufficient (i.e. the protein remains in the pellet instead of the supernatant) varying salt and detergent concentrations in the lysis buffer can be tried. Alternate sonication times and settings should be considered, and samples before and after sonication can be checked by microscopy to determine efficiency of lysis.
No precipitated GEF, even though the GST-protein is present on the beads: This may be due to technical issues during the precipitation assay, or by a real absence of activation of the studied GEF using the stimulus applied. Always use a known stimulus as a positive control to verify that your assay works. Use the prepared beads within a few days. If the precipitation assay captures undetectable amounts of the GEF studied in all conditions, verify that your GEF is present and is well detectable in the supernatant after centrifugation that is to be used for the assay (step 3.3). Make sure all buffers and protease inhibitors are fresh, and perform all steps on ice as fast as possible. Increase the amount of input protein (e.g. by using lysates from 2 plates/sample). If the precipitation assay shows basal precipitation of your GEF, but no difference is seen between the control and stimulated samples (Fig 4), start troubleshooting by verifying that the applied stimulus worked using other known effects (e.g. by detecting activation of other signaling pathways). Consider changing the treatment conditions and/or concentrations. Rely on data from the literature reporting how your stimulus activates Rho or other signaling to predict likely times and concentrations. When optimizing treatment time, use both short and long time points, as GEF activation might be best detectable at a time point prior to well detectable Rho activation. Finally, the same stimulus could result in a variable degree of activation due to a change in cell responsiveness caused by passage number, cell confluency, etc.
開示
The authors have nothing to disclose.
Acknowledgements
This work was funded by the Canadian Institute for Health Research (CIHR) (MOP-97774) and the Natural Sciences and Engineering Research Council of Canada (NSERC, grant nr: 480619). KS is a recipient of a KRESCENT New Investigator Award (a joint award of the Kidney Foundation of Canada, Canadian Nephrology Society and Canadian Institute of Health Research) and an Early Researcher Award from the Ontario Ministry of Research and Innovation. FW is supported by a Li Ka Shing scholarship.
Materials
| Name of reagent | Company | Catalogue number | Comments (optional) |
| pGEX-RhoA(G17A) | Addgene | Will be available soon | Generated in the lab of Dr. Keith Burridge (University of North Carolina) (ref 3) |
| DH5α | Invitrogen | 18258-012 | Store -80°C |
| SOC medium | BioShop | SOC001.10 | |
| LB | BioShop | LBL407.1 | Keep sterile |
| Glycerol | BioShop | GLY002.1 | |
| Ampicillin | BioShop | AMP201.25 | Store stock at -20°C |
| Agar | BioShop | AGR001.500 | |
| IPTG ( Isopropyl B-D-thiogalacto-pyranoside) | Calbio-chem | 420322 | Store stock solution at -20°C |
| Glutathione Sepharose beads | GE Healthcare | 17-0756-01 | |
| PBS 10x | SIGMA | D1408 | |
| Hepes | SIGMA | H4034 | |
| NaCl | BioShop | SOD001.10 | |
| MgCl2 | SIGMA | M-9272 | Add just before use |
| TX-100 | SIGMA-ALDRICH | X100 | |
| DTT | OmniPur EMD | 3860 | Add just before use |
| PMSF | SIGMA | P-7626 | Add just before use |
| Protease Inhibitor 50x | BD Baculo-Gold | 51-21426Z | Add just before use |
| Complete Mini, EDTA-free 10x | Roche | 11 836 170 001 | Add just before use |
| Laemmli Sample Buffer 2x | Bio-Rad | 161-0737 | |
| β-mercaptoethanol | SIGMA | M7154 | Add just before use |
| Coomassie Brilliant Blue | Bio-Rad | R-250 | |
| Acetic Acid | CALEDON | 1000-1 | |
| Methanol | CALEDON | 6701-7 | |
| Bio-Rad Protein Assay | Bio-Rad | 500-0114 | |
| BLUeye ladder | FroggaBio | PM008-0500 |
Table 1. Reagents used.
| Name of equipment | Company | Catalogue number | Comments (optional) |
| RC-5B centrifuge | Sorvall | SS-34 Rotor | Use at 4°C |
| Centrifuge | Sorvall-Thermo Scientific | ST-16R | Use at 4°C |
| Micro centrifuge | Eppendorf | 5417R | Use at 4°C |
| Bacterial shaker | INFORS HT Ecotron | AG CH-1403 Bottmingen | Use at 37°C or at 22°C |
| Sonicator | Branson | Sonifier-450 | Use at RT |
| Rotator | Glas-Col | 099A CR4012 | Use at 4°C |
Table 2. Specific equipments used.
参考文献
- Jaffe, A. B., Hall, A. R. h. o. GTPases: biochemistry and biology. Annu. Rev. Cell Dev. Biol. 21, 247-269 (2005).
- Rossman, K. L., Der, C. J., Sondek, J. GEF means go: turning on RHO GTPases with guanine nucleotide-exchange factors. Nat. Rev. Mol. Cell Biol. 6, 167-180 (2005).
- Arthur, W. T., Ellerbroek, S. M., Der, C. J., Burridge, K., Wennerberg, K. XPLN, a guanine nucleotide exchange factor for RhoA and RhoB, but not RhoC. J. Biol. Chem. 277, 42964-42972 (2002).
- Garcia-Mata, R. Analysis of activated GAPs and GEFs in cell lysates. Methods Enzymol. 406, 425-437 (2006).
- Kakiashvili, E. GEF-H1 Mediates Tumor Necrosis Factor-{alpha}-induced Rho Activation and Myosin Phosphorylation: role in the regulation of tubular paracellular permeability. J. Biol. Chem. 284, 11454-11466 (2009).
- Waheed, F. Extracellular signal regulated kinase and GEF-H1 mediate depolarization-induced Rho activation and paracellular permeability increase. Am. J. Physiol. Cell Physiol. , (2010).
- Kakiashvili, E. The epidermal growth factor receptor mediates tumor necrosis factor-alpha-induced activation of the ERK/GEF-H1/RhoA pathway in tubular epithelium. J. Biol. Chem. 286, 9268-9279 (2011).
- Garcia-Mata, R., Burridge, K. Catching a GEF by its tail. Trends Cell Biol. 17, 36-43 (2007).
- Dubash, A. D. The small GTPase RhoA localizes to the nucleus and is activated by Net1 and DNA damage signals. PLoS One. 6, e17380-e17380 (2011).
- Guilluy, G., Dubash, A. D., García-Mata, R. Analysis of RhoA and Rho GEF activity in whole cells and the cell nucleus. Nature Protocols. , (2011).
- Guilluy, C., Swaminathan, V., Garcia-Mata, R., O’Brien, E. T., Superfine, R., Burridge, K. The Rho GEFs LARG and GEF-H1 regulate the mechanical response to force on integrins. Nature Cell Biology. 13, 722-727 (2011).
- Dubash, A. D., Wennerberg, K., Garcia-Mata, R., Menold, M. M., Arthur, W. T., Burridge, K. A novel role for Lsc/p115 RhoGEF and LARG in regulating RhoA activity downstream of adhesion to fibronectin. J. Cell Sci. 120, 3989-3998 (2007).
- Arthur, W. T., Ellerbroek, S. M., Der, C. J., Burridge, K., Wennerberg, K. XPLN, a guanine nucleotide exchange factor for RhoA and RhoB, but not RhoC. J. Biol. Chem. 277, 42964-42972 (2002).

