Microelectrode Array-Based Assessment of Neuronal Networks in Mouse Spinal Cord Slices
Abstract
Source: Iredale, J. A., et al. Recording Network Activity in Spinal Nociceptive Circuits Using Microelectrode Arrays. J. Vis. Exp. (2022).
This video demonstrates a microelectrode array-based assay for studying neuronal network activity in mouse spinal cord sections. First, the electrophysiological activity from the superficial dorsal horn (SDH) of the slice is recorded. Then, a potassium channel inhibitor is introduced to prolong depolarization, resulting in synchronous rhythmic activity across the neuronal network.
Protocol
All procedures involving sample collection have been performed in accordance with the institute's IRB guidelines.
1. In vitro electrophysiology
- Preparation of solutions for spinal cord slice preparation and recording
- Artificial cerebrospinal fluid
NOTE: Artificial cerebrospinal fluid (aCSF) is used in an interface incubation chamber, where slices are stored until recording commences and during experiments as both perfusate and diluent for drugs. See Table 1 for the detailed composition.
- Artificial cerebrospinal fluid
- Prepare aCSF containing (in mM) 118 NaCl, 25 NaHCO3, 10 glucose, 2.5 KCl, 1 NaH2PO4, 1 MgCl2, and 2.5 CaCl2 by adding the required quantities of the above, excluding CaCl2, to 2 L of distilled water.
- Bubble the above solution with carbogen (95% O2, 5% CO2) for 5 min and add CaCl2.
NOTE: This step prevents CaCl2 precipitation, i.e., the solution should not turn cloudy. For drug application during experiments, dilute the drug stock solutions in aCSF to desired final concentrations.
2. Sucrose-substituted artificial cerebrospinal fluid
NOTE: Sucrose-substituted aCSF is used during dissection and spinal cord slicing. As indicated by the name, sucrose is substituted for NaCl to reduce neuronal excitation during these procedures while maintaining osmolarity. See Table 1 for the detailed composition.
- Prepare sucrose-substituted aCSF containing (in mM) 250 sucrose, 25 NaHCO3, 10 glucose, 2.5 KCl, 1 NaH2PO4, 1 MgCl2, and 2.5 CaCl2 by adding the required quantities of all of the above, excluding CaCl2, to 300 mL of distilled water.
- Bubble the solution with carbogen for 5 min and then add CaCl2.
- Store the solution in a -80 °C freezer for approximately 40 min or until the solution forms a slurry. Avoid freezing solid and use while in slurry consistency.
3. Microelectrode array preparation
NOTE: The contact surface of the MEA requires a pretreatment to make it hydrophilic.
- Before the experiment, fill the MEA well with either fetal bovine serum (FBS) or horse serum (HS) for 30 min.
- Remove the FBS or HS and thoroughly rinse MEA with approximately five washes of distilled water until the distilled water is no longer foamy. Fill the well with aCSF, ready for use.
4. Acute spinal cord slice preparation
- Deeply anesthetize the mouse with 100 mg/kg ketamine (i.p.) and then decapitate it using large surgical scissors.
- Remove the skin over the abdominal region by making a small cut in the skin at the level of the hips. Pull the skin on either side of the cut rostrally until all the skin is removed, i.e., from the top of the rib cage to the top of the pelvis (both ventrally and dorsally).
- Place the body on ice and use a ventral approach to expose the vertebral column by removing all the viscera and cutting through the ribs lateral to the sternum.
- Remove the ventral rib cage, both scapulae (cut off at approximately T2), and the lower limbs and pelvis (cut off at approximately the top of the sacrum).
- Transfer the vertebral column and rib preparation to a dissecting bath containing ice-cold sucrose aCSF. Pin all four corners of the preparation (ventral surface upwards) by placing pins through the lower back muscles and the attached upper ribs.
- Remove all muscle and connective tissue overlying the ventral surface of the vertebrae with rongeurs and identify the vertebral region over the lumbosacral enlargement, which lies approximately beneath the T12 to L2 vertebral bodies.
- Remove a vertebral body that is caudal to the lumbosacral enlargement region to provide access to the spinal cord as it sits in the vertebral canal.
- Using curved spring scissors, cut through the vertebral pedicles bilaterally while lifting and pulling the vertebral body rostrally to separate the ventral and dorsal aspects of the vertebrae and expose the spinal cord.
- Once the vertebral bodies are removed to reveal the lumbosacral enlargement, carefully clear the remaining roots that anchor the spinal cord with spring scissors until the cord floats free.
- Isolate the spinal cord with rostral and caudal cuts well above and below the lumbosacral enlargement, allowing the target region of the cord to 'float free.'
NOTE: The preferred slice orientation will determine how the cord is subsequently mounted for sectioning (Figure 1). - For transverse slices, lift the lumbosacral segment by an attached root and place it on a pre-cut polystyrene (Styrofoam) block (1 cm x 1 cm x 1 cm) with a shallow channel cut in the center. Use cyanoacrylate adhesive (see the Table of Materials) to attach the block and cord to the sectioning platform and place it in the cutting bath containing ice-cold sucrose aCSF (slurry).
NOTE: The shallow channel helps secure and orient the spinal cord, with the dorsal side exposed and the thoracic end of the cord at the bottom of the block. - For sagittal slices, lay a thin line of cyanoacrylate adhesive on the sectioning platform, lift the lumbosacral enlargement by an attached root, and place the cord along the line of glue, ensuring one lateral surface is in the adhesive and the other faces upwards. Place it in the cutting bath containing ice-cold sucrose aCSF (slurry).
- For horizontal slices, put a thin line of cyanoacrylate adhesive on the sectioning platform. Lift the lumbosacral enlargement by an attached root, and place the lumbosacral enlargement along the line of adhesive, ensuring the ventral surface is in the adhesive and the dorsal surface faces upwards. Use attached roots to position the cord. Place it in the cutting bath containing ice-cold sucrose aCSF (slurry).
5. Microelectrode array recordings
NOTE: The following steps detail how to use record data from MEA-based experiments on spinal cord slices. Several MEA designs can be used depending on the experiment. Design details for MEAs used in these experiments are shown in Table 2 and Figure 2. Detailed design information has been published by Egert et al. and Thiebaud et al. for planar and 3-dimensional (3D) MEAs, respectively. Both MEA types are composed of 60 titanium nitride electrodes, with a silicon nitride insulating layer and titanium nitride tracks and contact pads.
- Experimental setup
- Turn on the computer and interface board, and start the recording software.
- Load the pre-assembled recording template (Figure 3A). Name the files for the day in the recorder tab.
- Continuously bubble aCSF with carbogen (5% CO2, 95% O2) for the duration of the experiment.
- Turn the perfusion system on, which is controlled by a peristaltic pump. Place the inlet line into aCSF and the inlet end in a waste beaker. Prime the perfusion lines with aCSF.
- Prepare 4-AP and any other drug solutions by diluting stocks in 50 mL of aCSF to the required final concentration (e.g., 200 µM for 4-AP).
- 4-aminopyridine (4-AP) activity
- Following incubation, transfer a single slice from the incubator using a large-tip Pasteur pipette filled with aCSF.
- Place the slice in the MEA well and add additional aCSF.
- Position the slice over the 60-electrode recording array using a fine short hair paintbrush. Avoid contacting the electrodes with the paintbrush or dragging the tissue across the electrodes, especially if using 3D arrays.
NOTE: Depending on the MEA layout, this can be done with or without the assistance of a microscope for accurate positioning. - After positioning the slice, place a weighted net over the tissue to hold it in place and promote good contact with MEA electrodes.
NOTE: The slice may need repositioning following net placement. - Place the MEA in the recording headstage (Figure 2A, B).
- Check the position of the tissue over the electrodes using an inverted microscope (2x magnification) to confirm that as many electrodes as possible are under the superficial dorsal horn (SDH). Ensure that at least 2-6 electrodes do not contact the slice as these electrodes are important for subtracting noise and recording artefacts during analysis (Figure 2E).
- Turn on the camera, connect it to the device, and take a reference image of the slice relative to the MEA for use during analysis.
- Press Start DAQ (data acquisition) in the recording software and confirm that all electrodes are receiving a clear signal.
NOTE: If the signal is noisy, unclip the headstage, and clean both the MEA contact pads and gold spring contacts with 70% ethanol (use a laboratory wipe to ensure that the pads and contacts are dry after cleaning). If the signal is still noisy, turn off the malfunctioning electrodes in the recording software or note down for exclusion later during analysis. - Attach the perfusion inlet and outlet lines to the MEA-well (previously filled with aCSF) and turn the perfusion system on. Check the flow rate, ideally 4-6 bath volumes per minute, and ensure that the outflow is sufficient to prevent overflow of the superfusate.
- Allow the tissue to equilibrate for 5 min and then record 5 min of raw, unfiltered baseline data.
- Move the perfusion inlet line from aCSF to a 4-AP solution and wait for 12 min for the 4-AP-induced rhythmic activity to reach steady state (2 min for drugs to reach the bath and 10 min for the activity to peak and then plateau).
- Record 5 min of 4-AP-induced activity. Be prepared for subsequent recordings to test the drugs or to check the stability of 4-AP.
Table 1: Artificial Cerebrospinal Fluid compositions.
| Chemical | aCSF (mM) | aCSF (g/100 mL) | Sucrose-substituted aCSF (mM) | Sucrose-substituted aCSF (g/100 mL) | High-potassium aCSF (mM) | High-potassium aCSF (g/100 mL) |
| Sodium chloride (NaCl) | 118 | 0.690 | – | – | 118 | 0.690 |
| Sodium hydrogen carbonate (NaHCO3) | 25 | 0.210 | 25 | 0.210 | 25 | 0.210 |
| Glucose | 10 | 0.180 | 10 | 0.180 | 10 | 0.180 |
| Potasium chloride (KCl) | 2.5 | 0.019 | 2.5 | 0.019 | 4.5 | 0.034 |
| Sodium dihydrogen phosphate (NaH2PO4) | 1 | 0.012 | 1 | 0.012 | 1 | 0.012 |
| Magnesium cloride (MgCl2) | 1 | 0.01 | 1 | 0.01 | 1 | 0.01 |
| Calcium chloride (CaCl2) | 2.5 | 0.028 | 2.5 | 0.028 | 2.5 | 0.028 |
| Sucrose | – | – | 250 | 8.558 | – | – |
Table 2: Microelectrode array layouts.
| Microelectrode Array Layouts | ||||
| Microelectrode Array Model | 60MEA 200/30iR-Ti | 60-3DMEA 100/12/40iR-Ti | 60-3DMEA 200/12/50iR-Ti | 60MEA 500/30iR-Ti |
| Planar or 3-Dimensional (3D) | Planar | 3D | 3D | Planar |
| Electrode Grid | 8 x 8 | 8 x 8 | 8 x 8 | 6 x 10 |
| Electrode Spacing | 200 µm | 100 µm | 200 µm | 500 µm |
| Electrode Diameter | 30 µm | 12 µm | 12 µm | 30 µm |
| Electrode Height (3D) | N/A | 40 µm | 50 µm | N/A |
| Experiments | Transverse slice | Transverse slice | Sagittal + Horizontal | Sagittal + Horizontal |
Representative Results
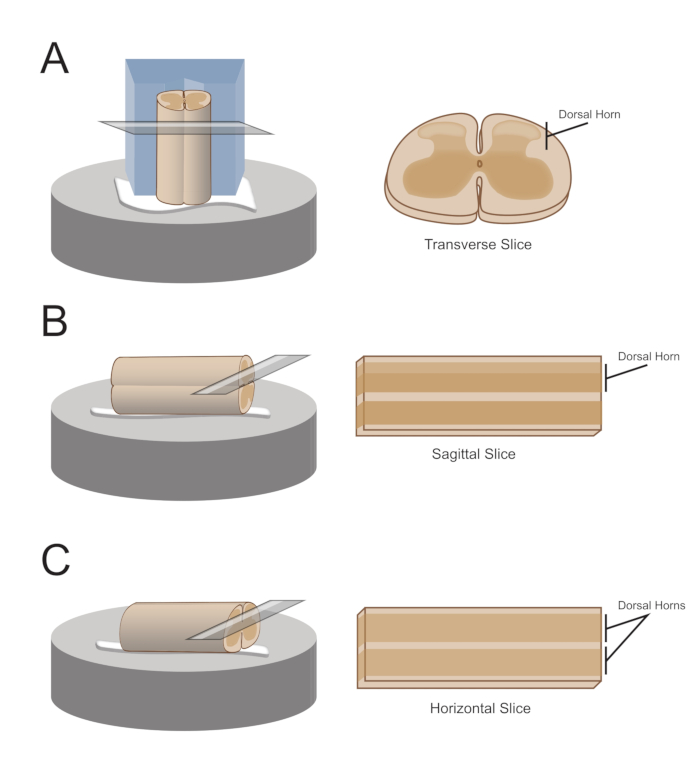
Figure 1: Spinal cord slice orientations, mounting and cutting methods. (A) Transverse slices require a Styrofoam cutting block with a supporting groove cut into it. The spinal cord is rested against the block in the support groove, the dorsal side of the cord facing away from the block. The block and cord are glued onto a cutting stage with cyanoacrylate adhesive. (B) Sagittal slices are prepared by placing a thin line of cyanoacrylate adhesive on the cutting stage and then positioning the spinal cord on its side on the glue. (C) Horizontal slices are prepared by placing a thin line of cyanoacrylate adhesive on the cutting stage and then positioning the spinal cord ventral side down on the glue.
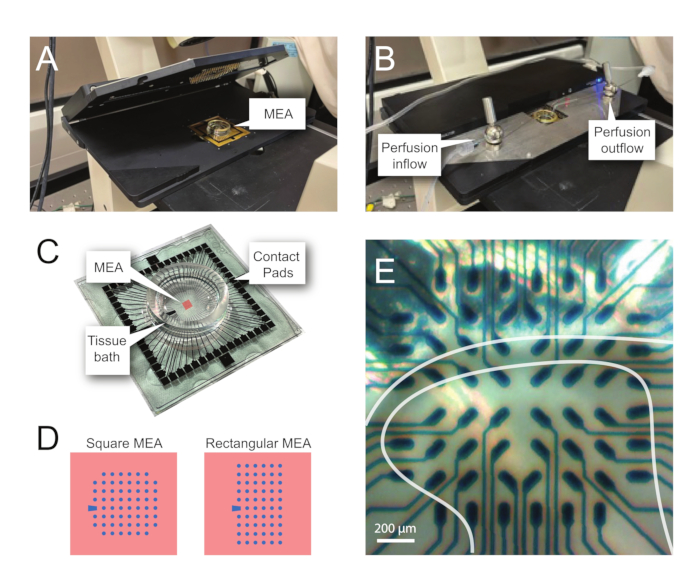
Figure 2: Tissue positioning on the microelectrode array. (A) Image shows an open MEA headstage with an MEA placed in position. (B) Same as A with MEA headstage closed for recordings and tissue perfusion system in place. (C) Image shows an MEA as supplied by the manufacturer. Contact pads, which interface with the gold springs of the headstage, and the MEA tissue bath that holds the tissue bathing solution and tissue slice are shown. The area highlighted by the red square in the center is the location of the electrode array. (D) Schematics show the two MEA electrode configurations used in this study, with further details presented in Table 2. The reference electrode is denoted by the blue trapezoid. The left MEA electrode layout shows a 60-electrode square configuration, used most in the presented work-models 60MEA200/30iR-Ti with 30 μm diameter electrodes spaced 200 μm apart, or 200 μm spaced and 100 μm spaced 3-dimensional MEAs (60MEA200/12/50iR-Ti and 60MEA100/12/40iR-Ti) with electrodes 12 μm in diameter and either 50 μm or 40 μm high, respectively. The left MEA electrode layout shows a 6 x 10 electrode rectangular layout-60MEA500/30iR-Ti. (E) High-magnification image of a 60MEA100/12/40iR-Ti square MEA with transverse spinal cord slice positioned for recording. The slice sits on electrode rows 3-8. The top row of electrodes, which do not contact any tissue, serve as reference electrodes. The SDH area appears as a semitransparent band. In this case, the SDH overlies electrodes in rows 4, 5, and 6 and columns 2, 3, 4, 5, and 7 of the MEA. Scale bar = 200 µm. Abbreviations: MEA = microelectrode array; SDH = superficial dorsal horn.
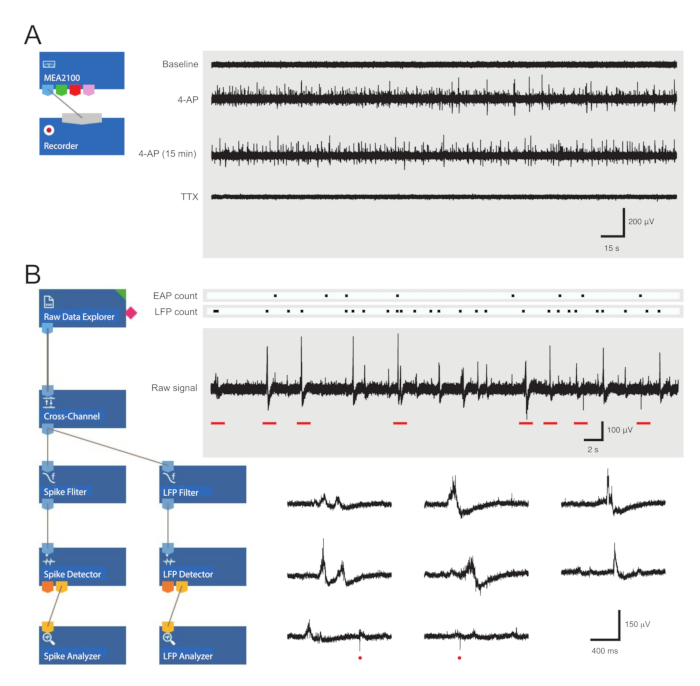
Figure 3: Data recording and analysis tool layouts and example microelectrode array recordings showing extracellular action potential and local field potential waveforms. (A) Schematic shows preconfigured recording templateused for the acquisition of MEA data. Linking the MEA2100 and the recording (headstage/amplifier) tool enables the data to be named and saved. Four example traces of raw data (right, 5-min epochs) were collected by one MEA channel showing activity at baseline, 12 min after 4-AP application, a further 15 min after established 4-AP activity, and following bath application of TTX (1 µM). Note, the addition of 4-AP (second trace) produces a clear increase in background noise and EAP/LFP activity. Importantly, the activity remains relatively stable for at least 15 min after 4-AP-induced activity is established (third trace). Addition of TTX (1 µM) abolishes all activity (bottom trace). (B) Schematic (left) shows analyzer software configuration for data analysis. The raw data explorer tool is used to import recordings collected by recording software. These data are then run through a cross-channel filter tool that subtracts the selected reference electrode(s) signal(s) from other electrodes to remove background noise. Data pass through the EAP filter and the LFP filter tools to optimize signal-to-noise relationships for each waveform. Following this step, the EAP path data enter the EAP detector tool, where thresholds are set. EAPs are detected and then sent to the EAP analyzer tool where the latencies of each event are recorded and exported as a txt. file. An identical workflow occurs for LFP data using a corresponding LFP toolkit. Right traces show data from a single MEA channel containing various extracellular waveforms. Location of EAP and LFP signals are highlighted in the above 'count rasters.' Lower traces are epochs from upper recording (denoted by red bars) showing waveforms on an expanded timescale, including various LFP signals (note the variety of appearances) and individual extracellular EAPs (red circles). Note, LFP/EAP waveform and polarity vary relative to the number of neurons producing these signals, their proximity to the recording electrode, and their location in relation to the nearby electrode(s). Abbreviations: MEA = microelectrode array; EAP = extracellular action potential; LFP = local field potential; 4-AP = 4-aminopyridine; TTX = tetrodotoxin.
Divulgazioni
The authors have nothing to disclose.
Materials
| 4-aminopyridine | Sigma-Aldrich | 275875-5G | |
| 100% ethanol | Thermo Fisher | AJA214-2.5LPL | |
| CaCl2 1M | Banksia Scientific | 0430/1L | |
| Carbonox (Carbogen – 95% O2, 5% CO2) | Coregas | 219122 | |
| Curved long handle spring scissors | Fine Science Tools | 15015-11 | |
| Custom made air interface incubation chamber | |||
| Foetal bovine serum | Thermo Fisher | 10091130 | |
| Forceps Dumont #5 | Fine Science Tools | 11251-30 | |
| Glucose | Thermo Fisher | AJA783-500G | |
| Horse serum | Thermo Fisher | 16050130 | |
| Inverted microscope | Zeiss | Axiovert10 | |
| KCl | Thermo Fisher | AJA383-500G | |
| Ketamine | Ceva | KETALAB04 | |
| Large surgical scissors | Fine Science Tools | 14007-14 | |
| Loctite 454 Instant Adhesive | Bolts and Industrial Supplies | L4543G | |
| MATLAB | MathWorks | R2018b | |
| MEAs, 3-Dimensional | Multichannel Systems | 60-3DMEA100/12/40iR-Ti, 60-3DMEA200/12/50iR-Ti | 60 titanium nitride (TiN) electrodes with 1 internal reference electrode, organised in an 8×8 square grid. Electrodes are 12 µm in diameter, 40 µm (100/12/40) or 50 µm (200/12/50) high and equidistantly spaced 100 µm (100/12/40) or 200 µm (200/12/50) apart. |
| MEA headstage | Multichannel Systems | MEA2100-HS60 | |
| MEA interface board | Multichannel Systems | MCS-IFB 3.0 Multiboot | |
| MEA net | Multichannel Systems | ALA HSG-MEA-5BD | |
| MEA perfusion system | Multichannel Systems | PPS2 | |
| MEAs, Planar | Multichannel Systems | 60MEA200/30iR-Ti, 60MEA500/30iR-Ti | 60 titanium nitride (TiN) electrodes with 1 internal reference electrode, organised in either a 8×8 square grid (200/30) or a 6×10 rectangular grid (500/30). Electrodes are 30 µm in diameter and equidistantly spaced 200 µm (200/30) or 500 µm (500/30) apart. |
| MgCl2 | Thermo Fisher | AJA296-500G | |
| Microscope camera | Motic | Moticam X Wi-Fi | |
| Multi Channel Analyser software | Multichannel Systems | V 2.17.4 | |
| Multi Channel Experimenter software | Multichannel Systems | V 2.17.4 | |
| NaCl | Thermo Fisher | AJA465-500G | |
| NaHCO3 | Thermo Fisher | AJA475-500G | |
| NaH2PO4 | Thermo Fisher | ACR207805000 | |
| Rongeurs | Fine Science Tools | 16021-14 | |
| Small spring scissors | Fine Science Tools | 91500-09 | |
| Small surgical scissors | Fine Science Tools | 14060-09 | |
| Sucrose | Thermo Fisher | AJA530-500G | |
| Superglue | cyanoacrylate adhesive | ||
| Tetrodotoxin | Abcam | AB120055 | |
| Vibration isolation table | Newport | VH3048W-OPT | |
| Vibrating microtome | Leica | VT1200 S |

