Isolation and Characterization of Extracellular Vesicles Produced by Iron-limited Mycobacteria
Summary
Mycobacterium tuberculosis shows increased production and release of extracellular vesicles in response to low iron conditions. This work details a protocol for generating low iron conditions and methods for the purification and characterization of mycobacterial extracellular vesicles released in response to iron deficiency.
Abstract
Mycobacteria, including Mycobacterium tuberculosis (Mtb), the causative agent of human tuberculosis, naturally release extracellular vesicles (EVs) containing immunologically active molecules. Knowledge regarding the molecular mechanisms of vesicle biogenesis, the content of the vesicles, and their functions at the pathogen-host interface is very limited. Addressing these questions requires rigorous procedures for isolation, purification, and validation of EVs. Previously, vesicle production was found to be enhanced when M. tuberculosis was exposed to iron restriction, a condition encountered by Mtb in the host environment. Presented here is a complete and detailed protocol to isolate and purify EVs from iron-deficient mycobacteria. Quantitative and qualitative methods are applied to validate purified EVs.
Introduction
Mycobacterial extracellular vesicles (MEVs) are membrane-bound nanoparticles, 60−300 nm in size, naturally released by fast- and slow-growing mycobacteria1. MEVs released by pathogenic mycobacteria constitute a mechanism to interact with the host via immunologically active proteins, lipids, and glycolipids secreted in a concentrated and protected manner2,3,4. To characterize MEVs and understand their biogenesis and functions, strict and efficient methods of vesicle purification and validation are crucial. Thus far, MEVs have been isolated from the culture filtrates of mycobacteria grown in an iron-rich medium1,5,6,7,8.
However, previous work demonstrated that iron limitation greatly stimulates vesicle release in Mtb,possibly to capture iron via mycobactin, a siderophore secreted in MEVs9. Although procedures for MEVs isolation from Mtb cultured in high iron medium have been described, an efficient methodology to obtain MEVs from low iron cultures has not been reported. Therefore, the goal of this method is to isolate, purify, and quantify MEVs obtained from low iron cultures so that they can be used for biochemical and functional assays and for the analysis of genetic determinants of vesicle production in mycobacteria.
Protocol
1. Preparation of Iron-depleted Defined Medium
- Prepare 1 L of minimal medium (MM) by dissolving 5 g of KH2PO4, 5 g of L-asparagine, 20 mL of glycerol, and 2 g of dextrose in 900 mL of deionized water in a plastic container. Avoid glass to prevent iron contamination. Adjust the pH to 6.8 with 5 N NaOH and the volume to 1 L with water.
- Add 50 g of metal chelating resin (MCR) and gently agitate using a magnetic stir bar for 24 h at 4 °C. Sterilize and remove the MCR by filtration through a 0.22 µm filter unit with a plastic receiver. To accelerate filtration and prevent filter clogging, let the resin sediment for about 30 min before filtration.
NOTE: This medium contains less than 2 µM residual iron, as determined by atomic absorption spectroscopy. - Supplement MM with 0.5 mg/L of ZnCl2, 40 mg/L of MgSO4, and 0.1 mg/L of MnSO4. Separately, prepare concentrated stocks (1,000x) of each of the metal supplements in deionized water and sterilize by filtration before MM supplementation. Iron-depleted, metal supplemented MM will be referred here as low iron MM (LIMM).
- From a 50 mM stock of FeCl3 dissolved in 10 mM HCl, add 1 mL to 1 L of LIMM (50 µM final concentration) to prepare high iron MM (HIMM).
2. Growing Mycobacteria in Iron-limited Conditions
- Thaw out 50 µL of a frozen 15% glycerol stock of Mtb and streak an agar plate supplemented with 10% ADN enrichment (5 g/L albumin, 2 g/L dextrose, and 0.85 g/L sodium chloride, 0.2% glycerol and 0.05% Tween-80)10. Incubate the plate at 37 °C until colonies are visible.
- Inoculate a single colony of Mtb10 in 2 mL of mycobacterial broth medium (Table of Materials) supplemented with ADN enrichment. Incubate with agitation at 37 °C.
- Let the Mtb culture grow to the late logarithmic phase (OD540 is ~0.8). To check this, measure the OD540 using a spectrophotometer.
- Spread 200 µL of the late logarithmic culture onto the mycobacterial agar plates (Table of Materials) supplemented with 0.2% glycerol, 0.05% Tween-80, and ADN. Inoculate at least 5 plates. Incubate the plates at 37 °C until bacterial growth is visible as a confluent layer. This takes ~1 week for Mtb.
- Wet a sterile cotton swab in LIMM. Use this swab to collect bacteria from the agar plates and inoculate 100 mL of LIMM to prepare a concentrated bacterial suspension with an OD540 of ~1.0.
- Dilute this suspension 10 times to 1 L with LIMM and divide it into two, 2 L sterile plastic bottles, each one containing 500 mL of culture.
- Take out 2 mL of culture and transfer it to a 5 mL culture tube. Add 10 µL of 10% vol/vol tyloxapol.
- Incubate the cultures at 37 °C standing for 14 days.
3. Collection of MEVs
- Measure the OD540 of the 2 mL culture at the time of collection. Make 1:10 serial dilutions of the culture and plate 100 µL of each dilution on agar plates with ADN and 0.05% Tween-80.
- Transfer the culture to five 225 mL conical centrifuge tubes and centrifuge at 2,850 x g for 7 min at 20 °C.
- Collect the culture supernatant with a 50 mL pipette and filter sterilize it through a 0.22 µm filter unit.
4. Isolation of MEVs
- Transfer the culture filtrate into a stirred cell ultrafiltration system placed at 4 °C and filter the concentrate at <50 psi through a 100 kDa cutoff membrane to ~50 mL.
- Centrifuge the concentrated culture filtrate at 15,000 x g for 15 min at 4 °C and collect the supernatant.
- Centrifuge the culture filtrate in polycarbonate ultracentrifugation tubes at 100,000 x g for 2 h at 4 °C.
- Resuspend the membranous pellets in a total of 1 mL of sterile phosphate buffered saline (PBS) by gentle pipetting.
- Mix 0.5 mL of the pellet suspension obtained in step 4.4 with 1.5 mL of 60% iodixanol solution, yielding a final iodixanol concentration of 45% wt/vol. Dispense this mix at the bottom of a 13 mm x 51 mm polypropylene thin-walled ultracentrifuge tube.
- Overlay the MEV-iodixanol 45% suspension with 1 mL of 40%, 35%, 30%, 25%, and 20% (vol/vol in PBS) iodixanol solutions and 1 mL of PBS at the top.
- Centrifuge at 100,000 x g for 18 h at 4 °C.
- Collect the 1 mL density gradient fractions starting from the top using a 1 mL Hamilton syringe.
- Dilute each collected fraction to 20 mL with PBS and centrifuge at 100,000 x g for 2 h at 4 °C.
- Remove the supernatant and suspend the pellet in 0.5 mL of PBS. Store this pellet at 4 °C.
5. Quantification of MEVs
- Measure the protein concentration in each fraction by a Bradford assay (Table of Materials), following the manufacturer's guidelines.
- Perform membrane lipid analysis.
- Incubate 10 µL of each gradient fraction with the fluorescent membrane probe 1-(4-Trimethylammoniumphenyl)-6-Phenyl-1,3,5-Hexatriene p-Toluenesulfonate (TMA-DPH) at a final concentration of 1 µg/mL in a final 50 µL volume of PBS in 96 well black plates.
- Incubate the plates at 33 °C for 20 min.
- Measure the fluorescence at 360 nm excitation and 430 nm emission.
6. Qualitative analysis of MEVs
- Perform protein electrophoresis.
- Mix approximately 1 µg of MEV samples in 16 µL with 4 µL of 5x sample loading buffer (10% w/v SDS, 10 mM dithiothreitol, 20% v/v glycerol 0.2 M Tris-HCl, pH 6.8 0.05% w/v bromophenol blue) and heat the samples in sample buffer at 85 °C for 5 min.
- Load in a 10% Tris/Glycine SDS-polyacrylamide gel11 and run at 10 V/cm in running buffer (25 mM Tris base, 190 mM glycine, 0.1% SDS) until the blue dye front reaches the bottom of the gel.
- Stain the gel with an ultrasensitive protein staining solution (Table of Materials).
- Perform the dot blot.
- Load 2 µL of a MEVs suspension with a concentration of approximately 0.5 µg/µL, and twofold serial dilutions on a nitrocellulose membrane and process for a dot blot according to the manufacturer's instructions.
- Use an antiserum raised in mice against a preparation of MEVs at a dilution of 1:5,000 as the primary antibody and a goat anti-mouse coupled to horseradish peroxidase (HRP) at a 1:10,000 dilution as the secondary antibody. Detect antigen-antibody complexes with an appropriate HRP substrate Blotting Detection Reagent and an imaging system.
- Perform negative staining and electron microscopy.
- Fix 250 µL of MEVs with 2% glutaraldehyde in 0.1 M cacodylate at room temperature for 2 h and incubate overnight in 4% formaldehyde, 1% glutaraldehyde, and 0.1% PBS.
- Stain the fixed samples with 2% osmium tetroxide for 90 min.
- Serially dehydrate the sample in ethanol and embed in Spurr's epoxy resin.
- Observe the MEVs under a transmission electron microscope.
Representative Results
MEVs were purified by differential sedimentation in a density gradient (Figure 1, Figure 2). Under the conditions described, MEVs separated mostly in gradient fraction 3 (F3), which corresponds to 25% iodixanol. This conclusion is based on the detection of protein, membrane lipid, microscopic visualization of intact MEVs, nanoparticle size distribution, and positive reactivity with an antivesicle antiserum (Figure 2, Figure 3). Protein and lipid concentration normalized to colony-forming units (CFUs) showed an approximately eightfold increase of MEV yield in low iron relative to high iron conditions (50 µM FeCl3) (Figure 3). Although the results of one representative experiment is presented, this is a highly reproducible result based on multiple (>10) isolations of MEVs. The pure MEV yield obtained from a 1 L low iron culture by this method was approximately 500 µg of protein.
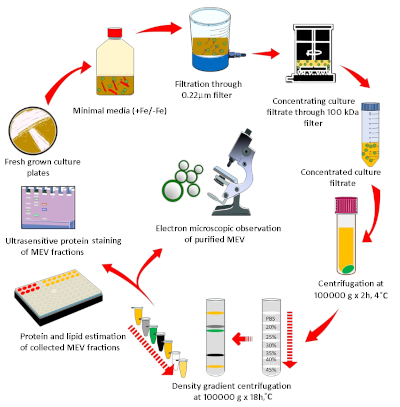
Figure 1: Diagrammatic representation of the methodology used for MEV purification and quantification. Mycobacteria grown in agar plates were used to inoculate iron-depleted minimal medium and grow Mtb for EV isolation. MEVs were purified by a discontinuous density gradient from the cell-free culture filtrate. A combination of membrane lipid and vesicle protein determination, microscopy, and nanoparticle analysis was implemented to characterize purified MEVs. Please click here to view a larger version of this figure.
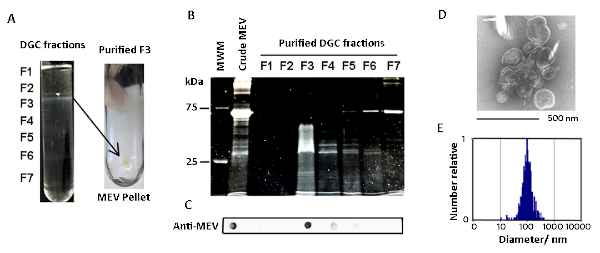
Figure 2: Characterization of purified MEVs. (A) Shown are photographs of an actual density gradient separation of crude MEVs and the pellet of purified MEVs collected by ultracentrifugation of gradient fraction 3 (F3). (B) SDS-gel stained showing the protein profile of the various density gradient fractions. (C) Dot blot analysis showing vesicle-associated proteins concentrated in F3. (D) MEVs present in F3 observed by negative staining. (E) MEV size distribution according to nanoparticle analysis (NTA). Please click here to view a larger version of this figure.
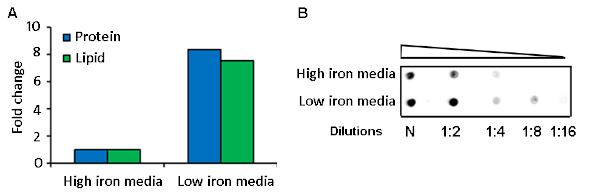
Figure 3: Comparative analysis of MEV yield in low and high iron cultures. A representative result of (A) protein and lipid quantification and (B) dot blot analysis of purified MEVs isolated from iron-limited and iron sufficient Mtb cultures. Please click here to view a larger version of this figure.
Discussion
Multiple methods to purify eukaryotic cell-derived exosomes have been developed12. In contrast, there is limited information on effective methods to purify bacteria-derived EVs7. Efficient isolation of Mtb-derived EVs needs to consider the intrinsic difficulties in growing this pathogenic mycobacterium. Mtb has a long division time (~24 h) and should be handled in biosafety level three (BSL-3) conditions. Therefore, it is important to optimize the efficiency of MEV isolation methods. Because mycobacteria release glycolipids and other hydrophobic molecules that aggregate and easily contaminate crude MEV preparations into the medium, it is important to purify and validate MEVs before conducting biochemical and functional studies. Based on previous observations that demonstrated that Mtb enhances the release of MEVs under conditions of iron limitation, a protocol was established for EV purification from iron-limited mycobacteria. It has also been confirmed that non-virulent M. smegmatis also increases release of EVs in response to low iron conditions (data not shown). Therefore, the same protocol could be used to purify EVs from this bacterium in BSL-2 conditions.
A critical step of this procedure is the preparation of the low iron medium. This medium should be prepared as described here and stored in a plastic container, not in glass, to prevent iron contamination. Supplements commonly used in Mtb growth medium to stimulate bacterial growth and prevent characteristic mycobacterial clumping, such as bovine serum albumin, Tween-80, or tyloxapol, must be avoided. These additives lead to lipoprotein complex artifacts that copurify with vesicles and reduce vesicle yield. For CFU determination, a small culture in medium supplemented with detergent (Tween-80 or tyloxapol) can be set in parallel to the detergent-free large culture. MEVs in the culture filtrate are stable at 4 °C for several days. Therefore, if not processed immediately, the culture filtrate can be stored refrigerated.
The total yield of purified MEV from 1 L of culture was around 500 µg/L protein, which is sufficient to conduct multiple analyses such as proteomics, lipidomics, and functional assays. Depending on the type of assay, sufficient MEVs can be isolated from smaller volumes (i.e., 250 mL). This facilitates comparative analysis of conditions and factors influencing MEV release.
This is an effective method to purify MEVs, but it has limitations. It is a long procedure with multiple ultracentrifugation steps. In the future, this method will be compared to gel filtration chromatography, and as molecular markers of MEVs are discovered, affinity capture methods could be implemented. The host environment is iron-limited, therefore MEVs produced by Mtb in a low iron medium are probably more closely related to MEVs produced during infection and could provide relevant insights about the role of MEVs in tuberculosis pathogenesis.
Divulgations
The authors have nothing to disclose.
Acknowledgements
We are grateful to Rafael Prados-Rosales for sharing the anti-MEV antisera and Navneet Dogra for performing nanoparticle tracking analysis.
Materials
| Amicon stirred cell Model 108 | EMD Milipore | UFSC40001 | Cell Ultrafiltration system |
| BD Polypropilene 225 ml conical tubes | Fisher | 05-538-61 | Conical centrifuge tubes |
| Biomax 100-kDa cut-off ultrafiltration membrane | EMD Milipore | PBHK07610 | Ultrafiltration membrane |
| Chelex-100 resin | Bio-Rad | 142-2842 | Metal chelating resin |
| Middlebrook 7H10 Agar | BD Difco | 262710 | Mycobacterial Agar plates |
| Middlebrook 7H9 Broth | BD Difco | 271310 | Mycobacterial broth medium |
| Nitro cellulose blotting membrane | GE Healthcare | 10600001 | Blotting Membrane |
| Optiprep | Sigma | D1556 | Iodixanol |
| Polycarbonate ultra centrifugation tubes 25 x 89 mm | Beckman Coulter | 355618 | Polycarbonate ultra centrifugation tubes 25 x 89 mm |
| Polypropylene thin walled centrifuge tube 13×15 mm | Beckman Coulter | 344059 | Polypropylene thin walled centrifuge tube 13×15 mm |
| Protein Assay dye | BioRad | 5000006 | Bradford Protein Staining |
| SYPRO Ruby | Molecular Probes | S12000 | Ultrasensitive protein stain |
| TMA-DPH | Molecular Probes | T204 | 1-(4-Trimethylammoniumphenyl)-6-Phenyl-1,3,5-Hexatriene p-Toluenesulfonate |
| Vacuum filtration flasks | CellPro | V50022 | Filter Unit |
References
- Prados-Rosales, R., et al. Mycobacteria release active membrane vesicles that modulate immune responses in a TLR2-dependent manner in mice. Journal of Clinical Investigation. 121, 1471-1483 (2011).
- Gupta, S., Rodriguez, G. M. Mycobacterial extracellular vesicles and host pathogen interactions. Pathogens and Disease. 76 (4), (2018).
- Athman, J. J., et al. Bacterial Membrane Vesicles Mediate the Release of Mycobacterium tuberculosis Lipoglycans and Lipoproteins from Infected Macrophages. Journal of Immunology. 195, 1044-1053 (2015).
- Athman, J. J., et al. Mycobacterium tuberculosis Membrane Vesicles Inhibit T Cell Activation. Journal of Immunology. 198, 2028-2037 (2017).
- Rath, P., et al. Genetic regulation of vesiculogenesis and immunomodulation in Mycobacterium tuberculosis. Proceedings of the National Academy of Science U.S.A. 110, E4790-E4797 (2013).
- White, D. W., Elliott, S. R., Odean, E., Bemis, L. T., Tischler, A. D. Mycobacterium tuberculosis Pst/SenX3-RegX3 Regulates Membrane Vesicle Production Independently of ESX-5 Activity. mBio. 9, pii 00778 (2018).
- Dauros Singorenko, P., et al. Isolation of membrane vesicles from prokaryotes: a technical and biological comparison reveals heterogeneity. Journal of Extracellular Vesicles. 6, 1324731 (2017).
- Prados-Rosales, R., Brown, L., Casadevall, A., Montalvo-Quiros, S., Luque-Garcia, J. L. Isolation and identification of membrane vesicle-associated proteins in Gram-positive bacteria and mycobacteria. MethodsX. 1, 124-129 (2014).
- Prados-Rosales, R., et al. Role for Mycobacterium tuberculosis membrane vesicles in iron acquistion. Journal of Bacteriology. 196, 1250-1256 (2014).
- Sanders, E. Aseptic Laboratory Techniques: Plating Methods. Journal of Visualized Experiments. 63, e3064 (2012).
- Harlow, E., Lane, L. . Antibodies. A laboratory manual. , (1988).
- Lotvall, J., et al. Minimal experimental requirements for definition of extracellular vesicles and their functions: a position statement from the International Society for Extracellular Vesicles. Journal of Extracellular Vesicles. 3, 26913 (2014).

