Whole-mount Immunostaining and Automatic Counting of Mouse Retinal Ganglion Cells
Summary
This protocol describes the procedure for isolating the whole-mount mouse retina and performing immunostaining to label all retinal ganglion cells (RGCs). The process is followed by imaging and automatically counting RGCs using AI-based software, providing a simple, fast, and accurate method for quantifying RGCs in the entire mouse retina.
Abstract
Glaucoma is a leading cause of blindness globally, characterized by a complex pathogenic mechanism that makes vision restoration challenging. Mice serve as valuable animal models for studying the pathogenesis and treatment of glaucoma due to their relatively homogenous genetic background and retinal ganglion cells (RGCs), which structurally resemble those in humans. Accurate assessment of RGC damage and treatment outcomes in mouse glaucoma models requires determining the RGC number across the entire retina. This protocol outlines a comprehensive method involving the isolation of the whole retina, labeling RGCs with specific antibodies, and rapid, accurate automatic counting of RGCs using an AI-based program. The streamlined approach allows for efficient and precise quantification of RGC numbers in mouse retinas, facilitating the evaluation of RGC degeneration and potential therapeutic interventions. By enabling researchers to assess the extent of RGC damage, this protocol contributes to a deeper understanding of glaucoma pathogenesis and aids in developing effective treatment strategies to manage and prevent vision loss.
Introduction
Glaucoma is characterized by the progressive death of ganglion cells, which poses a significant challenge for sight restoration1,2. This disease is a major focus of ophthalmic research due to its prevalence and impact on vision3. Mouse models are indispensable in glaucoma
research due to their homogeneous genetic background, high reproductive capacity, and the similarity of their ganglion cell properties to humans4. The primary goal of this method is to accurately quantify retinal ganglion cells (RGCs) in mouse models, which is essential for understanding the pathogenesis of glaucoma and developing relevant treatments.
The rationale behind developing this technique stems from the need for a reliable and efficient method to assess RGC degeneration in mouse models. Traditional methods, such as labeling RGCs in retinal sections, often provide unreliable results due to the non-uniform distribution of RGCs in the retina5. Quantifying RGCs across the entire retina better reflects changes in their numbers and is crucial for evaluating disease progression and therapeutic interventions.
This method offers several advantages over alternative techniques. For instance, manual counting of retinal ganglion cells (RGCs) in a normal adult mouse retina, which contains 40,000 to 60,000 RGCs, can be time-consuming and error-prone6,7,8. The software for automatic RGC counting that we developed allows for accurate counting in less than 3 min, potentially saving researchers a significant amount of time. Additionally, the AI-based software used for automatic counting minimizes bias and enhances reproducibility.
Furthermore, this technique provides a standardized approach to assess RGC degeneration across different mouse models and experimental conditions, contributing valuable data to the field of glaucoma research. The method aligns with other studies that emphasize the importance of whole-mount retina analysis for understanding retinal changes in diseases9.
To help readers determine whether this method is suitable for their application, it is important to note that this technique is particularly advantageous for researchers studying retinal ganglion cell (RGC) degeneration in mouse models of glaucoma or other retinal diseases. The method is adaptable to various experimental setups and offers a high degree of accuracy and efficiency in RGC counting, making it ideal for both small-scale and large-scale studies. Additionally, the protocol's straightforward design and the availability of user-friendly software make it accessible to researchers with varying levels of expertise in retinal analysis.
Protocol
The procedure complied with the guidelines of the Association for Research in Vision and Ophthalmology for using animals in research and was approved by the Institutional Animal Care and Use Committee (IACUC) of Sichuan Provincial People's Hospital. C57Bl/6J male mice (2 months old) were used in this study. Figure 1 illustrates the overall procedure described here. The details of the reagents and equipment used are listed in the Table of Materials.
1. Isolation of whole-mount retinas
- Animal sacrifice
- Sacrifice the mice using cervical dislocation (following institutionally approved protocols) (Figure 2A). Prior to cervical dislocation,
anesthetize the mice by inhaling isoflurane (3%-5% in oxygen) or by intraperitoneal injection of the ketamine/xylazine mixture.
- Sacrifice the mice using cervical dislocation (following institutionally approved protocols) (Figure 2A). Prior to cervical dislocation,
- Enucleation and eyeball processing
- Mark the edge of the cornea on the dorsal side of the eyeball with a blue marker to remember the orientation during subsequent dissection.
- Remove residual muscle tissues attached to the eyeball with scissors. Transfer the enucleated eyeball to a 2 mL round-bottom microcentrifuge tube filled with 1x PBS for a brief rinse (Figure 2C).
- Fixation
- Transfer the eyeball to another 2 mL round-bottom microcentrifuge tube containing freshly made 4% paraformaldehyde (PFA) for fixation at 4 °C for 10 min (Figure 2D).
- Make a small incision (approximately 1-2 mm) on the cornea using fine scissors under a dissecting microscope.
- Place the eyeball back into the fixative for further fixation at 4 °C for 3 h to preserve tissue morphology (Figure 2D).
- Removal of cornea and lens1,9
- Make a small incision of approximately 1 mm around the corneal limbus, the border between the cornea and the sclera, using fine surgical scissors.
- Carefully cut around the circumference of the cornea, excising it entirely to expose the underlying structures.
- Use fine forceps to gently grasp and carefully lift the lens from the surrounding tissues to prevent damage to the retina. Maintain the structural integrity of the retina with delicate operation.
- Transfer the eyecup to a microcentrifuge tube for further processing.
NOTE: Excise the cornea and extract the lens more efficiently using the "cross-slit" technique10. Make two perpendicular slits across the cornea and use forceps to extract the lens.
- Scleral incision
- Cut a precise slit in the sclera with fine ophthalmic scissors to facilitate subsequent dissection steps (Figure 2J).
- Retina separation
- Insert forceps with teeth into the incision between the sclera and the retina. Gently grasp the edge of the sclera, avoiding excessive force to prevent tearing the tissue.
- Gradually enlarge the incision by moving the forceps along the boundary between the sclera and the retina with slow, steady hand movements. Use the toothed part of the forceps to carefully separate the sclera from the retina, ensuring the retina is not damaged (Figure 2J).
- Retina cutting
- Divide the retina into four roughly equal-sized flaps using sharp scissors. Ensure cuts are approximately halfway towards the optic nerve and oriented at 90° from each other.
- Retina cleaning
- Gently unfold the dissected retina using a soft-bristled brush, and meticulously remove debris from its surface (Figure 2K).
- Final fixation
- Transfer the isolated retina to a microcentrifuge tube containing fresh 4% PFA using a disposable Pasteur pipette. Allow fixation on ice for an additional 5 h to overnight for tissue preservation.
2. Immunostaining
- Washing
- Wash the isolated whole-mount retina in 1x PBS with gentle agitation for 5 min. Discard the PBS solution. Repeat this process twice to remove residual fixative and debris.
- Blocking
- Immerse the washed retina in a blocking buffer solution (1x PBS with 4% normal donkey serum (NDS) and 0.2% Triton X-100) for 30 min to prevent nonspecific antibody binding and enhance permeabilization (Figure 3A).
- Primary antibody incubation
- Replace the blocking buffer with primary antibody BRN3A (diluted 1:200 in the blocking buffer). Incubate the retina overnight at 4 °C to allow specific antigen-antibody binding (Figure 3B).
- Washing
- Wash the retina three times with 1x PBS for 5 min each to remove unbound primary antibodies and reduce background staining (Figure 3C).
- Secondary antibody incubation
- Incubate the retina with Alexa594- or Alexa488-conjugated anti-rabbit secondary antibody (diluted 1:300) for 2 h at room temperature to visualize target antigens (Figure 3D).
- Washing
- Perform three consecutive washes with 1x PBS for 5 min each to remove excess secondary antibodies and minimize nonspecific binding.
- Mounting
- Gently transfer the immunostained retina onto a glass microscope slide, carefully flattening it to minimize folding or distortion.
- Apply a small drop of anti-fade mounting medium to the center of the retina, ensuring even distribution across the tissue.
- Carefully position a coverslip over the mounted retina, avoiding air bubbles (Figure 3E,F).
3. Image processing
NOTE: Import the image into the RGC automatic counting software and start counting. The number of BRN3A-positive cells for the entire retina can be obtained in minutes. Download the AutoCount software from GitHub (https://github.com/MOEMIL/Intelligent-quantifying-RGCs). Follow the detailed installation steps for the software below:
- Download the code from the GitHub link. One will receive a file named "Intelligent-quantifying-RGCs.zip". Unzip this file.
- Download the necessary files from the cloud disk (see Table of Materials) and unzip them. The downloaded file is named "yolov5_cpu". Unzip this package to the current folder to avoid unexpected issues.
- Locate the Config file from step 1 and modify its content to point to the "yolov5_cpu" path.
NOTE: For example, if the downloaded "yolov5_cpu" is placed in "D:1meyolov5_cpu_", change the "config.ini" file in step 1 to "D:/1me/yolov5_cpu" (use / instead of ). (Figure 4A). - Open the graphical interface by clicking on userinterface.exe. The initial loading may be slow (run as administrator) (Figure 4B).
- After opening the software, verify whether the "pmse_plus.pt" model file is present in the root directory of drive C. If it is not present, copy it to this directory.
NOTE: The "pmse_plus.pt" model file is located in the weights folder of the "yolov5" folder, found in the extracted Intelligent-quantifying RGCs folder (Figure 4C). - Prepare the images to be detected.
NOTE: The software allows only one or five pictures to be read at a time. Reading more will result in an error.
4. Automated cell counting
- Open the counting software (Figure 5A).
- Click on OPEN IMAGE to import the image (Figure 5B).
- Click on RUN to allow the software to automatically count the cells (Figure 5C).
NOTE: The software will mark the counted cells with a red square box and display the cell count in the top right corner (Figure 5D).
Representative Results
This protocol details the methodology for whole-mount immunostaining of mouse retinas, ensuring meticulous tissue preparation, precise antibody incubation, and reliable automated cell counting. The procedure facilitates robust labeling and quantification of retinal ganglion cells (RGCs), enabling accurate assessment of cellular populations in various experimental contexts. The method was used to count RGCs in wild-type and glaucoma-modeled mice, yielding consistent results. A representative result shows that the number of RGCs in a normal adult mouse at 2 months of age is approximately 45,000 (Figure 6).
To evaluate whether the AI software can accurately quantify BRN3a-labeled RGCs in normal and disease models, we generated an N-methyl-D-aspartate (NMDA)-induced mouse model of glaucoma7. The whole-mount retina from this model was immunostained with anti-BRN3A, and the RGCs were quantified using the automatic counting software. The result indicated a relatively high background staining (Figure 7). Nonetheless, the software was able to accurately identify almost all the RGCs. A total of 15,231 RGCs were identified in this retina, compared to approximately 45,000-50,000 RGCs in a normal mouse retina, indicating severe degeneration. Thus, the AI software can also be effectively used to count RGCs in retinas with less optimal staining quality.
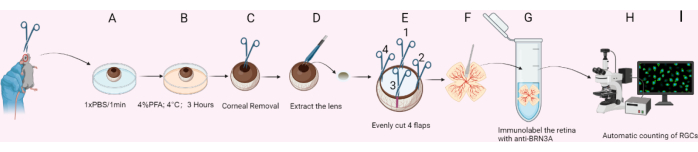
Figure 1: Schematic diagram for RGC preparation. (A) The eyeball is removed and placed in 1x PBS. (B) The eyeball is fixed in 4% PFA on ice for 3 h. (C) Removal of the cornea. (D) Extraction of the lens. (E) The eyecup is divided into 4 flaps using scissors. (F) The sclera is peeled off, and debris is gently removed from the retina surface with a soft brush. (G) The retina is immunolabeled with anti-BRN3A. (H) Fluorescent signals are imaged using a confocal microscope. (I) Automatic counting of RGCs. Please click here to view a larger version of this figure.
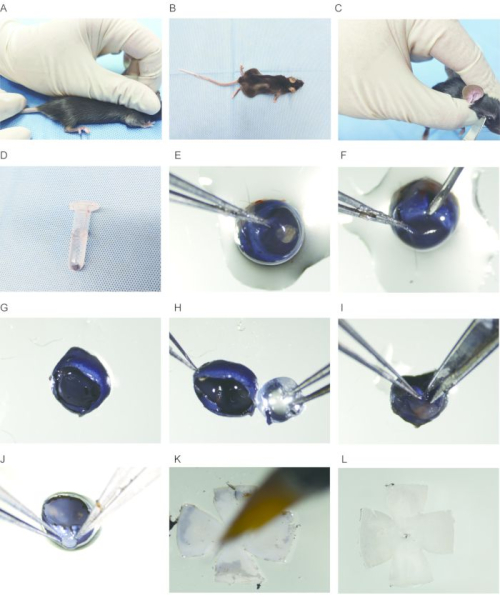
Figure 2: Isolation of the whole-mount mouse retina. (A) The mouse is euthanized by cervical dislocation. (B) The mouse is placed on a flat surface. (C) The eyeball is carefully removed with scissors. (D) The eyeball is gently placed in 1x PBS. (E) The eyeball is fixed with forceps at the edge of the cornea. (F) A small incision is made on the edge of the cornea with a needle. (G) The bowl-shaped structure is removed after removing the cornea. (H) The lens is carefully removed with forceps. (I) A small incision is made along the edge of the cornea towards the optic nerve on the sclera, avoiding damage to the retina. (J) The sclera is torn open along the incision with forceps. (K) The retina is flattened and cleaned with a paintbrush. (L) The retina is flattened and rapidly fixed with ice-cold methanol. Please click here to view a larger version of this figure.
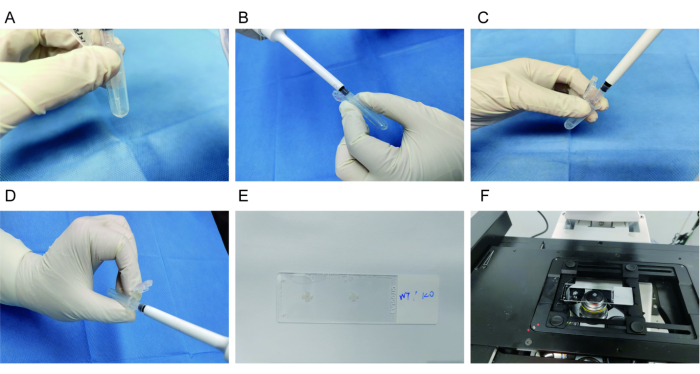
Figure 3: Immunostaining of the whole-mount mouse retina. (A) After washing with 1x PBS, the retina is blocked with a blocking buffer. (B) The retina is incubated with a primary antibody at 4 °C overnight. (C) The retina is washed with 1x PBS three times. (D) The retina is incubated with a secondary antibody for 2 h at room temperature. (E) The retina is mounted on a glass slide with RGCs facing upwards. (F) Images are acquired using a laser confocal microscope. Please click here to view a larger version of this figure.
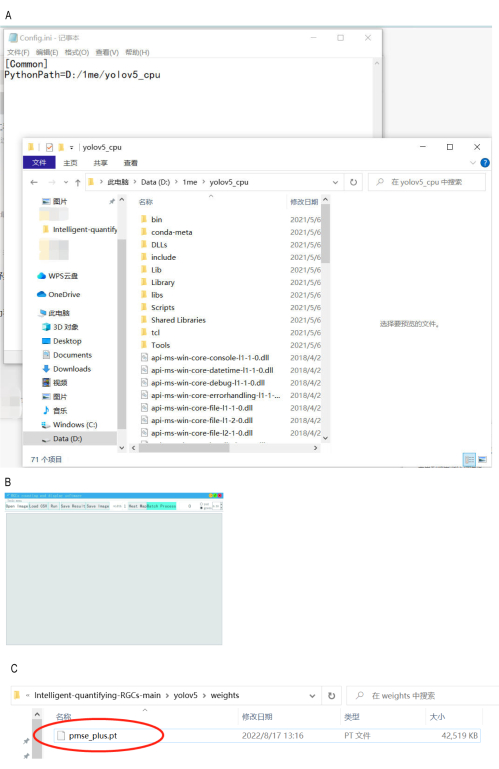
Figure 4: Installation of automatic counting software. (A) Step 1 involves updating the file path to the "yolov5_cpu" directory. (B) The graphical interface is launched. (C) Verification of the model file's presence in the root directory of drive C. Please click here to view a larger version of this figure.
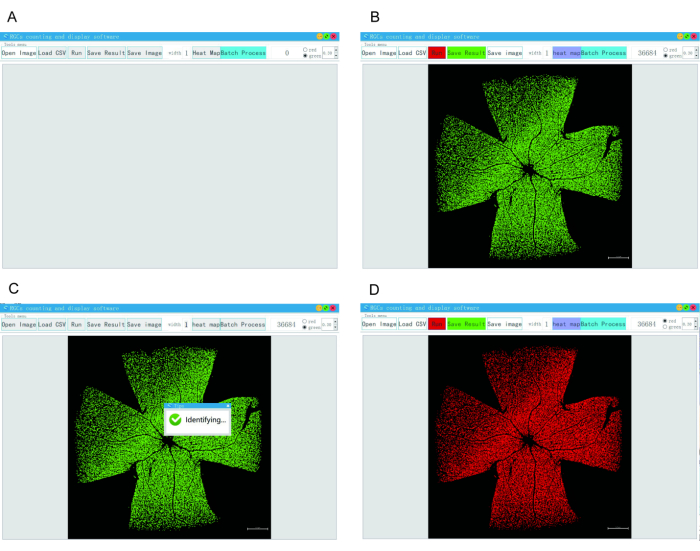
Figure 5: Operation of automated RGC counting software. (A) The counting software is opened. (B) The OPEN IMAGE button is clicked to import the image. (C) The RUN button is clicked to initiate automatic counting. (D) The counted cells are marked with a red square box, and the cell number is displayed at the top right corner. Please click here to view a larger version of this figure.
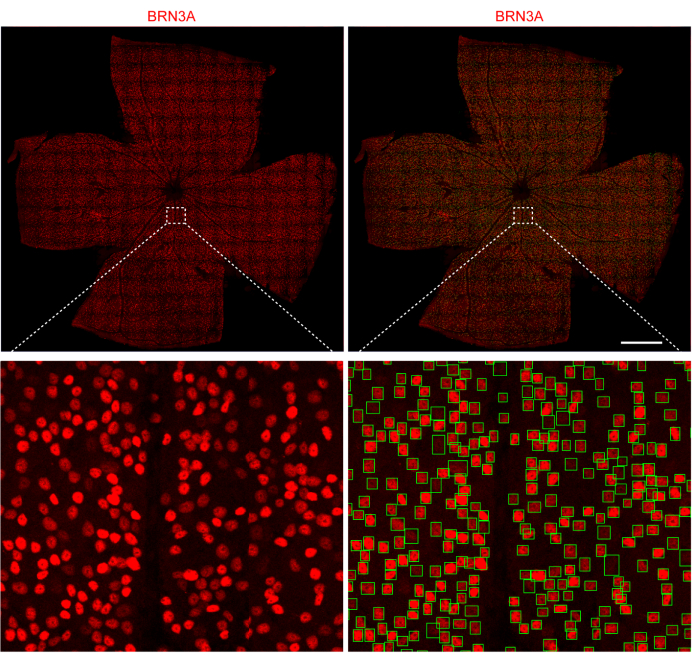
Figure 6: Automated counting of mouse RGCs. Top panel (left) shows representative results of flat-mount retina immunostained with anti-BRN3A; the top panel (right) shows automated counting of RGCs with previously developed software. Scale bar: 100 µm. Insets from the top panels are enlarged and shown in the bottom panel, respectively. Please click here to view a larger version of this figure.
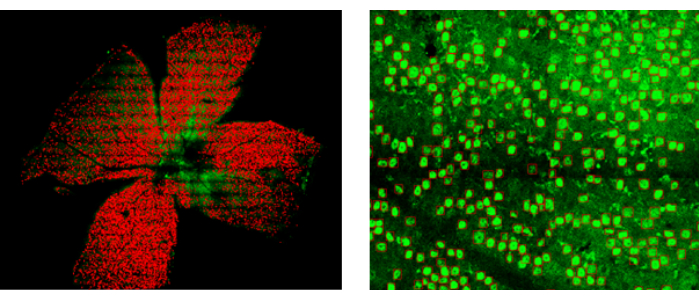
Figure 7: RGC counting in an NMDA-induced mouse model of glaucoma. Left panel: Overview of the whole-mount retina, showing RGCs recognized by the AI software. Right panel: Enlarged view of a partial area of the retina. Please click here to view a larger version of this figure.
Discussion
This protocol provides a method for determining all retinal ganglion cells (RGCs) in a mouse retina, which can be used to monitor the progression of RGC degeneration in mouse models for glaucoma studies. The mouse retina is a delicate nerve tissue5, and isolating whole-mount retinas from mouse eyes requires repeated practice. During experimentation, it was found that the fixation time significantly affects retina morphology. For mice around 2 weeks of age, a fixation time of 20 min is sufficient to obtain good results due to the thin and weak sclera. However, for adult mice older than 2 months, 20 min of fixation is inadequate. A fixation time of 3 h is optimal for obtaining intact whole-mount retinas.
In the process of retina isolation, the most challenging step is separating the sclera from the retina10. The sclera can be relatively easily peeled off from the retina using two sets of forceps with toothed tips. By using the two sets of forceps to grip the sclera and pull apart, the sclera is torn into small pieces and separated from the retina.
A high-quality image of retinal ganglion cells (RGCs) is critical for accurate counting. Since the retina is three-dimensional, not all RGCs are on the same level in a flat-mounted retina8. When the retina is mounted on a glass slide, it should be spread as flat as possible to facilitate subsequent imaging. During imaging, images are acquired at multiple focusing levels to capture all RGCs across different areas of the entire retina.
In summary, this protocol provides a standardized procedure for the automatic counting of all RGCs in a flat-mounted mouse retina. This method enables scientists to accurately assess RGC degeneration in mouse models, enhancing the efficiency and accuracy of RGC quantification. Such precision is crucial for understanding and treating glaucoma and related diseases.
Divulgations
The authors have nothing to disclose.
Acknowledgements
This research project was supported by the National Natural Science Foundation of China (82371059 (H.Z.)), the Department of Science and Technology of Sichuan Province, China (2023JDZH0002 (H.Z.)), the Chengdu Science and Technology Bureau (2022-YF05-01984-SN (H.Z.)), and Sichuan Provincial People's Hospital (30320230095 (J.Y.), 30420220062 (J.Y.)).
Materials
| 1× PBS | Servicebio | G4202 | |
| Alexa594-conjugated Goat Anti-Rabbit IgG (H+L) | ThermoFisher | A-11012 | |
| Anti-BRN3A antibody [EPR23257-285] | Abcam | ab245230 | |
| AutoCount software | https://github.com/MOEMIL/Intelligent-quantifying-RGCs | ||
| Cloud disk | Google drive link | https://drive.google.com/file/d/1yOEsBvil6KEdZFa5ENQxB6 | |
| Cloud disk | Baidu link | Extraction code: g44k | https://pan.baidu.com/s/1lccg1OVbeudsp2VtnqxWZg |
| Marker | Sharpie | ||
| Normal Donkey Serum | Biosharp, Labgic | 25030081 | |
| Paraformaldehyde | Macklin | P804536 | |
| ProClean 300 | Beyotime | ST853 | |
| Sucrose | BBI, Sangon | A610498 | |
| Triton X-100 | BioFroxx, neoFroxx | 1139ML100 |
References
- Quigley, H. A., Broman, A. T. The number of people with glaucoma worldwide in 2010 and 2020. Br J Ophthalmol. 90 (3), 262-267 (2006).
- Gallo Afflitto, G., Swaminathan, S. S. Racial-ethnic disparities in concurrent rates of peripapillary & macular OCT parameters among a large glaucomatous clinical population. Eye. , (2024).
- Atkinson, M. J., et al. A new measure of patient satisfaction with ocular hypotensive medications: the Treatment Satisfaction Survey for Intraocular Pressure (TSS-IOP). Health Qual Life Outcomes. 1, 67 (2003).
- Guo, J., et al. A new mouse-fixation device for IOP measurement in awake mice. Vision Res. 219, 108397 (2024).
- Almasieh, M., Wilson, A. M., Morquette, B., Cueva Vargas, J. L., Di Polo, A. The molecular basis of retinal ganglion cell death in glaucoma. Prog Retin Eye Res. 31 (2), 152-181 (2012).
- Jeon, C. J., Strettoi, E., Masland, R. H. The major cell populations of the mouse retina. J Neurosci. 18 (21), 8936-8946 (1998).
- Zhang, J., et al. Automatic counting of retinal ganglion cells in the entire mouse retina based on improved YOLOv5. Zool Res. 43 (5), 738-749 (2022).
- Al-Khindi, T., et al. The transcription factor Tbx5 regulates direction-selective retinal ganglion cell development and image stabilization. Curr Biol. 32 (19), 4286-4298 (2022).
- Claes, M., Moons, L. Retinal ganglion cells: Global number, density and vulnerability to glaucomatous injury in common laboratory mice. Cells. 11 (17), 2689 (2022).
- Zhang, N., Wang, Z., Lin, P., Xing, Y., Yang, N. Methanol-based whole-mount preparation for the investigation of retinal ganglion cells. J Vis Exp. (194), e65222 (2023).

.
