A Non-random Mouse Model for Pharmacological Reactivation of Mecp2 on the Inactive X Chromosome
Summary
Here, we describe a protocol to generate a viable female murine model with non-random X chromosome inactivation, i.e., the maternally-inherited X chromosome is inactive in 100% of the cells. We also describe a protocol to test feasibility, tolerability, and safety of pharmacological reactivation of the inactive X chromosome in vivo.
Abstract
X chromosome inactivation (XCI) is the random silencing of one X chromosome in females to achieve gene dosage balance between the sexes. As a result, all females are heterozygous for X-linked gene expression. One of the key regulators of XCI is Xist, which is essential for the initiation and maintenance of XCI. Previous studies have identified 13 trans acting X chromosome inactivation factors (XCIFs) using a large-scale, loss-of-function genetic screen. Inhibition of XCIFs, such as ACVR1 and PDPK1, using short-hairpin RNA or small molecule inhibitors, reactivates X chromosome-linked genes in cultured cells. But the feasibility and tolerability of reactivating the inactive X chromosome in vivo remains to be determined. Towards this goal, a XistΔ:Mecp2/Xist:Mecp2-Gfp mouse model has been generated with non-random XCI due to deletion of Xist on one X chromosome. Using this model, the extent of inactive X reactivation was quantitated in the mouse brain following treatment with XCIF inhibitors. Recently published results show, for the first time, that pharmacological inhibition of XCIFs reactivates Mecp2 from the inactive X chromosome in cortical neurons of the living mouse brain.
Introduction
X chromosome inactivation (XCI) is a process of dosage compensation that balances X-linked gene expression by silencing one copy of the X chromosome in females1. As a result, the inactive X chromosome (Xi) accumulates characteristic features of heterochromatin including DNA methylation and inhibitory histone modifications, such as histone H3-lysine 27 trimethylation (H3K27me3) and histone H2A ubiquitination (H2Aub)2. The master regulator of X chromosome silencing is the X-inactivation center (Xic) region, around 100−500 kb, which controls the counting and pairing of the X chromosomes, the random choice of the X chromosome for inactivation, and the initiation and spreading of silencing along the X chromosome3. The process of X inactivation is initiated by X inactive specific transcript (Xist) that coats the Xi in cis to mediate chromosome-wide silencing and remodel the three-dimensional structure of the X chromosome4. Recently, several proteomic and genetic screens have identified additional regulators of XCI, such as Xist interacting proteins5,6,7,8,9,10,11,12. For example, a previous study using an unbiased genome-wide RNA interference screen identified 13 trans-acting XCI factors (XCIFs)12. Mechanistically, XCIFs regulate Xist expression and therefore, interfering with XCIFs function causes defective XCI12. Together, recent advances in the field have provided important insights into the molecular machinery that is required to initiate and maintain XCI.
Identification of XCI regulators and understanding their mechanism in XCI is directly relevant to X-linked human diseases, such as Rett syndrome (RTT)13,14. RTT is a rare neurodevelopmental disorder caused by a heterozygous mutation in the X-linked methyl-CpG binding protein 2 (MECP2) that affects predominantly girls15. Because MECP2 is located on the X chromosome, RTT girls are heterozygous for MECP2 deficiency with ~50% cells expressing wild-type and ~50% expressing mutant MECP2. Notably, RTT mutant cells harbor a dormant but wild-type copy of Mecp2 on the Xi, providing a source of the functional gene, which if reactivated, could potentially alleviate symptoms of the disease. In addition to RTT, there are several other X-linked human diseases, for which reactivation of Xi represents a potential therapeutic approach, such as DDX3X syndrome.
Inhibition of XCIFs, 3-phosphoinositide dependent protein kinase-1 (PDPK1), and activin A receptor type 1 (ACVR1), either by short hairpin RNA (shRNA) or small molecule inhibitors, reactivates Xi-linked genes12. Pharmacological reactivation of Xi-linked genes is observed in various ex vivo models that include mouse fibroblast cell lines, adult mouse cortical neurons, mouse embryonic fibroblasts, and fibroblast cell lines derived from an RTT patient12. However, whether pharmacological reactivation of Xi-linked genes is feasible in vivo remains to be demonstrated. One limiting factor is the lack of effective animal models to accurately measure the expression of genes from reactivated Xi. Towards this goal, a XistΔ:Mecp2/Xist:Mecp2-Gfp mouse model has been generated that carries a genetically labeled Mecp2 on Xi in all the cells due to heterozygous deletion in Xist on the maternal X chromosome16. Using this model, the expression of Mecp2 from Xi has been quantitated following treatment with XCIFs inhibitors in the brain of living mice. Here, the generation of the XistΔ:Mecp2/Xist:Mecp2-Gfp mouse model and methodology to quantitate Xi reactivation in cortical neurons using immunofluorescence-based assays is described.
Protocol
Work involving mice was approved by the University of Virginia Institutional Animal Care and Use Committee (IACUC; #4112).
1. Generate a Non-random XCI Mouse Model with Genetically Labeled Mecp2 on Xi
NOTE: Mouse strains used in the study were as follows: Mecp2-Gfp/Mecp2-Gfp (Mecp2tm3.1Bird, Table of Materials) and Xist/ΔXist (B6;129-Xist<tm5Sado>; provided by Antonio Bedalov, Fred Hutchinson Cancer Center, Seattle). Breeding strategies among the respective strains have been designed to expand the mouse colonies for each strain.
- Perform PCR-genotyping on all mice strains and respective progenies obtained after breeding using gene specific primers listed in Table 1.
2. Design the Mouse Breeding Strategy to Generate XistΔ:Mecp2/Xist:Mecp2-Gfp
- Set up a breeding pair by housing a Mecp2-Gfp/Y male and a XistΔ-Mecp2/Xist-Mecp2 female together (12 h/12 h: light / dark cycle) (Figure 1A). Ideally, set up at least 5 breeding pairs at a time using viable and fertile mice. After the pregnant female gives birth, allow her to raise her first litter with the male.
NOTE: Because paternal Xist knockout impairs imprinted XCI, dosage compensation, and differentiation pathways17, the use of XistΔ-Mecp2/Y mice in the breeding will fail to produce the female pups with required genotype. - After weaning the litter at post-natal day 21 (P21), identify and tag the XistΔ:Mecp2/Xist:Mecp2-Gfp female pups using a PCR-based genotyping assay (Figure 1B).
NOTE: In terms of the number of animals needed per group, for the results reported below, 5 breeding pairs were set up which generated approximately 10 female XistΔ:Mecp2/Xist:Mecp2-Gfp mice. - Use female mouse models for all the proposed experiments.
NOTE: Sex is an important biological variable for XCI studies, and the male model does not account for the confounding effects of random XCI. - To be consistent throughout the animal studies and rule out any effects of the animal age, perform all the experiments in 5−8-week-old female XistΔ:Mecp2/Xist:Mecp2-Gfp mice.
3. Isolate Female XistΔ:Mecp2/Xist:Mecp2-Gfp Mouse Embryonic Fibroblasts (MEFs)
- Set up timed mating between the Mecp2-Gfp/Y male and XistΔ-Mecp2/Xist-Mecp2 female. Set up at least 3−4 mating cages to increase the likelihood of pregnancy.
- After confirming the vaginal plug the morning after mating, separate the female and this day is considered embryonic day 0.5 (E0.5). Monitor the weight gain of the prospective pregnant female and visually inspect the abdomen of the mice to confirm pregnancy. On E14.5−15.5, euthanize the pregnant female mice via cervical dislocation.
- Under a laminar hood, wipe the abdomen of the pregnant female with 70% ethanol. Using sterile scissors, dissect the abdominal cavity and remove the uterine horns containing the embryos. Using forceps, gently take out the embryos while cutting off the remaining abdominal tissue (6−12 embryos is usually expected).
- Place the uterine horns containing the embryos in a 10 mm tissue-culture dish. Using sterile scissors and forceps, make an incision along the uterine horns to isolate individual sacs carrying an embryo. Carefully, place the uterine horns containing the rest of the embryos in 30 mL of sterile Hanks’ balanced salt solution (HBSS) on ice.
- Gently cut open the sac and isolate the embryo using sterile scissors and forceps. Remove the placenta by cutting the umbilical cord.
- Decapitate the embryo.
NOTE: While the rest of the embryonic tissue will be further processed, the tissue from the embryo head will be used to isolate DNA for genotyping. - Open the abdomen by cutting the midline of the embryo using sterile scissors and forceps. Remove the visceral organs, such as the heart, liver, and lungs.
- Transfer the remaining embryonic tissue into a sterile 60 mm tissue-culture plate and cut into small pieces using scissors or a razor blade. To break open the cell clumps, add 3 mL of trypsin-EDTA (0.05%) and incubate at 37 °C for 15 min.
- Neutralize trypsin-EDTA by adding 5 mL of Dulbecco’s modified Eagle medium (DMEM) with 10% fetal bovine serum (FBS) and 10 µg/mL penicillin/streptomycin (pen/strep) to the plate and dissociate the tissue by repetitive pipetting (approximately 20−30 times).
- Spin down cells at 300 x g for 5 min and re-suspend the cell pellet in 4 mL of DMEM with 10% FBS and 10 µg/mL pen/strep. Plate the cells on a 60 mm culture dish, and culture the cells at 37 °C in the presence of 5% CO2.
- Carry out steps 3.5−3.10 for each embryo.
- Culture MEFs obtained in step 3.11 for at least 3−4 days.
NOTE: At this stage, MEFs can also be cryopreserved for future experiments. - To determine the sex and genotype of each embryo, carry out genotyping-PCR using DNA isolated from the heads of the embryos using primers and PCR conditions listed in Table 1.
4. Confirm the Lack of Green Fluorescent Protein (GFP) Expression in the Brain of XistΔ:Mecp2/Xist:Mecp2-Gfp Mice Using a Ffluorescence Activated Cell Sorting-based Assay
- Using scissors, separate the cerebral cortex from the rest of the brain hemispheres, and place in 1 mL of ice-cold nuclei isolation media (NIM) buffer containing 250 mM sucrose, 25 mM potassium chloride (KCl), 5 mM magnesium chloride (MgCl2), 10 mM Tris-Cl, supplemented with 2% paraformaldehyde (PFA), and 0.1% nonionic surfactant (Table of Materials).
- Homogenize using an ice-cold glass homogenizer.
- Spin down homogenized tissue at 600 x g, 4 °C for 5 min.
- Remove supernatant and resuspend pellet in 1 mL of 25% iodixanol in NIM solution.
- Add 1 mL of 29% lodixanol in NIM solution to a 4 mL ultracentrifuge tube (store on ice until samples are ready) and carefully layer 1 mL of sample (in 25% lodixanol).
- Centrifuge at 9,000 x g, 4 °C for 10 min in an ultracentrifuge.
- Aspirate supernatant and resuspend pellet in 500 μL of fluorescence activated cell sorting (FACS) buffer (phosphate-buffered saline [PBS] supplemented with 1 mM EDTA, 0.05% sodium azide, and 2% bovine serum albumin [BSA]).
NOTE: Samples can be stored at 4 °C or up to 1 week. - Add 5 μL of 7-amino-actinomycin D (7-AAD; 50 mg/mL) for 5 min at room temperature to stain DNA.
- Analyze samples for 7-AAD and green fluorescent protein (GFP) signal using flow cytometry.
5. Determine Feasibility of the XistΔ:Mecp2/Xist:Mecp2-Gfp Mouse Model for Xi Reactivation
- Seed 1 x 105 cells/mL female XistΔ:Mecp2/Xist:Mecp2-Gfp MEFs, Mecp2/Mecp2-Gfp and Xist-Mecp2/Y MEFs obtained in step 3.12 in a 6-well format and in chamber slides, in DMEM with 10% FBS and 10 µg/mL pen/strep, at 37 °C in the presence of 5% CO2.
NOTE: Mecp2/Mecp2-Gfp and Xist-Mecp2/Y MEFs are used as positive and negative controls in the experiments. - Add fresh medium to the cells after 24 h. For XistΔ:Mecp2/Xist:Mecp2-Gfp MEFs, add medium supplemented with XCIFs inhibitors (e.g., 0.5 μM LDN193189 and 2.5 μM GSK650394), or vehicle alone. Replace medium supplemented with fresh inhibitor or vehicle alone every 2 days. For Mecp2/Mecp2-Gfp and Xist-Mecp2/Y MEFs, add fresh medium every 2 days.
- Post 1 week of inhibitor treatment, harvest MEFs either for RNA isolation (6-well plate) or fix cells for immunofluorescence (chamber slides). Use MEFs isolated from Mecp2/Mecp2-Gfp embryos as positive and Xist-Mecp2/Y embryos as negative controls respectively.
- For RNA isolation, isolate total RNA by guanidinium thiocyanate-based RNA extraction reagent, and reverse transcribe using reverse transcriptase. Measure Mecp2-Gfp expression by quantitative reverse transcriptase-PCR (qRT-PCR) using Mecp2-WT and Mecp2-GFP, and primers listed in Table 1, as described previously12.
- For immunofluorescence, stain MEFs with an anti-GFP primary antibody (1:100), as described previously12,16. Measure GFP intensity by quantitative immunofluorescence in drug treated XistΔ:Mecp2/Xist:Mecp2-Gfp MEFs, as described previously18.
6. Demonstrate the Pharmacological Xi Reactivation in the Brain of the XistΔ:Mecp2/Xist:Mecp2-Gfp Mouse Model
- Prepare drugs and vehicle control.
- Prepare a fresh, sterile solution of vehicle (0.9% NaCl, 0.5% methylcellulose, 4.5% dimethyl sulfoxide [DMSO]) for brain injections.
- Prepare chemical inhibitors including 1.5 mM LDN193189 (small molecule inhibitor of ACVR1) and 1.6 mM GSK650394 (small molecule inhibitor of SGK1, a downstream effector of PDPK1) re-suspended in vehicle (0.9% NaCl, 0.5% methylcellulose, 4.5% DMSO), or vehicle alone. The total volume of chemical inhibitors or vehicle injected is 10 µL per dose per animal.
- Prepare animal for brain injections.
- Prepare the surgical area by wiping the bench and heating pad with disinfectant (10% sodium hypochlorite solution).
- Anesthetize the 4 week old mouse with an intraperitoneal injection of ketamine/xylazine mixture at a dose of 140 mg/kg and 10 mg/kg, respectively. Use the pedal withdrawal reflex to determine the level of anesthesia.
- Apply ophthalmic ointment to the eyes following induction of anesthesia to prevent corneal drying.
- Shave off the fur from the neck to the top of the head of the mouse.
- Position the mouse in the stereotactic platform by hooking the mouse’s incisor teeth in the bite bar of the snout restrainer and tightening the nose clamp over the snout while ensuring that the mouse’s head is on a level plane.
- Adjust the height of the ear bars, as necessary, to reach the caudal portion of the ear canal, securing them such that the mouse’s head is in a level plane and immobilized on finger touch.
- Disinfect the head of the mouse with alternating wipes of a topical antiseptic, such as povidone-iodine and 70% ethanol.
- Administer drugs.
- Using a sterile scalpel, make a 0.75 cm horizontal incision in the mid-scalp.
- Using a 0.45 mm burr, drill two symmetrical holes above the right and left cortical hemispheres (2 mm from the sagittal suture and 2 mm from the lambdoid suture, approximately the middle of parietal bone).
- Attach a 10 μL syringe to the stereotactic platform firmly.
- Mix the solution of chemical inhibitors, and draw 10 μL of solution into the syringe. Avoid any air bubbles in the syringe.
- Advance the syringe needle into the burr hole maintaining the needle perpendicular (90°) to the skull. When the needle traverses the skull, zero out the coordinates on the stereotactic digital display and then advance the tip of the needle until it reaches a depth of 2.5 mm.
- Withdraw the needle 0.5 mm to the depth for 2 mm.
- Slowly inject 10 μL of solution (~1 min). After injection is complete, leave the needle in the brain for ~1 min and then withdraw the needle.
- Repeat the injection for the second hemisphere (vehicle only control).
- Using sutures or “skin glue” close the skin of the mouse (if wound is > 0.5cm both sutures and “skin glue” should be used).
- Loosen the ear bars and remove the mouse from the stereotactic apparatus.
- Administer analgesics (Buprenorphine SR 0.5 mg/kg IP) and place the mouse on a heating pad set to 37 °C until the animal regains consciousness.
- Once the mouse is alert and responsive, transfer the animal back to its original cage.
- Repeat the procedure every 2 days for 20 days. Repeat drilling of the area is not required for subsequent injections.
- Isolate the mouse brain.
- Once the dose regimen is completed, euthanize the mouse in a CO2 chamber.
- Immobilize the mouse on a surface using needles.
- Make a lateral incision through the integument and abdominal wall just beneath the rib cage using scissors and forceps. Carefully separate the liver from the diaphragm.
- Using scissors, cut the diaphragm and continue cutting along the entire length of the rib cage to expose the pleural cavity.
- Using scissors, make an incision to the posterior end of the left ventricle.
- Immediately, start injecting the right heart chamber with ~15 mL of PBS over ~2 min. Liver color change from red to pale pink is indicative of good perfusion.
- Inject the right chamber of the mouse heart with ~10 mL of 4% paraformaldehyde in PBS over ~2 min.
- Decapitate the mouse, and use scissors to make a midline incision of the scalp to expose the skull.
- Place one tip of the scissors into the foramen magnum, and cut laterally into the skull toward the eye. Repeat for the other side. Try to keep the end of the scissors as superficial as possible to avoid injury of the brain.
- Use scissors to cut the region between the eyes and above the nose of the mouse.
- Use forceps to gently peel the cranial bones from the brain hemispheres.
- Lift the brain with a spatula, and use scissors to carefully dissect the cranial nerve fibers that fix it to the skull.
- Place the brain on a plastic dish, cut out the cerebellum and olfactory bulbs, and place the brain into a 15 mL tube filled with 4% PFA is PBS.
- Cryo-section the mouse brain.
- Fix brain in 4% paraformaldehyde in PBS at 4 °C overnight.
- Rinse the brain with PBS at 4 °C at least 3x for 5 min each.
- Label the disposable molds for cryopreserving the tissues.
- Cut out the front of the brain using scissors and forceps (start making ~5 mm sections from injection sites).
- Transfer the brain to the cryomold with the front of the brain facing down to obtain coronal sections. Submerge the mold containing brain in the optimal cutting temperature compound (OCT).
- Pour liquid nitrogen into a 10 mm plastic petri dish and place the brain in the cryo-mold into the nitrogen.
NOTE: It is important to orient the tissue as described in step 6.5.5 to guide the sectioning of the brain. - When the OCT is solid white, place the frozen brain into an -80 °C freezer for storage.
- Equilibrate the brain to -20 °C for at least 30 min prior to sectioning with cryostat and cryosection (5−6 μm thickness) the brain, mounting 2−3 sections per slide. Slides can be stored at -80 °C for later use.
- Determine Xi reactivation using an immunofluorescence-based approach.
- Before starting the staining procedure, dry the brain sections overnight at 4 °C.
- Immerse slides in antigen retrieval solution (0.1 M citric acid, 0.1 M Tris-base, pH = 6.0) on a 100 °C heat block for 5 min.
- Wash slides 4x with 1x PBS for 5 min each at room temperature.
- Immerse slides in blocking solution (0.1 M NH
4Cl/PBS/0.2% gelatin/0.05% nonionic surfactant) for 20 min at room temperature. - Wash slides 3x with wash buffer (PBS/0.2% gelatin) at room temperature for 5 min each.
- Stain brain sections with anti-GFP-AlexaFluor647 (1:100) and anti-MAP2 (1:1,000) antibodies in incubation medium (PBS/0.2% gelatin/1% BSA) and incubate at 4 °C overnight.
- Collect primary antibodies (can be reused). Wash slides 4x with wash buffer for a total of 30 min at room temperature.
- Incubate brain sections with goat anti-chicken Fluorescein isothiocyanate (FITC)-labeled secondary antibody (1:1,000) in incubation medium and incubate 1−2 h at room temperature in the dark.
- Wash slides 4x with wash buffer for a total of 30 min at room temperature.
- Place a drop of mounting medium with 4′,6-diamidino-2-phenylindole (DAPI) on a 22 mm x 50 mm coverslip, then invert the coverslip onto a slide, covering all tissue sections.
- Image on a microscope and adjust images for contrast and brightness. Capture images and quantify the number of GFP-positive cells for both drug-treated and vehicle treated XistΔ:Mecp2/Xist:Mecp2-Gfp mouse brain hemispheres.
Representative Results
To demonstrate the feasibility of the XistΔ:Mecp2/Xist:Mecp2-Gfp mouse model for Xi reactivation studies, XCIF inhibitor-mediated reactivation of Xi-linked Mecp2-Gfp was tested in mouse embryonic fibroblasts (MEFs). Female MEFs were isolated from day 15.5 XistΔ:Mecp2/Xist:Mecp2-Gfp embryos as described in section 3 (Figure 1A). The genotypes of female XistΔ:Mecp2/Xist:Mecp2-Gfp MEFs were confirmed by genotyping-PCR, as described previously19 (Figure 1B), and FACS-based assay (Figure 1C). MEFs were treated with either DMSO or the two drugs LDN193189 and GSK650394 (0.5 μM and 2.5 μM, respectively) for 7 days. Following drug treatment, the expression of Mecp2-Gfp was monitored by qRT-PCR. As shown in Figure 1D, the drug treatment, but not DMSO, reactivated expression of Xi-Mecp2-Gfp. Next, quantitative immunofluorescence was carried out to determine the extent of Xi-Mecp2-Gfp expression in individual MEFs, as described in step 5.3.2. Signal from negative-control MEFs isolated from male embryos (Mecp2/Y) was set as the background, and ~66% of positive control Mecp2-Gfp/Mecp2 MEFs had a nuclear GFP signal. As expected, XistΔ:Mecp2/Xist:Mecp2-Gfp MEFs treated with DMSO had a very low level of nuclear GFP (~3%). By contrast, ~31% of drug-treated XistΔ:Mecp2/Xist:Mecp2 MEFs were positive for nuclear GFP (Figure 1E). Together, these results demonstrate that XCIF inhibitors reactivate Xi-linked Mecp2 in MEFs, but the extent of Xi reactivation varies in the cell population.
To assess the feasibility of pharmacological Xi reactivation-based approach in vivo, whether drug treatment reactivates Xi-linked Mecp2 in the brain of XistΔ:Mecp2/Xist:Mecp2-Gfp female mice was investigated. 10 μL of vehicle or 10 μL of XCIF inhibitors (1.5 mM LDN193189 and 1.6 mM GSK650394) was administered in opposite brain hemispheres of 4-week-old Xi-Mecp2-Gfp female mice by intracerebroventricular injection using stereotactic surgical procedures (Figure 2A,B). The procedure was repeated every second day (Figure 2C), and three weeks later, animals were euthanized, and brains were isolated. One subset was used for qRT-PCR and another was analyzed by immunohistochemistry. The expression of wild-type Mecp2 and Xi-Mecp2-Gfp in the vehicle- and drug-infused hemispheres was determined by qRT-PCR (sequences of primers listed in Table 1). As shown in Figure 2D, drug treatment reactivated Xi-Mecp2-Gfp in ~30% of cells in the drug-infused brain hemisphere, whereas Xi- Mecp2-Gfp was not detected in the vehicle-infused hemisphere. A large number of MAP2, a neuronal marker, positive neurons were also GFP positive (~45%) in the drug-treated hemisphere, indicative of Xi-Mecp2-Gfp expression. Approximately 20% of MAP2 negative brain cells expressed GFP, confirming Xi-Mecp2-Gfp reactivation in non-neuronal cells (Figure 2E).
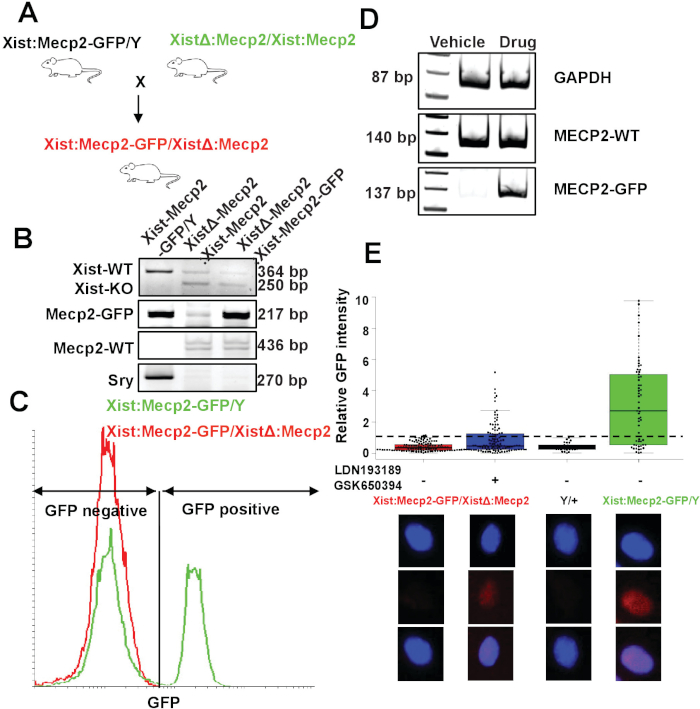
Figure 1: Generation and validation Xi-Mecp2 mouse model. (A) Schematic of the breeding strategy for generating XistΔ:Mecp2/Xist:Mecp2-Gfp mice. (B) PCR genotyping of Xist:Mecp2-Gfp/Y, XistΔ:Mecp2/Xist:Mecp2 and XistΔ:Mecp2/Xist:Mecp2-Gfp mice. Mice were monitored for the presence of Mecp2-Gfp, Mecp2 and sex-determining region Y (SRY). (C) Flow cytometry analysis of nuclei isolated from the mouse cortex. Mecp2/Mecp2-Gfp mouse cortex show ~50% of GFP-positive nuclei while XistΔ:Mecp2/Xist:Mecp2-Gfp shows no GFP-positive nuclei. (D) qRT-PCR analysis monitoring the expression of Mecp2-Gfp and wild-type Mecp2 transcripts in female XistΔ:Mecp2/Xist:Mecp2-Gfp MEFs following treatment with DMSO or drug (LDN193189 and GSK650394). Gapdh was monitored as a loading control. (E) Quantitative immunofluorescence monitoring GFP intensity in female XistΔ:Mecp2/Xist:Mecp2-Gfp MEFs following treatment with DMSO or the drugs LDN193189 and GSK650394. MEFs isolated from Mecp2/Y or Mecp2/Mecp2-Gfp mice were used as negative and positive controls, respectively. Each dot represents an MEF, and the dashed line indicates the maximum background signal obtained in Mecp2/Y, which was set to 1. Lower panel shows representative pictures of nuclei. This figure has been modified from Przanowski et al.16. Please click here to view a larger version of this figure.
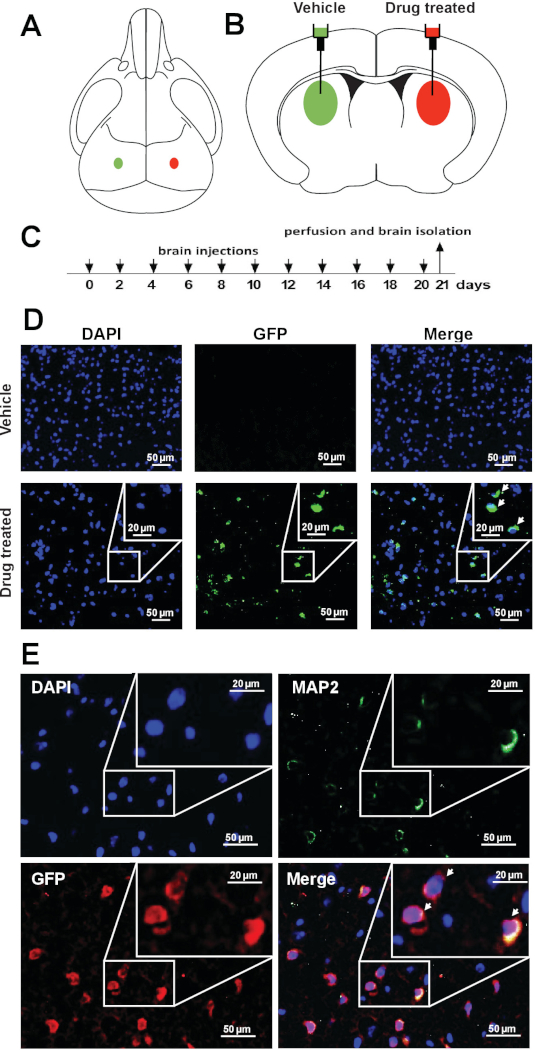
Figure 2: Pharmacological reactivation of X-linked Mecp2 in cerebral cortical neurons of living mice. (A) Schematic of a mouse skull and (B) brain showing the site of injection for vehicle or drug in the left or right hemispheres of the brain. (C) Schematic of the drug regimen. (D) Representative immunofluorescence images showing endogenous GFP signal (green) in coronal brain sections from vehicle- or drug-treated hemispheres. DAPI staining is shown in blue. (E) Representative immunofluorescence images of the coronal brain sections monitoring the expression of GFP (anti-GFP; red) and MAP2 (green) in drug-treated hemisphere. DAPI staining is shown in blue. This figure has been modified from Przanowski et al.16. Please click here to view a larger version of this figure.
| Reverse primer (5’ -> 3’) | Anealing temperature/PCR product length |
| GGCATGGACTGTGGTCATGAG | 60 °C/87 bp |
| GCTGAACTTGTGGCCGTTTA | 62 °C/137 bp |
| TGTCAGAGCCCTACCCATAAG | 62 °C/140 bp |
| GCACAACCCCGCAAATGCTA | 62 °C/364 bp |
| GCACAACCCCGCAAATGCTA | 62 °C/250 bp |
| AATTGCCCTAACGAGCACAC | 62 °C/436 bp |
| GAACTTCAGGGTCAGCTTGC | 62 °C/217 bp |
| CTCCTCTGTGACACTTTAGCCCTCCGA | 66 °C/270 bp |
Table 1: List of primers used for genotyping and quantitative real-time RT-PCR.
Discussion
Previously, XCIFs that are selectively required for silencing of Xi-linked genes in mammalian female cells were identified12. We further optimized potent small molecule inhibitors to target XCIFs, such as ACVR1 and downstream effectors of PDPK1, which efficiently reactivate Xi-linked Mecp2 in mouse fibroblast cell lines, mouse cortical neurons, and a human fibroblast cell line derived from a RTT patient. These results suggest that Xi reactivation is a plausible therapeutic approach to rescue the gene deficiencies in X-linked disease patients; however, the in vivo feasibility remains to be determined. Recently, XCIF inhibitors were shown to reactivate Xi-linked Mecp2 in vivo, for which a non-random XistΔ:Mecp2/Xist:Mecp2-Gfp mouse model was generated.
An attractive feature of the XistΔ:Mecp2/Xist:Mecp2-Gfp model is that it allows an accurate quantitation of the Xi-linked Mecp2 reactivation for several reasons. First, due to the deletion of Xist on the maternal X chromosome, the XistΔ:Mecp2/Xist:Mecp2-Gfp mouse has non-random XCI. As a result, the genetically labeled Mecp2 is silent in 100% of cells (Figure 1C), unlike an expected 50:50 expression of X-linked genes in random XCI mice models, such as Xist:Mecp2/Xist:Mecp2-Gfp. Therefore, the results are not precluded by the mosaic expression of GFP, and 100% cells carry Mecp2-Gfp on Xi in the XistΔ:Mecp2/Xist:Mecp2-Gfp model. Second, the genetic labeling of Mecp2 permits direct visualization of individual neurons with reactivated GFP, thereby minimizing the experimental manipulations in neuronal analysis.
A recent study found that intracerebroventricular injection of the XCIF inhibitors in the mouse brain hemisphere reactivates Xi-linked Mecp2 using immunofluorescence analysis of the mouse brain16. Importantly, drug treatment had no adverse effect on the general health, such as weight, grooming, or mobility16. Moreover, there is no toxicity detected by drug treatment in the liver or spleen. Together, this study provides an essential proof-of-principle to demonstrate that interfering with the function of XCIFs leads to de-repression of Xi in vivo.
In summary, a sensitive mouse model can be used to evaluate the reactivation of Xi. This animal model design can also be adapted for generating an improved RTT mouse model that harbors Mecp2 mutations (probably less symptomatic) on the active X chromosome and wild-type Mecp2 on the Xi in all cells. Due to non-random XCI, while the phenotypic symptoms may be more pronounced, it is expected that this model will also allow for better evaluation and accurate assessment of the reversal of symptoms due to Xi reactivation. Additionally, XistΔ:Mecp2/Xist:Mecp2-Gfp can also be modified to study Xi reactivation in other X-linked disease models, such as the DDX3X syndrome.
Divulgazioni
The authors have nothing to disclose.
Acknowledgements
The authors thank Antonio Bedalov for providing reagents; University of Virginia Tissue Histology Core for cryosectioning; University of Virginia Flow Cytometry Core for flow cytometry analysis; Christian Blue and Saloni Singh for technical assistance with genotyping. This work was supported by a Double Hoo Research Grant to Z.Z., and a Pilot Project Program Award from the University of Virginia-Virginia Tech Seed Fund Award and the Hartwell Foundation Individual Biomedical Research Award to S.B.
Materials
| MICE | |||
| Mecp2tm3.1Bird | The Jackson Laboratory | #014610 | |
| B6;129-Xist (tm5Sado) | provided by Antonio Bedalov, Fred Hutchinson Cancer Center, Seattle | ||
| REAGENTS | |||
| 22×22 mm coverslip | FISHERfinest (Fisher Scientific) | 125488 | |
| 32% Paraformaldehyde | Electron Microscopy Sciences | 15714-S | |
| 50 ml syringe | Medline Industries | NPMJD50LZ | |
| 60mm culture dish | CellStar | 628160 | |
| 7-AAD | BioLegend | 420403 | |
| ammonium chloride (NH4Cl) | Fisher Chemical | A661-3 | |
| anti-GFP-AlexaFluor647 | Invitrogen | A-31852 | |
| anti-MAP2 | Aves Labs | MAP | |
| BSA | Promega | R396D | |
| Buprenorphine SR | Zoopharm | ||
| citric acid | Sigma | C-1857 | |
| DMSO | Fisher Bioreagents | BP231-100 | |
| Dulbecco's Modified Eagle Medium (DMEM) | Corning Cellgro | 10-013-CV | |
| Ethanol | Decon Labs | 2701 | |
| fetal bovine serum (FBS) | VWR Life Science | 89510-198 | |
| gelatin | Sigma-Aldrich | G9391 | |
| glass slides | Fisherbrand | 22-034-486 | |
| goat anti-chicken FITC-labeled secondary antibody | Aves Labs | F-1005 | |
| GSK650394 | ApexBio | B1051 | |
| hamilton 10μl syringe | Hamilton Sigma-Aldrich | 28615-U | |
| Hank's Balanced Salt Solution (HBSS) | Gibco | 14025-092 | |
| Ketamine | Ketaset | NDC 0856-2013-01 | |
| Large blunt/blunt curved scissors | Fine Science Tools | 14519-14 | |
| LDN193189 | Cayman Chemicals | 11802 | |
| lodixanol | Sigma | 1343517 | |
| magnesium chloride (MgCl2) | Fisher Chemical | M35-212 | |
| Methylcelulose | Sigma | M0262-100G | |
| mounting medium with DAPI | Vectashield | H-1200 | |
| Needle tip, 26 GA x 1.25" | PrecisionGlide | 305111 | |
| ophthalmic ointment | Refresh Lacri-Lube | 93468 | |
| optimal cutting temperature (O.C.T.) | ThermoFisher | ||
| PCR mix | |||
| Penicillin/Streptomycin (Pen/Strep) | Corning | 30-002-Cl | |
| Phosphate buffered saline pH 7.4 (PBS) | Corning Cellgro | 46-103-CM | |
| Potassium chloride (KCl) | Fisher Scientific | P330-500 | |
| scalpel blades | |||
| Shallow glass or plastic tray | |||
| skin glue/tissue adhesive | 3M Vetbond | 1469SB | |
| sodium azide | Fisher Scientific | CAS 26628-22-8 | |
| Sodium chloride (NaCl) | Fisher Chemical | S642-212 | |
| standard hemostat forceps | Fine Science Tools | 13013-14 | |
| Standard tweezers | Fine Science Tools | 11027-12 | |
| Straight iris scissors | Fine Science Tools | 14058-11 | |
| sucrose | Fisher Scientific | BP220-1 | |
| Tris-base | Fisher Bioreagents | BP152-5 | |
| Triton X-100 | Fisher Bioreagents | BP151-500 | |
| Trypsin-EDTA | Gibco | 15400-054 | |
| Xylazine | Akorn | NDC: 59399-111-50 | |
| EQUIPMENT | |||
| Zeiss AxioObserver Live-Cell microscope | Zeiss | Zeiss AxioObserver | |
| 0.45mm burr | IDEAL MicroDrill | 67-1000 | |
| BD FACScalibur | |||
| centrifuge | |||
| glass homogenizer | |||
| cell culture incubator | Thermo Scientific HERACELL VIOS 160i | 13-998-213 | |
| Leica 3050S research cryostat | |||
| stereotactic platform | |||
| thermocycler | |||
| Timer | |||
| ultracentrifuge | Beckman Coulter Optima L-100 XP | ||
| Water bath (37 ºC) | Fisher Scientific Isotemp 2239 |
Riferimenti
- Lyon, M. F. X-chromosome inactivation as a system of gene dosage compensation to regulate gene expression. Progress in Nucleic Acid Research and Molecular Biology. 36, 119-130 (1989).
- Heard, E. Delving into the diversity of facultative heterochromatin: the epigenetics of the inactive X chromosome. Current Opinion in Genetics Development. 15 (5), 482-489 (2005).
- Augui, S., Nora, E. P., Heard, E. Regulation of X-chromosome inactivation by the X-inactivation centre. Nature Review Genetics. 12 (6), 429-442 (2011).
- Pontier, D. B., Gribnau, J. Xist regulation and function explored. Human Genetics. 130 (2), 223-236 (2011).
- Barnes, C., Kanhere, A. Identification of RNA-Protein Interactions Through In Vitro RNA Pull-Down Assays. Methods in Molecular Biology. 1480, 99-113 (2016).
- McHugh, C. A., et al. The Xist lncRNA interacts directly with SHARP to silence transcription through HDAC3. Nature. 521 (7551), 232-236 (2015).
- Minajigi, A., et al. Chromosomes. A comprehensive Xist interactome reveals cohesin repulsion and an RNA-directed chromosome conformation. Science. 349 (6245), (2015).
- Mira-Bontenbal, H., Gribnau, J. New Xist-Interacting Proteins in X-Chromosome Inactivation. Current Biology. 26 (8), R338-R342 (2016).
- Mira-Bontenbal, H., Gribnau, J. New Xist-Interacting Proteins in X-Chromosome Inactivation. Curren Biology. 26 (10), 1383 (2016).
- Ridings-Figueroa, R., et al. The nuclear matrix protein CIZ1 facilitates localization of Xist RNA to the inactive X-chromosome territory. Genes and Development. 31 (9), 876-888 (2017).
- Sunwoo, H., Colognori, D., Froberg, J. E., Jeon, Y., Lee, J. T. Repeat E anchors Xist RNA to the inactive X chromosomal compartment through CDKN1A-interacting protein (CIZ1). Proceedings of National Academy of Sciences of the United States of America. , (2017).
- Bhatnagar, S., et al. Genetic and pharmacological reactivation of the mammalian inactive X chromosome. Proceedings of National Academy of Sciences of the United States of America. 111 (35), 12591-12598 (2014).
- Zoghbi, H. Y., Percy, A. K., Schultz, R. J., Fill, C. Patterns of X chromosome inactivation in the Rett syndrome. Brain Development. 12 (1), 131-135 (1990).
- Anvret, M., Wahlstrom, J. Rett syndrome: random X chromosome inactivation. Clinical Genetics. 45 (5), 274-275 (1994).
- Amir, R. E., et al. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nature Genetics. 23 (2), 185-188 (1999).
- Przanowski, P., et al. Pharmacological reactivation of inactive X-linked Mecp2 in cerebral cortical neurons of living mice. Proceedings of Natlional Academy of Sciences of the United States of America. 115 (31), 7991-7996 (2018).
- Borensztein, M., et al. Xist-dependent imprinted X inactivation and the early developmental consequences of its failure. Nature Structural and Molecular Biology. 24 (3), 226-233 (2017).
- Jensen, E. C. Quantitative analysis of histological staining and fluorescence using ImageJ. Anatomical Record (Hoboken). 296 (3), 378-381 (2013).
- Cseke, L. J., Talley, S. M. A PCR-based genotyping method to distinguish between wild-type and ornamental varieties of Imperata cylindrica. Journal of Visualized Experiments. (60), (2012).

