Comparative Analysis of Experimental Methods to Quantify Animal Activity in Caenorhabditis elegans Models of Mitochondrial Disease
Summary
This study presents protocols for two semi-automated locomotor activity analysis approaches in C. elegans complex I disease gas-1(fc21) worms, namely, ZebraLab (a medium-throughput assay) and WormScan (a high-throughput assay) and provide comparative analysis among a wide array of research methods to quantify nematode behavior and integrated neuromuscular function.
Abstract
Caenorhabditis elegans is widely recognized for its central utility as a translational animal model to efficiently interrogate mechanisms and therapies of diverse human diseases. Worms are particularly well-suited for high-throughput genetic and drug screens to gain deeper insight into therapeutic targets and therapies by exploiting their fast development cycle, large brood size, short lifespan, microscopic transparency, low maintenance costs, robust suite of genomic tools, mutant repositories, and experimental methodologies to interrogate both in vivo and ex vivo physiology. Worm locomotor activity represents a particularly relevant phenotype that is frequently impaired in mitochondrial disease, which is highly heterogeneous in causes and manifestations but collectively shares an impaired capacity to produce cellular energy. While a suite of different methodologies may be used to interrogate worm behavior, these vary greatly in experimental costs, complexity, and utility for genomic or drug high-throughput screens. Here, the relative throughput, advantages, and limitations of 16 different activity analysis methodologies were compared that quantify nematode locomotion, thrashing, pharyngeal pumping, and/or chemotaxis in single worms or worm populations of C. elegans at different stages, ages, and experimental durations. Detailed protocols were demonstrated for two semi-automated methods to quantify nematode locomotor activity that represent novel applications of available software tools, namely, ZebraLab (a medium-throughput approach) and WormScan (a high-throughput approach). Data from applying these methods demonstrated similar degrees of reduced animal activity occurred at the L4 larval stage, and progressed in day 1 adults, in mitochondrial complex I disease (gas-1(fc21)) mutant worms relative to wild-type (N2 Bristol) C. elegans. This data validates the utility for these novel applications of using the ZebraLab or WormScan software tools to quantify worm locomotor activity efficiently and objectively, with variable capacity to support high-throughput drug screening on worm behavior in preclinical animal models of mitochondrial disease.
Introduction
Caenorhabiditis elegans is widely recognized as an outstanding model in neuroscience based on it having 302 neurons that coordinate all worm behaviors, including mating, feeding, egg-laying, defecation, swimming, and locomotion on solid media1. These hermaphroditic nematodes are also widely used to understand a wide array of human disease mechanisms, made possible by its well-characterized genome and high homology of ~80% genes between C. elegans and humans2,3,4. C. elegans have long been used to interrogate human mitochondrial disease5,6,7,8,9,10, which is a highly genetically and phenotypically heterogeneous group of inherited metabolic disorders that share impaired capacity to generate cellular energy and often clinically present with substantially impaired neuromuscular function, exercise intolerance, and fatigue11,12,13,14.To this end, the use of C. elegans models enable preclinical modeling of quantitative aspects of animal activity and neuromuscular function in different genetic subtypes of mitochondrial disease, as well as their response to candidate therapies that may improve their neuromuscular function and overall activity.
Neuromuscular activity in C. elegans is objectively measurable by a range of experimental methodologies, including both manual and semi-automated approaches that allow functional analyses in either solid or liquid media (Table 1)1,15. Accurate quantitation of C. elegans activity has proven important to enable discoveries related to function and development of the muscular and nervous system16,17,18. This study summarizes and compares experimental requirements, advantages, and limitations of 17 different assays that can be performed in research laboratories to evaluate neuromuscular function and activity on four key outcomes in C. elegans diseases models, both at the baseline at a range of developmental stages and ages as well as in response to candidate therapies (Table 1). Indeed, the study provides a detailed overview of the range of available experimental approaches to characterize rates of C. elegans thrashing (body bends per minute), locomotor activity, pharyngeal pumping, and chemotaxis-in each case specifying the experimental and analytic methodology used, the advantages and limitations of each method, the equipment and software needed to perform and analyze each assay, and the throughput capacity of each method to support its use for high-throughput genetic or drug screening purposes. The throughput capacity of each assay is described as low, medium, or high based on the experimental protocol complexity, including worm maintenance, processing time, the use of single or multi-well plates, and/or experimenter time needed to complete the experimental setting and data analyses.
Manual analyses of thrashing19, locomotor activity20, pharyngeal pumping17,21, and chemotaxis22,23 are well-established methodologies to evaluate worm activity that require a stereomicroscope24. While measuring thrashing activity of worms requires analysis in liquid media to determine the frequency of body bends per minute, worm locomotor activity may be measured either on solid media or in liquid media. However, manual analyses of individual worm activity are inherently time-consuming and involves unavoidable user-generated bias. Automation of worm activity analyses minimizes user-generated bias and can greatly increase experimental throughput25. Video recordings of worm thrashing activity in liquid media can be analyzed using wrMTrck, an ImageJ plugin26. However, the original experimental settings that were developed for wrMTrck limited its utility, since too many worms in a single liquid drop led to overlapping of worms that made accurate tracking difficult. While this experimental limitation has been resolved27, the wrMTrck method is not able to support high-throughput screening.
A range of methods exist to quantify worm locomotor activity at baseline and in response to candidate therapies in C. elegans mitochondrial disease models. These include ZebraLab (ViewPoint Life Sciences), Tierpsy Tracker28, wide field-of-view nematode tracking platform (WF-NTP)29, WormMotel, WormWatcher30, WormLab31, Infinity Chip32, and WMicrotracker One33 (Table 1). These methods enable concurrent analysis of locomotion in multiple worm strains or conditions, typically on multi-well plates, thereby supporting higher-throughput drug screening applications. Some of these methods have unique considerations that may limit or enhance their general utility, such as the need for expensive equipment versus open-access software, and varying ease of performing experimental protocols. Overall, no single experimental system or protocol is ideally suited to all C. elegans locomotor activity experiments. Rather, it is important to carefully choose which method is best suited to the specific investigator's experimental goals and requirements.
Pharyngeal pumping represents another important outcome to assess neuromuscular activity in C. elegans. The C. elegans pharynx is composed of 20 muscle cells, 20 neurons, and 20 other cells that enable ingestion of Escherichia coli (E. coli) at the anterior end of the worm's alimentary tract34,35,36. Several manual methods have been established to determine pharyngeal pumping rates17,21,37,38. Most methods are based on the use of a stereomicroscope and camera to visualize and record pharyngeal pumping frequency with direct counting by the experimental observer21. Automated pharyngeal pumping rate analysis is possible by performing an extracellular recording termed electropharyngeogram (EPG), which provides additional information on the duration of each pump39. Pharyngeal pumping rate analysis is also possible in a microfluidic system, WormSpa, where individual worms are confined in chambers40,41. A commercial method available to facilitate analysis of the pharyngeal pump rate is the ScreenChip System (InVivo Biosystems), which measures, visualizes, and analyzes the neuromuscular aspects of feeding behavior in a single worm that is immobilized in a custom chip. This pharyngeal pumping quantitation approach can be used to assess both neuronal and physiological responses to drugs, aging, and other factors42,43,44,45.
Chemotaxis describes the movement of C. elegans in response to an odorant placed away from the worms in a defined area of the nematode growth media (NGM) plate. Assessing the chemotaxis response provides an integrated measure of worm neuronal and neuromuscular activity that is quantifiable by observing and measuring the physical distance traveled by worms toward the odorant in a defined time period46. The Multi-Worm Tracker is an automatic method that can be used to improve the experimental efficiency of quantifying the distance traveled by worms toward an attractant or from a repellant47.
Here, the detailed protocol for two novel, semi-automated methods established for quantifying worm activity is described. The first approach utilizes ZebraLab a commercial software that was originally developed to study swimming activity of Danio rerio (zebrafish), for a novel medium-throughput application to quantify overall locomotor activity in liquid media of C. elegans based on pixel changes during movement (Table 1, Figure 1). Data output is quickly obtained from a large number of concurrent conditions and samples analyzed on a glass slide, although this method is not suitable to a multi-well plate format. The second approach is a novel adaptation of the WormScan methodology48,49 (Figure 2), which uses a flatbed scanner to create a differential image of two sequential scans that can variably be used with open-source software to enable semi-automated quantitative analysis of integrated physiologic outcomes such as fecundity and survival. Here, a novel high-throughput adaptation of the WormScan methodology to quantify worm locomotor activity in liquid media in populations of fifteen larval-stage 4 (L4) worms per well of a 96-well, flat-bottom plate was developed. This semi-automated and low-cost WormScan methodology can be readily adapted to high-throughput drug screens, as well as to analyses of various animal stages and ages48,49.
Here, the protocol and efficacy of analyzing C. elegans locomotor activity using both ZebraLab and WormScan semi-automated methods is demonstrated in a well-established C. elegans model for mitochondrial complex I disease, gas-1(fc21). gas-1 (K09A9.5 gene) is an ortholog of human NDUFS2 (NADH: ubiquinone oxidoreductase core (iron-sulfur protein) subunit 2) (Figure 3). The C. elegans gas-1(fc21) mutant strain carries a homozygous p.R290K missense mutation in the human ortholog of NDUFS250, causing significantly decreased fecundity and lifespan, impaired respiratory chain oxidative phosphorylation (OXPHOS) capacity51, as well as decreased mitochondrial mass and membrane potential with increased oxidative stress5,8. Despite its well-established use over the past two decades to study mitochondrial disease, locomotor activity of gas-1(fc21) mutants was not previously reported. Here, ZebraLab and WormScan methods were applied to independently quantify the locomotor activity of gas-1(fc21) as compared to wild-type (WT, N2 Bristol) worms, both as a way to validate the methods as well as to demonstrate their comparative utility and efficiency of the experimental protocols and informatics analyses. ZebraLab software allowed rapid quantitation of several concurrent conditions of worm locomotor activity in C. elegans mitochondrial disease models, with potential application for targeted drug screening or validation studies. WormScan analysis, in particular, is well-suited to readily enable high-throughput drug screens of compound libraries and prioritize leads that improve the animal neuromuscular function and the locomotor activity in preclinical C. elegans models of primary mitochondrial disease.
Protocol
1. Worm locomotor activity analysis in liquid media on glass slides using ZebraLab software
- Nematode growth and handling
- Grow C. elegans on Petri plates containing nematode growth media (NGM) and spread with Escherichia coli OP50 as food source. Maintain worm culture at 20 °C, as previously described8.
- Synchronize worms performing a timed egg lay52 and study worms at the desired stage. In this protocol, L4 stage worms were analyzed.
- Grow control and mutant worm strains on NGM plates with and without drug treatments to be tested or buffer control. To evaluate the drug treatment effects, prepare the desired drug stock concentration in S. basal solution; spread the calculated specific volume onto the NGM plates and allow it to dry. Transfer the worms at a specific larval or adult stage and maintain on the drug treatment plate for the desired duration before analysis.
- Experimental set-up of worms for locomotor activity video recording and ZebraLab analysis
- Pick 5 synchronized L4 worms per strain and condition using a worm pick. Pipette a single 20 µL drop of S. basal solution onto a glass slide located under a stereomicroscope connected to a camera and transfer 5 worms into it (Figure 1A,B). Transfer the 5 worms from the Petri dish containing NGM and E. coli OP50 to the liquid drop only at the moment preceding the recording.
NOTE: Continue to maintain the other worms on the Petri dish until the prior video is recorded. This will avoid the damage caused on the other worms due to the drying up of the 20 µl drops during the procedure (dry time ~15-20 min). - Pipette multiple drops on one slide to obtain multiple technical replicates (Figure 1A). Select worms from different NGM plates (biological replicate). Do not use cover slip.
- Adjust the microscope's working distance to visualize the complete area of a single drop. Set and maintain a low video resolution (<1024 x 768) to upload the files in the software.
- Allow worms to acclimate on the slide at room temperature for 1 min before recording.
- Record the worm swim activity in 1 drop for 1 min at 15 frames per second (fps). Repeat the imaging for each additional drop on the plate.
- Pick 5 synchronized L4 worms per strain and condition using a worm pick. Pipette a single 20 µL drop of S. basal solution onto a glass slide located under a stereomicroscope connected to a camera and transfer 5 worms into it (Figure 1A,B). Transfer the 5 worms from the Petri dish containing NGM and E. coli OP50 to the liquid drop only at the moment preceding the recording.
- C. elegans locomotor activity recording analysis in ZebraLab software
- Use the option ZEBRALAB AVI to upload videos to the software. Click on the option Quantization with AVI Files (Figure 1C).
- To create a new protocol, select File > Generate Protocol, and then add the number of areas selected for the analysis. Choose Location Count: 1.
- Open Protocol Parameters and select 1 min in the Experiment Duration window. Select a different experimental duration for different experimental durations. Select or deselect the window No Time Bin and choose Integration Period, depending on the data output desired. In this study No Time Bin was selected (Figure 1D).
NOTE: Time bin is the time over which the activity will be averaged. - If a protocol was already created, select Open Protocol and select the saved protocol (in .vte format).
- Select File and Open a Movie to upload each individual video file that was previously recorded.
- Select the Area icon indicated in Figure 1E (black arrow) to build a single area of detection and create an area around the whole liquid drop where worms are located. Click on Select, then the green circle icon (gray arrow) under Areas > Build > Clear marks.
NOTE: The activity of all worms in the defined drop will be detected in the selected area (Figure 1E,F). - Go to Calibration > Draw scale (Figure 1E) and draw a horizontal line from the left to the right of the video area. Indicate the real distance to calibrate. Then select Apply to Group.
- Unselect the icon selected to build the area (arrow in Figure 1E) and select or deselect Transparent.
NOTE: In this study, Transparent was selected and gave better results. - Adjust Detection Sensitivity and Activity Threshold to allow the detection of all the different C. elegans worm strains analyzed.
NOTE: In this experiment, the Detection Sensitivity was set at 8 with Burst and Freezing values of 15 and 2, respectively (Figure 1F). - Set Display Scale at 70 to visualize the track made by the animal while activity analysis is underway. Then select Apply to Group (Figure 1F).
- Click on Experiment > Execute > Save as, and then on Start. A window opens. Choose Do you want to process the video media at maximum computer speed? to analyze the video quickly (e.g., a 1 min video recording is analyzed by the in ZebraLab software in 5 s).
- Another window opens: Running Experiment; click on Start to proceed with the experiment.
- After the video recording is complete, the analysis stops. Click on Experiment > Stop. This saves the activity analyzed from a single drop in a spreadsheet.
- Repeat the analysis for each video of individual drop. Each drop is one technical replicate.
- Output and analysis of ZebraLab data
NOTE: After the experiment, data from each video is individually saved as separate spreadsheets in the chosen folder. In the data output file, the integrated activity level of all worms moving in an individual drop is recorded as pixel changes under actinteg.- Open each spreadsheet obtained from the analysis of each video. Compile them manually into a single file.
Normalize the mutant and wild-type data to a percent of control. Here, statistical analyses were performed to compare mean activity levels between groups.
- Open each spreadsheet obtained from the analysis of each video. Compile them manually into a single file.
2. Worm locomotor activity analysis in liquid media in 96-well plate format by WormScan software analysis
- Nematode growth and handling
- Grow C. elegans as described in section 1.1.1.
- Synchronize worms as described in section 1.1.2.
- Grow worms on specific media as described in section 1.1.3 until L4 stage or day 1 adult.
- Experimental set-up of worms in 96-well plate for WormScan activity analysis
- Add 50 µL of 2% weight per volume of E. coli OP50 in liquid suspension in S. basal medium to each well of a 96-well, clear, flat-bottom, microplate, as previously described49,53.
- Under a stereomicroscope, manually pick 15 synchronized worms at L4 stage or day 1 adult from their NGM plates into liquid media within each experimental well of the 96-well microplate. Allow the worms to acclimate to the liquid media for 20 min before scanning.
NOTE: Other animal stages and ages can be readily substituted for study.
- WormScan activity analysis in 96-well plate and data export to spreadsheet.
- Scan each 96-well, clear, flat-bottom, microplate twice sequentially using a standard flatbed scanner, with less than 10 s between scans.
NOTE: Here, the photo scanner with resolution of 1,200 dots/in and 16-bit grayscale was used to produce jpeg images. Time required to scan four 96-well plates using the photo scanner is less than 10 min. - Align the two sequential scans (Figure 2A) using open source software49.
NOTE: The software generates a difference image to evaluate pixel changes between the two sequential images for a region of interest (Figure 2B) and a relative WormScan Score. This WormScan Score is equivalent to changes in locomotor response based on the light intensity produced by the scanner when set to a pixel threshold of 5 (Figure 2C). - Export the data from WormScan as a spreadsheet. Save the spreadsheet containing the data to the local computer. Normalize the data as percentage of control (POC) and compare across biological replicate experiments for diverse mutant or treatment conditions. Perform statistical analysis to compare mutant and control means using student t-test.
- Scan each 96-well, clear, flat-bottom, microplate twice sequentially using a standard flatbed scanner, with less than 10 s between scans.
Representative Results
Analysis of C. elegans locomotor activity in the liquid media could easily capture an integrated phenotype of mitochondrial disease worm models that may not be easily quantifiable on solid media. ZebraLab was used to quantify locomotor activity of the well-established mitochondrial complex I disease gas-1(fc21) strain relative to WTworms in liquid media at the L4 larval stage. The activity of 5 worms in a single liquid drop was recorded over 1 min, with a total of 19 videos (technical replicates) recorded for each strain, resulting in total the analysis of 95 worms per strain. Four biological replicate experiments were obtained per strain. Worm activity is displayed as pixel change (Figure 3A), and as percent of control (POC) when normalized to N2 Bristol WT control (Figure 3B). The gas-1(fc21) worms (62% ± 16% pixel change, mean ± SD, n = 19) had a significant 38% decrease (p < 0.001, t-test) in their locomotor activity at L4 stage as compared to WT worms (100% ± 11.35%, mean ± SD, n = 95 worms per condition in 19 technical replicates over 4 biological replicates).
WormScan analysis was also performed to quantify the locomotor activity of L4 stage gas-1(fc21) and WT worms in liquid media. Data was collected for three biological replicate experiments, where each biological replicate plate was evaluated by two sequential images scanned using a standard flatbed scanner. Worm activity of the differential images was compared as pixel change and normalized to concurrent N2 Bristol WT control. Similarly, as was seen by the Zebrafish behavior screening method, WormScan based analysis demonstrated that the gas-1(fc21) worms (65.9 ± 6.1, mean ± SD, n = 13 wells) had a significant decrease in locomotor activity by 34% (p < 0.001, t-test) compared to N2 Bristol wild-type worms (100% ± 4.8%, mean ± SEM, n = 12 wells) (Figure 3C). Analysis using WormScan on day 1 adult gas-1(fc21) worms (50.1% ± 10.7%, mean ± SD, n = 7 wells) demonstrated a decrease in locomotor activity by 49% (p < 0.001, t-test) compared to WT worms (100% ± 16.2%, mean ± SD, n = 6 wells) (Figure 3D).
Table 1: Comparative overview of experimental assays available to evaluate C. elegans neuromuscular activity. A detailed overview is provided of a wide array of 16 different experimental techniques that can be used to quantify worm neuromuscular activity on the phenotypic outcomes of thrashing, locomotion, pharyngeal pumping, and/or chemotaxis in C. elegans. Read format, methodology, experimental throughput capacity, software and/or equipment requirements, as well as advantages and limitations of each assay are detailed. References and relevant websites for each assay and software tool are also provided. The throughput capacity of each assay is described as low, medium, or high, as based on the experimental complexity, the use of single or multi-well plates, and/or experimenter time needed to complete the experimental setting and data analyses. * Indicates that the methodologies can also be used for evaluation of locomotion. Please click here to download this Table.

Figure 1: C. elegans locomotor activity analysis using ZebraLab software. (A,B) Experimental protocol for worm video recordings. Five worms were introduced per drop (20 µL) of S. basal solution, with four drops placed on a single glass slide under a stereomicroscope. Each drop of 5 worms represented a technical replicate experiment and was recorded for 1 min in a separate movie using a charged-coupled device (CCD) camera. (C–F) Experimental settings in ZebraLab as adapted to evaluation of locomotor activity in C. elegans. (C) Selection of Quantization with AVI files to quantify worm locomotor activity of each recorded video. (D) Protocol parameter settings, with 1 min selected as experiment duration. (E) Build area to select the area of interest. The area was selected and built around 1 drop of solution in which 5 worms were placed. (F) Detection was determined based on gray-scale thresholding to detect the whole body of each worm (red). In the threshold section, burst and freezing values were selected to analyze worm activity as pixel changes. Please click here to view a larger version of this figure.
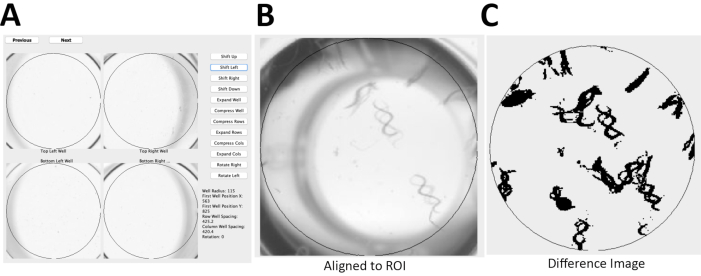
Figure 2: C. elegans locomotor activity analysis using WormScan methodology. (A) Using an Epson v800 flatbed scanner, two immediately sequential scans of a 96-well plate were captured with a resolution of 1,200 dots/in and 16-bit grayscale to produce jpeg images. (B) These two sequential images of a 96-well plate were then aligned to a reference region of interest (ROI), of WT worms. (C) Image analysis is based on a difference image score calculated for each ROI with 15 worms/well for N2 Bristol. The difference image was normalized and reported as percentage of control (POC). Please click here to view a larger version of this figure.
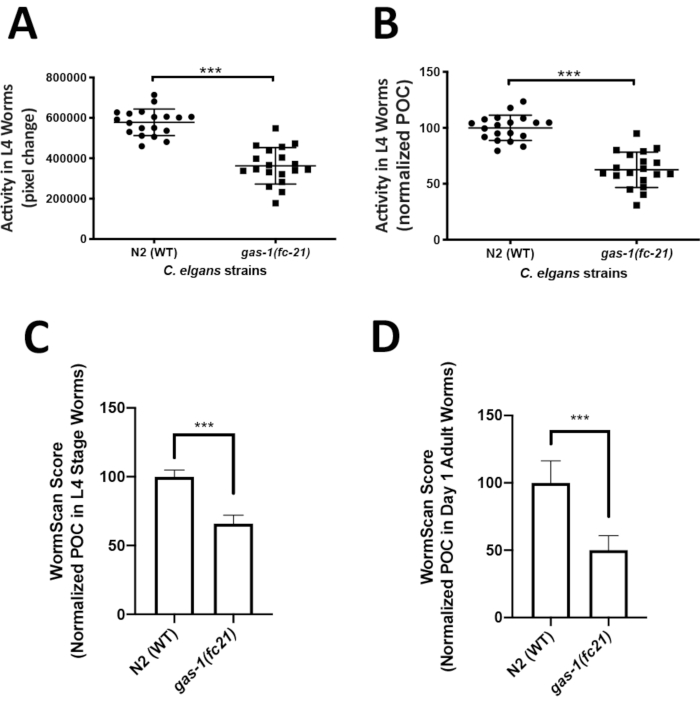
Figure 3: Comparative analysis of locomotor activity by ZebraLab and WormScan software assays in gas-1(fc21) mitochondrial disease worms relative to N2 Bristol wild-type worms. (A,B) WT and gas-1(fc21) worm activity in liquid drops (5 worms/drop) was video recorded for 1 min and quantified as (A) pixels change or (B) percentage of wild-type control using the ZebraLab software. Overall, ZebraLab-based worm activity analysis demonstrated a significant decrease by 38% in gas-1(fc21) L4 stage worms as compared to wild-type controls (*** p < 0.001). The graph displays mean ± SD of all data, where each dot conveys the overall activity of five worms per S. basal drop. Each drop represents a technical replicate, with a total of four biological replicates studied per condition. A total of 19 videos were recorded (one video for each drop of 5 worms), across a total of 95 individual worms studied per condition. Statistical analysis was performed using the student t-test in Prism -GraphPad v6. (C) WT and gas-1(fc21) worms at L4 stage were analyzed by flatbed scanning to produce two sequential images that were analyzed in WormScan software to yield a difference image. Three biological replicate experiments were performed with 15 worms per well in a 96 well-plate. The activity of WT worms was used as the baseline to normalize percentage of control (POC). gas-1(fc21) activity was decreased by 34% as compared to wild-type control (*** p < 0.001). Bar graphs convey mean and standard deviation across three biological replicate experiments. (D) N2 and gas-1(fc21)worms at adult day 1 stage were analyzed similarly as detailed for panel C. gas-1(fc21) activity in day 1 adults was decreased by 49.1% relative to wild-type control worms (*** p < 0.001). Bar graphs convey mean and standard deviation of pixel changes in one biological replicate comparing N2 (n = 6 wells of 15 worms/well) and gas-1(fc21) (n = 7 wells of 15 worms/well). Please click here to view a larger version of this figure.
Discussion
Here, the study summarized detailed information and rationales for studying C. elegans neuromuscular activity at the level of diverse outcomes, including worm thrashing, locomotion, pharyngeal pumping, and chemotaxis. The comparison of 16 different activity analysis methodologies was performed in terms of the relative throughput, advantages, and limitations of quantify nematode activities in a single worm or worm populations at different ages and experimental durations. Among these, two novel adaptations and applications of semi-automated analyses were highlighted to demonstrate significant reduction in locomotor activity in larval-stage worms at the L4 larval developmental stage and in day 1 young adults of a well-established mitochondrial complex I disease C. elegans strain, gas-1(fc21) relative to WT controls.
In particular, C. elegans neuromuscular function and locomotor activity has been extensively studied on solid media since WT worm movement is very regular in sinusoidal wave patterns. Abnormalities of their regular movement path and the speed can be microscopically detected and manually scored by the experimental observer, in assays that are often low-throughput and tedious. To increase experimental throughput, automated and high-throughput methods should be selected. Impaired activity of worms can be quantifiable in liquid media, where overall locomotor activity of worms in droplets on glass slides or in multi-well plates can be video recorded and quantified in semi-automated or automated fashion with different software tools. Indeed, our data highlight the utility of objectively and efficiently measuring nematode locomotor activity both by a novel application of ZebraLab software to quantify locomotor activity in videos of worms in liquid drops on glass slides (a medium-throughput screening capacity approach), as well as by utilizing WormScan software to quantify worm locomotor activity in differential flatbed scanning images of worms in a 96-well plate liquid media approach54,55,56 (a high-throughput screening capacity approach). The ZebraLab software approach is considered as a medium-throughput assay since it requires that single plates be used for each condition studied, without current developed protocol for multi-well plate formats. While using the ZebraLab software approach requires minimal time when analyzing C. elegans activity in a few conditions, the experimental time increases when applied to multiple conditions. Here, the experimental time was approximately 2 h to transfer worms into liquid droplets and record videos of their activity, considering 18 technical replicates for each condition. The time spent for the analysis of these videos using the ZebraLab software was approximately 1 h. By comparison, the WormScan method is high-throughput because it incorporates a multi-well plate format that permits concurrent analysis of four 96-well plates in less than 10 min and setting up a 96-well plate with a COPAS Bisoter is also less than 10 min.
Both methodologies showed a similarly reduced activity in L4 larval stage gas-1(fc21) mitochondrial disease mutant worms relative to WT worms, thus validating both distinct approaches for quantifying differences in worm behavior. Further, WormScan analysis was used to readily demonstrate that progressive reduction in animal locomotor activity occurred with age in the gas-1(fc21) worms as was evident by the day 1 adult stage.
The main advantages of our adapting the ZebraLab software that was developed for zebrafish swimming analysis to C. elegans activity analysis is that it is experimentally simple and inexpensive to objectively capture worm movement in videos, with semi-automated quantitative analysis in movie files uploaded to ZebraLab software requiring only seconds per technical replicate and removes investigator-based bias that is present in manual quantitation methodologies. Further, having this one software tool in the research laboratory is useful to quantify locomotor activity in two animal model species, namely, zebrafish and C. elegans. The disadvantage is that this is a commercial software that requires purchase and worm videos need to be manually uploaded into the software, although the upload process is straightforward and software analysis time is relatively quick. Overall, the novel application described here of using ZebraLab software to quantify C. elegans locomotor activity holds direct potential to evaluate drug effects on worm behavior, although its throughput remains low-to-medium given its high-resolution requirements necessitate that movies be captured of worms moving in media drops placed on glass slides.
We also adapted WormScan software to efficiently quantify worm locomotor capacity of worms in liquid media in a 96-well plate. This approach offers a high-throughput and low-cost experimental method that uses a standard flatbed scanner to objectively quantify animal fecundity and survival and has previously been used for high-throughput screens in C. elegans49. The main advantages of this technique are that it is very amenable to high-throughput screening, enabling parallel comparisons of a large number of conditions at any stage or age, with ease of use, low setup cost, and rapid analysis in an objective fashion by the WormScan software that is free and publicly available49. The disadvantage of the WormScan is that it can only interrogate the change that occurs between the sequential scans, which in some mutations or conditions may not be sufficiently sensitive to detect small degrees of phenotypic change. In addition, as both ZebraLab and WormScan methods exclusively rely on image pixel changes to assay animal activity, substantial differences in worm size that may occur between strains or in response to a specific therapy over time may need to be considered and/or used as a normalization factor for both methods, to more specifically enable evaluation and comparison of mutation and/or treatment effects on animal locomotor activity.
Overall, a wide array of experimental methods can be used to assess nematode neuromuscular activity on integrated phenotypic outcomes of thrashing, locomotion, pharyngeal pumping, and/or chemotaxis. We compared 16 of these methods (Table 1), highlighting their specific experimental and analytic requirements, advantages, limitations, and throughput capacity. Among these, we provided detailed experimental protocols for two novel applications of existing software tools, ZebraLab (a medium-throughput approach) and WormScan (a high-throughput approach), which are particularly useful to semi-automatically, objectively, and quickly quantify worm locomotion activity in liquid media. Both experimental approaches revealed a similarly reduced degree of locomotion activity occurred in mitochondrial disease (gas-1(fc21)) relative to WT C. elegans strains at the L4 stage, with progressive decline in locomotor activity by the young adult stage in gas-1(fc21) worms. This data demonstrates the validity of these experimental approaches that yield internally consistent data. Furthermore, this array of methods is highly versatile, enabling a wide-range of worm locomotor activity metrics in diverse disease etiologies, animal stages and ages, and in response to candidate therapeutic modeling or high-throughput drug screens that are useful for preclinical evaluation of lead targets for human disease.
Divulgations
The authors have nothing to disclose.
Acknowledgements
We are grateful to Anthony Rosner, PhD., with his organizational support for the early preparation of this project, and to Erin Haus for contributing to protocol analysis. This work was funded by the Juliet's Cure FBXL4 Mitochondrial Disease Research Fund, the Jaxson Flynt C12ORF65 Research Fund, and the National Institutes of Health (R01-GM120762, R01-GM120762-08S1, R35-GM134863, and T32-NS007413). The content is solely the responsibility of the authors and does not necessarily represent the official views of the funders or the National Institutes of Health.
Materials
| C. elegans wild isolate | Caenorhabditis Genetics Center (CGC) | N2 Bristol | |
| Camera | Olympus | DP73 | |
| gas-1(fc-21) | CGC | CW152 | |
| Microscope slides | ThermoFisher | 4951PLUS | |
| Nematode Growth Medium (NGM) | Research Products International Corp. | N81800-1000.0 | |
| OP50 Escherichia coli | CGC | Uracil auxotroph E. coli strain | |
| Petri dishes (60 mm) | VWR international | 25373-085 | |
| S. Basal | VWR 5.85 g NaCl, 1 g K2 HPO4, 6 g KH2PO4, and 5 mg cholesterol, in 1 l H2O | VWR 101175-162, 103467-156, EM1.09828.1000, 97061-660 | |
| Scanner | EPSON | V800 | |
| Stereomicroscope | Olympus | MVX10 microscope | |
| 96-well flat bottom | VWR international | 29442-056 | |
| WormScan software | Mathew et al. 45 | S1 Standalone Java platform | Software for automation of difference image of scanned plates |
| ZebraLab software | ViewPoint | Software for automated quantization and tracking of zebrafish behavior, designed by ViewPoint (http://www.viewpoint.fr/en/p/software/zebralab-zebrafish-behavior-screening) and here applied to C. elegans. This system is applicable for high-throughput behavioral analysis |
References
- Husson, S. J., Costa, W. S., Schmitt, C., Gottschalk, A. Keeping track of worm trackers. WormBook. , 1-17 (2013).
- Shaye, D. D., Greenwald, I. OrthoList: a compendium of C. elegans genes with human orthologs. PLoS One. 6 (5), 20085 (2011).
- van Ham, T. J., et al. C. elegans model identifies genetic modifiers of alpha-synuclein inclusion formation during aging. PLoS Genetics. 4 (3), 1000027 (2008).
- Kim, W., Underwood, R. S., Greenwald, I., Shaye, D. D. OrthoList 2: A new comparative genomic analysis of human and Caenorhabditis elegans genes. Génétique. 210 (2), 445-461 (2018).
- Dingley, S., et al. Mitochondrial respiratory chain dysfunction variably increases oxidant stress in Caenorhabditis elegans. Mitochondrion. 10 (2), 125-136 (2010).
- Polyak, E., Zhang, Z., Falk, M. J. Molecular profiling of mitochondrial dysfunction in Caenorhabditis elegans. Methods in Molecular Biology. 837, 241-255 (2012).
- McCormick, E., Place, E., Falk, M. J. Molecular genetic testing for mitochondrial disease: from one generation to the next. Neurotherapeutics. 10 (2), 251-261 (2013).
- McCormack, S., et al. Pharmacologic targeting of sirtuin and PPAR signaling improves longevity and mitochondrial physiology in respiratory chain complex I mutant Caenorhabditis elegans. Mitochondrion. 22, 45-59 (2015).
- Polyak, E., et al. N-acetylcysteine and vitamin E rescue animal longevity and cellular oxidative stress in pre-clinical models of mitochondrial complex I disease. Molecular Genetics and Metabolism. 123 (4), 449-462 (2018).
- Guha, S., et al. Pre-clinical evaluation of cysteamine bitartrate as a therapeutic agent for mitochondrial respiratory chain disease. Human Molecular Genetics. 28 (11), 1837-1852 (2019).
- Gorman, G. S., et al. Prevalence of nuclear and mitochondrial DNA mutations related to adult mitochondrial disease. Annals of Neurology. 77 (5), 753-759 (2015).
- Mancuso, M., Orsucci, D., Filosto, M., Simoncini, C., Siciliano, G. Drugs and mitochondrial diseases: 40 queries and answers. Expert Opinion on Pharmacotherapy. 13 (4), 527-543 (2012).
- Gai, X., et al. Mutations in FBXL4, encoding a mitochondrial protein, cause early-onset mitochondrial encephalomyopathy. American Journal of Human Genetics. 93 (3), 482-495 (2013).
- Dillin, A., et al. Rates of behavior and aging specified by mitochondrial function during development. Science. 298 (5602), 2398-2401 (2002).
- Yemini, E., Jucikas, T., Grundy, L. J., Brown, A. E., Schafer, W. R. A database of Caenorhabditis elegans behavioral phenotypes. Nature Methods. 10 (9), 877-879 (2013).
- Bargmann, C. I., Avery, L. Laser killing of cells in Caenorhabditis elegans. Methods in Cell Biology. 48, 225-250 (1995).
- Avery, L., Horvitz, H. R. Effects of starvation and neuroactive drugs on feeding in Caenorhabditis elegans. Journal of Experimental Zoology. 253 (3), 263-270 (1990).
- Chalfie, M., et al. The neural circuit for touch sensitivity in Caenorhabditis elegans. Journal of Neuroscience. 5 (4), 956-964 (1985).
- Ghosh, R., Emmons, S. W. Episodic swimming behavior in the nematode C. elegans. Journal of Experimental Biology. 211 (23), 3703-3711 (2008).
- Rankin, C. H., Beck, C. D., Chiba, C. M. Caenorhabditis elegans: a new model system for the study of learning and memory. Behavioural Brain Research. 37 (1), 89-92 (1990).
- Avery, L. Motor neuron M3 controls pharyngeal muscle relaxation timing in Caenorhabditis elegans. Journal of Experimental Zoology. 175, 283-297 (1993).
- Ward, S. Chemotaxis by the nematode Caenorhabditis elegans: identification of attractants and analysis of the response by use of mutants. Proceedings of the National Academy of Sciences of the United States of America. 70 (3), 817-821 (1973).
- Bargmann, C. I., Thomas, J. H., Horvitz, H. R. Chemosensory cell function in the behavior and development of Caenorhabditis elegans. Cold Spring Harbor Symposia on Quantitative Biology. 55, 529-538 (1990).
- Anne, C. H. Behavior. WormBook: The Online Review of C. elegans Biology. 2005-2018, (2006).
- Biston, M. C., et al. An objective method to measure cell survival by computer-assisted image processing of numeric images of Petri dishes. Physics in Medicine & Biology. 48 (11), 1551-1563 (2003).
- Nussbaum-Krammer, C. I., Neto, M. F., Brielmann, R. M., Pedersen, J. S., Morimoto, R. I. Investigating the spreading and toxicity of prion-like proteins using the metazoan model organism C. elegans. Journal of Visualized Experiments: JoVE. (95), e52321 (2015).
- Shi, W., Qin, J., Ye, N., Lin, B. Droplet-based microfluidic system for individual Caenorhabditis elegans assay. Lab on a Chip. 8 (9), 1432-1435 (2008).
- Javer, A., et al. An open-source platform for analyzing and sharing worm-behavior data. Nature Methods. 15 (9), 645-646 (2018).
- Koopman, M., et al. Assessing motor-related phenotypes of Caenorhabditis elegans with the wide field-of-view nematode tracking platform. Nature Protocols. 15 (6), 2071-2106 (2020).
- Churgin, M. A., et al. Longitudinal imaging of Caenorhabditis elegans in a microfabricated device reveals variation in behavioral decline during aging. eLife. 6, 26652 (2017).
- Angstman, N. B., Kiessling, M. C., Frank, H. G., Schmitz, C. High interindividual variability in dose-dependent reduction in speed of movement after exposing C. elegans to shock waves. Frontiers in Behavioral Neuroscience. 9, 12 (2015).
- Rahman, M., et al. NemaLife chip: a micropillar-based microfluidic culture device optimized for aging studies in crawling C. elegans. Scientific Reports. 10 (1), 16190 (2020).
- Bianchi, J. I., Stockert, J. C., Buzzi, L. I., Blazquez-Castro, A., Simonetta, S. H. Reliable screening of dye phototoxicity by using a Caenorhabditis elegans fast bioassay. PLoS One. 10 (6), 0128898 (2015).
- Albertson, D. G., Thomson, J. N. The pharynx of Caenorhabditis elegans. Philososophical Transactions of the Royal Society of London. Series B, Biological Sciences. 275 (938), 299-325 (1976).
- Raizen, D. M., Avery, L. Electrical activity and behavior in the pharynx of Caenorhabditis elegans. Neuron. 12 (3), 483-495 (1994).
- Avery, L., You, Y. J. C. elegans feeding. WormBook. , 1-23 (2012).
- Morck, C., Rauthan, M., Wagberg, F., Pilon, M. pha-2 encodes the C. elegans ortholog of the homeodomain protein HEX and is required for the formation of the pharyngeal isthmus. Biologie du développement. 272 (2), 403-418 (2004).
- Song, B. M., Avery, L. Serotonin activates overall feeding by activating two separate neural pathways in Caenorhabditis elegans. TheJournal of Neuroscience. 32 (6), 1920-1931 (2012).
- Avery, L., Raizen, D., Lockery, S. Electrophysiological methods. Methods in Cell Biology. 48, 251-269 (1995).
- Kopito, R. B., Levine, E. Durable spatiotemporal surveillance of Caenorhabditis elegans response to environmental cues. Lab in a Chip. 14 (4), 764-770 (2014).
- Lee, K. S., et al. Serotonin-dependent kinetics of feeding bursts underlie a graded response to food availability in C. elegans. Nature Communications. 8, 14221 (2017).
- Brinkmann, V., Ale-Agha, N., Haendeler, J., Ventura, N. The Aryl Hydrocarbon Receptor (AhR) in the aging process: Another puzzling role for this highly conserved transcription factor. Frontiers in Physiology. 10, 1561 (2019).
- Huang, C., et al. Intrinsically aggregation-prone proteins form amyloid-like aggregates and contribute to tissue aging in Caenorhabditis elegans. eLife. 8, 43059 (2019).
- Zhu, B., et al. Functional analysis of epilepsy-associated variants in STXBP1/Munc18-1 using humanized Caenorhabditis elegans. Epilepsia. 61 (4), 810-821 (2020).
- Weeks, J. C., Robinson, K. J., Lockery, S. R., Roberts, W. M. Anthelmintic drug actions in resistant and susceptible C. elegans revealed by electrophysiological recordings in a multichannel microfluidic device. International Journal of Parasitology. Drugs and Drug Resistance. 8 (3), 607-628 (2018).
- Haroon, S., et al. Multiple molecular mechanisms rescue mtDNA disease in C. elegans. Cell Reports. 22 (12), 3115-3125 (2018).
- Swierczek, N. A., Giles, A. C., Rankin, C. H., Kerr, R. A. High-throughput behavioral analysis in C. elegans. Nature Methods. 8 (7), 592-598 (2011).
- Mathew, M. D., Mathew, N. D., Ebert, P. R. WormScan: a technique for high-throughput phenotypic analysis of Caenorhabditis elegans. PLoS One. 7 (3), 33483 (2012).
- Mathew, M. D., et al. Using C. elegans forward and reverse genetics to identify new compounds with anthelmintic activity. PLoS Neglected Tropical Diseases. 10 (10), 0005058 (2016).
- Kayser, E. B., Morgan, P. G., Hoppel, C. L., Sedensky, M. M. Mitochondrial expression and function of GAS-1 in Caenorhabditis elegans. Journal Biological Chemistry. 276 (23), 20551-20558 (2001).
- Falk, M. J., Kayser, E. B., Morgan, P. G., Sedensky, M. M. Mitochondrial complex I function modulates volatile anesthetic sensitivity in C. elegans. Current Biology. 16 (16), 1641-1645 (2006).
- Kwon, Y. J., Guha, S., Tuluc, F., Falk, M. J. High-throughput BioSorter quantification of relative mitochondrial content and membrane potential in living Caenorhabditis elegans. Mitochondrion. 40, 42-50 (2018).
- Hirsh, D., Oppenheim, D., Klass, M. Development of the reproductive system of Caenorhabditis elegans. Biologie du développement. 49 (1), 200-219 (1976).
- Steele, W. B., Mole, R. A., Brooks, B. W. Experimental protocol for examining behavioral response profiles in larval fish: Application to the Neuro-stimulant caffeine. Journal of Visualized Experiments: JoVE. (137), e57938 (2018).
- Carlsson, G., Blomberg, M., Pohl, J., Orn, S. Swimming activity in zebrafish larvae exposed to veterinary antiparasitic pharmaceuticals. Environmental Toxicology and Pharmacology. 63, 74-77 (2018).
- Yang, X., et al. High-throughput screening in larval zebrafish identifies novel potent sedative-hypnotics. Anesthesiology. 129 (3), 459-476 (2018).

