Immobilization of Caenorhabditis elegans to Analyze Intracellular Transport in Neurons
Summary
Caenorhabditis elegans (C. elegans) is a good model to study axonal and intracellular transport. Here, I describe a protocol for in vivo recording and analysis of axonal and intraflagellar transport in C. elegans.
Abstract
Axonal transport and intraflagellar transport (IFT) are essential for axon and cilia morphogenesis and function. Kinesin superfamily proteins and dynein are molecular motors that regulate anterograde and retrograde transport, respectively. These motors use microtubule networks as rails. Caenorhabditis elegans (C. elegans) is a powerful model organism to study axonal transport and IFT in vivo. Here, I describe a protocol to observe axonal transport and IFT in living C. elegans. Transported cargo can be visualized by tagging cargo proteins using fluorescent proteins such as green fluorescent protein (GFP). C. elegans is transparent and GFP-tagged cargo proteins can be expressed in specific cells under cell-specific promoters. Living worms can be fixed by microbeads on 10% agarose gel without killing or anesthetizing the worms. Under these conditions, cargo movement can be directly observed in the axons and cilia of living C. elegans without dissection. This method can be applied to the observation of any cargo molecule in any cells by modifying the target proteins and/or the cells they are expressed in. Most basic proteins such as molecular motors and adaptor proteins that are involved in axonal transport and IFT are conserved in C. elegans. Compared to other model organisms, mutants can be obtained and maintained more easily in C. elegans. Combining this method with various C. elegans mutants can clarify the molecular mechanisms of axonal transport and IFT.
Introduction
Live cell imaging is an essential tool for analyzing intracellular transport. In neuronal cell biology, analyses of axonal transport with live cell imaging are essential for understanding neuronal function and morphogenesis1. Defects in axonal transport underlie several neurodegenerative disorders2. Kinesin superfamily proteins and dynein carry out axonal transport anterogradely and retrogradely, respectively1,2.
Cilia are another cellular compartment in which the microtubule network and trafficking machinery are highly developed3. Protein synthesis machinery is not localized in cilia, which means that ciliary proteins must be transported from the cytoplasm to the cilia tips. Cilia-specific kinesin and dynein, called kinesin-2 and cytoplasmic dynein-2, respectively, transport the components of cilia4, in a phenomenon called intraflagellar transport (IFT)5. Impairment of IFT causes a spectrum of diseases called ciliopathies6. Thus, analysis of the IFT mechanism by live cell imaging is required to understand basic mechanisms of ciliary formation and pathogenesis.
Caenorhabditis elegans (C. elegans) is a good model to study axonal transport and IFT7,8,9. To observe IFT, Chlamydomonas has been widely used as a model organism5,6. As Chlamydomonas is a unicellular organism, the relationship of IFT with aging, neuronal function, and behavior would be difficult to analyze. In addition, essential genetic techniques such as CRISPR/Cas9 have not been applied to Chlamydomonas. In higher model organisms, such as mice and Drosophila, disruption of axonal transport and IFT often causes lethal phenotypes because axonal transport and IFT are essential for the morphogenesis and homeostasis of the animals 10,11. In the case of mice, cell culture and transfection is generally needed to observe axonal transport and IFT, which requires many skills and extensive time12,13. In addition, a lot of important physiological context may be lost in cultured cells and cell lines. Because the nervous system is not essential for the survival of the worms, C. elegans mutants in which axonal transport or IFT are disrupted are often not lethal7,9,14. Axonal transport and IFT can be directly observed in vivo without dissection because C. elegans is transparent and it is therefore easy to observe GFP-tagged markers.
There are several protocols to immobilize C. elegans, such as using a microfluidic device15, agarose pads with anesthesia16, or microbeads17. The inclusion of anesthesia may inhibit the trafficking events in neurons15. A clear drawback of the microfluidic-device method is that preparing a microfluidic device is not always easy. Instead, immobilization by agarose pads and microbeads is a convenient and easy way to perform time lapse imaging in C. elegans. Here, I describe this basic protocol to immobilize C. elegans and visualize axonal transport and IFT in vivo de C. elegans. Compared to the other methods, the method described here does not require special equipment and is much cheaper and easier to perform.
Protocol
1. Sample Preparation
- Generate a transgenic C. elegans strain of interest using transgenic methodologies18. Alternatively, obtain appropriate strains from the Caenorhabditis Genetics Center (CGC). To visualize axonal transport of synaptic vesicle precursors, use the transgenic line wyIs251 [Pmig-13::gfp::rab-3]7. To observe IFT, obtain the transgenic line mnIs17 [osm-6::gfp]19. These strains are used in this protocol.
NOTE: Either extrachromosomal array, genomic integration, or even GFP knock-in works. To reduce background signals, use cell-specific promoters, rather than pan-neuronal promoters.
NOTE: Maintenance of worms is described in other protocols20. Strains were maintained on lawns of Escherischia coli OP50 feeder on nematode growth medium (NGM) plates (1.7% (w/v) agarose, 50mM NaCl, 0.25% (w/v) peptone, 1 mM CaCl2, 5 mg/mL cholesterol, 25 mM KH2PO4, 1 mM MgSO4 on 60 mm diameter plastic dishes) at 20 °C. - Grow worms at 20 °C on NGM plates to any life cycle stage of the worms.
NOTE: One can observe both axonal transport and IFT at any stage. Late L4 to young adult is a good positive control because multiple publications have used these stages and visualized trafficking events14,21,22,23.
2. Preparation of 10% Agarose
NOTE: Many vendors provide similar products such as agar, agar powders, and agarose. Use electrophoresis grade agarose (gel strength >1200 g/cm2). Cheap agar powders do not work because the resulting gel is not strong enough to immobilize worms.
- Mix 0.4 g agarose in 4 mL distilled water (DW) in a glass tube (1.5 cm x 10.5 cm). Put a lid on the glass tube to avoid evaporation.
- Put the glass tube on a 95 °C heat block for 90 min. In addition, prepare another glass tube (1.5 cm x 10.5 cm) filled with DW and keep it at 95 °C. Put a Pasteur pipet in the 95 °C DW (Figure 1A). A lot of small bubbles will be seen in the agarose tube and the solution should be turbid. Leave it until the solution becomes clear. Then, mix by pipetting up and down until the agarose solution becomes homogeneous.
NOTE: Microwaves cannot be used to prepare 10% agarose solution when made from DW and agarose. Small bubbles caused by microwaves prevent agarose melting. However, solidified 10% agarose gel can easily be melted again using a microwave. Glass tubes containing 10% agarose gel can be prepared ahead of time and stored at 4 °C for at least 6 months. If doing so, melt the stock using a microwave when needed and start from step 3 in this protocol.
NOTE: Agarose gradually loses the ability to solidify if incubated at 95 °C for a long time or after repeated solidification and melting. If this happens, prepare new 10% agarose.
3. Preparation of Agarose Pad
- Using the warmed Pasteur pipet prepared in step 2.2, place 2 drops of 10% agarose on a 76 x 22 mm slide.
- Quickly cover with a second 76 x 22 mm2 slide before the agarose drop solidifies as shown in Figure 1B, and push down on the second slide to flatten the agarose solution. The thickness should be 0.5-1 mm.
- Quickly flush the Pasteur pipette by pipetting 95 °C DW up and down until the agarose solution is washed out. Otherwise, the pipet will become clogged by agarose gel. Leave the pipette standing in the 95 °C DW.
- Wait for 1 min. The agarose pad forms and becomes cloudy. Then, slide the slides apart by hand (Figure 1C).
4. Mounting of Worms
NOTE: Even small amounts of worm movement prevent good observation. Levamisole has traditionally been used to prevent worm movement on the agarose pad24,25. However, Levamisole inhibits neuronal receptors in C. elegans, and therefore may affect trafficking events in neurons15. Using polystyrene microbeads described hereis a good alternative17.
- Under a stereo microscope equipped with a slidable mirror transillumination, transfer ~30 transgenic worms expressing the appropriate markers to a new NGM plate without bacteria using a platinum wire pick.
NOTE: A platinum wire pick is made of platinum wire and Pasteur pipette (5 inches), and the tip of the platinum wire was flattened. The preparation of the platinum wire picks and handling of worms with these are described in the Wormbook (http://www.wormbook.org). - Leave the plate for 1 min. Bacteria are automatically removed from the surfaces of worms as the worms move on NGM agarose gel without bacteria.
- Put a 0.5-1.0 µL drop of 2.6% 100 nm diameter polystyrene microbeads in water onto the agar pad (Figure 1D). Transfer 10-20 worms from the bacteria-free NGM plate to the microbeads. Drop and spread the worms using the platinum wire pick to avoid the overlapping of worm bodies (Figure 1E).
- Apply a 22 x 40 mm2 coverslip (Figure 1F). Push gently to remove air bubbles, but avoid crushing worms. The sample is now ready for imaging.
NOTE: Because no anesthesia is administered, worms are fixed only by the frictional force generated by the agar pad, polystyrene microbeads, and coverslip17. The presence of feeder bacteria and/or too much solution will make the frictional force weaker.
NOTE: Different sizes of coverslips (e.g., 22 x 22 mm2, 18 x 18 mm2) work. However, larger coverslips can prevent immersion water or oil from the objective lens leaking into samples during the observation.
5. Observation
NOTE: Appropriate imaging parameters (laser power, gain, binning, etc.) will differ for each microscope system and camera. Here, a widefield microscope equipped with a spinning disk confocal scanner and a digital CCD camera is used.
- Set the temperature control of the stage to 20 °C, or adjust the room temperature to 20 °C.
- Put the sample slide on the microscope stage. Use 100x (numerical aperture = 1.3 or better) objective lens.
- Look for the areas indicated in Figure 2A (for wyIs251 and axonal transport) or Figure 2B (for mnIs17 and IFT) as positive controls. Other areas can be analyzed as well22.
- Set parameters (CCD Gain = 200, Binning = 1 or 2, laser power at 488 nm = 1.0-1.3 mW). Perform time lapse recording at 4 frames/s for up to 1 min. Save files as mutli-TIFF format.
NOTE: If GFP signals disappear and it is difficult to continue to observe GFP::RAB-3 or OSM-6::GFP for 30 s, the laser power is too strong. In that case, reduce the laser power. Conversely, if no vesicular movement is recorded, the laser power is too weak. In that case, increase the laser power. - Observe a different worm and repeat 5.3 and 5.4. The observations can last for up to 20 min, but each worm should be recorded only once to avoid the effects of photo damage.
NOTE: Worms have been observed for at least 20 min without significant defects7,27,28. Longer observation may be possible but haven't been tested yet. A clear sign of neuronal death is the change of neuronal morphology such as neurite swelling. As weak GFP signals can be seen in axons and dendrites in both wyIs251 and mnIs17, neurite morphology can be easily checked by just looking at axons or dendrites. Neuronal morphology is shown in Figure 2. - Generate movie files.
- Open the multi-TIFF file using Fiji software. Go to File>Open then select the multi-TIFF file saved in step 5.4.
NOTE: Fiji can be downloaded at jttps://fiji.sc. Image J and plugins can also be used to generate kymographs. If doing so, follow the relevant instructions for the chosen plugin. - Click the "Rectangular selection" button (Figure 3A, arrow) and select the area of interest by drawing a rectangle with a computer mouse (yellow rectangle in Figure 3A). Go to Image>Stacks>Tools>Make Substacks and select the slice numbers that are included in the movie. A substack is generated.
- Activate the substack window by clicking and go to Image>Adjust>Brightness/Contrast. A window to adjust the brightness and contrast shows up. In the window, slide the top bar (Minimum) to the right so that the background signal disappears and the second top bar (Maximum) to the left so that the moving vesicles and particles can be seen quite well over the background.
- Go to Image>Stacks>Time Stamper. As the movie is recorded at 4 frames/s in step 5.4, the time interval is 0.25 s. Thus, type "0.25" in Interval.
- Generate a movie file by selecting Save as>AVI from the File menu (Movie 1 and 2).
NOTE: By using appropriate plugins, you can save the file as other formats such as "mpeg4" or "mov" files.
- Open the multi-TIFF file using Fiji software. Go to File>Open then select the multi-TIFF file saved in step 5.4.
- Generate kymographs.
- Open the original movie files using FIji software again and repeat steps 5.6.2 and 5.6.3 to adjust the brightness and contrast.
- Click "segmented line" (Figure 3B, arrow). Using a mouse, draw a segmented line along the cilia or axon (yellow line, in Figure 3B).
- Go to Analyze>Multi Kymograph>Multi Kymograph (Figure 3C). Line width window appears (Figure 3D). Type "3" in the Linewidth window and Click OK (Figure 3D).
- Kymograph window is created. Save the image in TIFF or JPEG format.
Representative Results
Axonal transport in DA9 neuron
Using the wyIs251 line, both the anterograde and retrograde axonal transport of GFP::RAB-3 can be simultaneously recorded in the DA9 motor neuron. The average speed of anterograde and retrograde transport in the proximal dorsal axon of the DA9 neuron is about 1.8 and 2.6 μm/s, respectively22. The number of moving vesicles is about 0.03 and 0.018 per μm of axon per s. Thus, for a 30 s observation of 10 μm of DA9 axon using a 100x lens, one can find about 9 and 5 moving vesicles in the axon (Figure 4 and Movie 1). If no vesicular movement can be observed, it is possible that the laser power is too weak. In addition to long range runs, short range movement of vesicle pools can be recorded (Movie 1). Some vesicles stop at these vesicle pools, while others dissociate from there7,23. These parameters can be monitored and used to analyze mutant phenotypes.
IFT in tail neurons
Cilia are localized to the tip of dendrites in C. elegans (Figure 2B). The mnIs17 strain expresses GFP-tagged OSM-6, one of the IFT subunits29. OSM-6 is expressed in both head and tail neurons19. While many head cilia are labeled by mnIs17, only two cilia are clearly observed in the tail neuron. Thus, observing these cilia is easier than head cilia (see also Discussion). OSM-6::GFP is diffuse in the neuronal cytoplasm such as that found in the axon, cell body, and dendrite. However, in cilia, OSM-6::GFP is concentrated to cilia and incorporated in moving particles. The length of PHA and PHB cilia are about 6.5 μm. Anterograde IFT is biphasic 8,9. The speed of anterograde IFT is about 0.7 and 1.3 μm/s in the middle and distal segments of the cilia, respectively (Figure 5 and Movie 2).
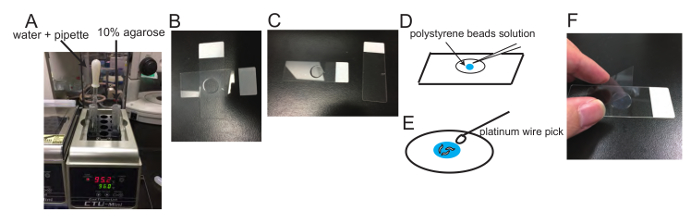
Figure 1: Demonstration of the sample preparation. (A) Hot DW, Pasteur pipet and 10% agarose on heat block. (B and C) Preparation of agarose pads: An agarose pad is formed between slides (B). The upper slide is removed after an agarose pad forms (C). (D) Schematic drawing showing how polystyrene bead solution is put on the agarose pad. (E) Schematic drawing showing how worms are put in the polystyrene bead solution using a platinum wire pick. (F) A coverslip (22 x 44 mm2) is put on the agarose pad. Please click here to view a larger version of this figure.

Figure 2: Imaging area. (A) Schematic drawing of DA9 and VA12 neurons and a representative image of wyIs251. In DA9 neurons, the ventral axon, commissure, and proximal axon does not have mature synapses. But synaptic vesicle precursors are transported from the cell body to synapses via these axonal regions. The region that should be observed is shown by boxes. (B) Schematic drawing of PHA and PHB neurons and their cilia. The region that should be observed is shown by boxes. Scale bar = 10 μm. Please click here to view a larger version of this figure.
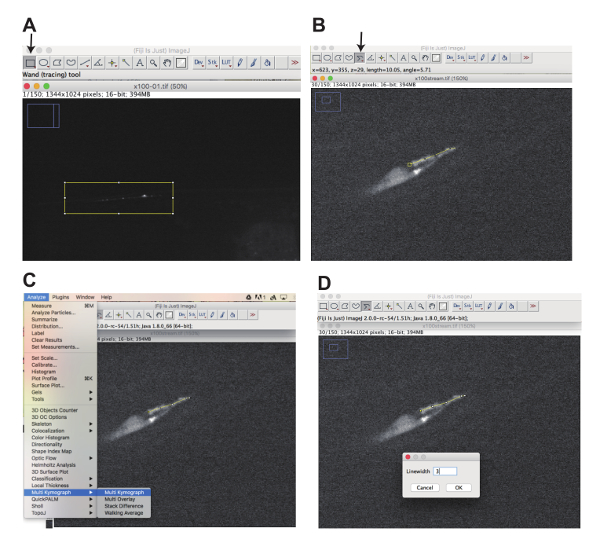
Figure 3: Generation of movies and kymographs using Fiji software. (A) Click the "Rectangular selection" button (arrow) and select the area one would like to focus on by drawing a rectangle (Yellow box) to generate substacks. (B) Click the segmented line button (arrow) and draw a segmented line along the cilia using a mouse (Yellow line). (C) Select "Analyze", "Multi Kymograph", and "Multi Kymograph" to start a plugin to generate kymographs. (S) Input linewidth, click "OK" and generate kymographs (Figure 5 and Figure 7). Please click here to view a larger version of this figure.
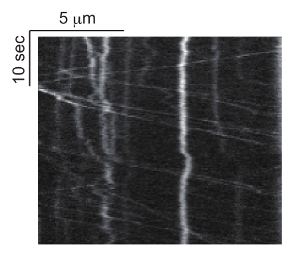
Figure 4: Representative kymograph of axonal transport of synaptic vesicle precursors. GFP::RAB-3 movement is observed at the proximal asynaptic region of DA9 neuron and the kymograph was generated with Multi Kymograph of Fiji. Horizontal and vertical bars represent length and time, respectively. Please click here to view a larger version of this figure.
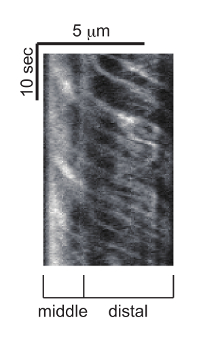
Figure 5: Representative kymograph of IFT. OSM-6::GFP is observed in PHA and PHB cilia and this kymograph is generated from the trafficking event in either one of them. The kymograph was generated with Multi Kymograph of Fiji. Horizontal and vertical bars represent length and time, respectively. Please click here to view a larger version of this figure.

Movie 1: Representative movie of axonal transport of synaptic vesicle precursors. GFP::RAB-3 movement is observed at the proximal asynaptic region of the DA9 neuron. GFP::RAB-3 is incorporated into synaptic vesicle precursors and transported. Both anterograde and retrograde axonal transport are recorded. Some vesicles stop but restart again. The movie was recorded at 4 frames/s and plays at 15 frames/s. Scale bar = 5 μm. Please click here to view this video. (Right-click to download.)

Movie 2: Representative movie of IFT. OSM-6::GFP is observed in PHA and PHB cilia. OSM-6::GFP is incorporated into the IFT complex and moves along the cilia. The movie was recorded at 4 frames/s and plays at 15 frames/s. Scale bar = 5 μm. Please click here to view this video. (Right-click to download.)
Discussion
Limitation with respect to existing methods
The method described here is optimized to observe fast events such as axonal transport and IFT. Thus, immobilization is more prioritized than longer incubation. While we have been able to observe trafficking events for at least 20 min without significant perturbation, this method may not be always suitable to observe slow events requiring longer observations, such as axon elongation and cell migration. For longer observations, one needs to optimize the conditions by reducing the percentage of the agarose (i.e., increasing the water to avoid drying up and reducing the pressure that potentially causes damage). Alternatively, the microfluidic device described above15 or immobilization of worms using agarose pads and anesthesia may be more suitable16 while side effects should be carefully assessed.
Critical steps within the protocol
For adequate observation, markers are critical. Markers need to be incorporated into vesicles or particles and be of sufficient brightness. Either extrachromosomal arrays or integrated lines can be used. For visualization of the axonal transport of synaptic vesicle precursors in DA9 neurons, the wyIs251 strain is useful7,23. jsIs821 has been used to observe axonal transport in mechanosensory neurons in some studies26,27. When other neurons are analyzed, synaptic vesicle proteins should be expressed in interested neurons using cell specific promoters. These promoters should be strong enough. RAB-3 and SNB-1 are widely used to label synaptic vesicle precursors22,23,26. To observe IFT, the components of the IFT complex should be labeled. As a positive control, the mnIs17 strain is a good choice29. Cilia emanating from 8 cells (ASE, ASG, ASI, ASL, ASH, ASK, ADF, and ADL) are bundled together in the head (amphid) while cilia from 2 cells (PHA and PHB) are bundled in the tail (phasmid). Because OSM-6::GFP is expressed in all ciliated neurons in mnIs17, it may be difficult to analyze the IFT in detail in head cilia. However, in the tail region, only PHA and PHB neurons are labeled and IFT in each cell can easily be resolved. Other IFT proteins labeled by fluorescent proteins can be used to observe IFT21,28. If one wishes to analyze head neurons, cell-specific promoters would be useful. While head cilia have varying morphologies, transport events can be recorded29. By crossing mutants with these markers, the functions of any genes in axonal transport and IFT can be analyzed in vivo. The parameters that can be analyzed have been described in previous work22,23,26,27. GFP is the best and primary choice. mCherry and tdTomato can also be used22, however, mCherry is much dimmer than GFP. tdTomato is a tandem dimer and larger than monomeric fluorescent proteins, which sometimes affects the dynamics of fusion proteins30. Therefore, the results should be analyzed and interpreted cautiously when tdTomato is used. mScarlet, a bright monomeric red fluorescent protein which has recently been developed, is an alternative choice31.
While spinning disk confocal microscopy is widely used to observe axonal transport and IFT7,21, the equipment is expensive and may be difficult to access in some research environments. However, these phenomena can also be recorded and analyzed in C. elegans using standard fluorescent microscopes, if equipped with high NA lenses, because C. elegans is a small and transparent animal and easy to observe.
Future application
By the similar immobilization method described here, IFT was observed at the single molecule resolution in vivo32. Moreover, several types of super-resolution microscopy have been developed. One can directly apply the methods described here to super-resolution microscopic analysis. In my experience, the fast mode of Airyscan33 works very well to analyze IFT. Theoretically, a spinning disk super-resolution microscope (SDSRM) would be a good choice as well34. In conclusion, by combining mutant libraries and these new techniques, the method described here should be useful for clarifying the molecular mechanisms of axonal transport and IFT in vivo.
Divulgations
The authors have nothing to disclose.
Acknowledgements
The author deeply thanks Dr. Asako Sugimoto (Tohoku University) for her helpful discussion. wyIs251 was a generous gift from Dr. Kang Shen (Stanford University). mnIs17 was provided by the CGC, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440). This work was supported by JSPS KAKENHI grant #17H05010 and #16H06536 and Daiichi Sankyo foundation, Brain Science Foundation and Naito Foundation.
Materials
| Slideglass (76 x 26 mm) | Matsunami | S1111 | |
| Coverglass (22 x 40 mm) | Matsunami | C024401 | |
| Agarose | Wako | 318-01195 | |
| Polystylene microbeads 0.1 micron | Polysciences | #00876 | |
| Heat block | TAITEC | 0063288-000 | CTU-mini |
| Microscope | Olympus | IX-71 | widefield microscope |
| Spinning disk Scanner | Yokogawa | CSU-X1 | spinning disc confocal scanner |
| Digital CCD camera | Hamamatsu Photonics | C10600-10B | ORCA-R2 degital CCD camera |
| Objective lens (x100, NA1.4) | Olympus | UPLSAPO 100XO | |
| Pasteur pipette (5 inch) | IWAKI | IK-PAS-5P | |
| Glass tube (1.5 cm diameter x 10.5 cm) | IWAKI | 9820TST15-105NP | |
| TV9211: wyIs251 | Laboratory of Kang Shen | N/A | |
| OTL11: mnIs17 | Ref. 27, 29 | N/A | SP2101 was backcrossed with wild type for 6 times |
| Stereo microscope | Carl Zeiss | 435064-9000-000 | STEMI 508 |
| Mirror transillumination unit | Carl Zeiss | 435425-9010-000 | |
| platinum wire (0.2 mm) | Nilaco Corporation | m78483501 | |
| 60 mm plastic dish | Falcon | #351007 | |
| Fiji | N/A | N/A | https://fiji.sc/ |
| nematode growth medium (NGM) | 1.7% (w/v) agarose, 50mM NaCl, 0.25% (w/v) Peptone, 1 mM CaCl2, 5 mg/mL Cholesterol, 25 mM KH2PO4, 1 mM MgSO4 |
References
- Hirokawa, N., Niwa, S., Tanaka, Y. Molecular motors in neurons: transport mechanisms and roles in brain function, development, and disease. Neuron. 68 (4), 610-638 (2010).
- Holzbaur, E. L., Scherer, S. S. Microtubules, axonal transport, and neuropathy. N Engl J Med. 365 (24), 2330-2332 (2011).
- Kamiya, R. Functional diversity of axonemal dyneins as studied in Chlamydomonas mutants. Int Rev Cytol. 219, 115-155 (2002).
- Scholey, J. M. Intraflagellar transport motors in cilia: moving along the cell’s antenna. J Cell Biol. 180 (1), 23-29 (2008).
- Rosenbaum, J. L., Witman, G. B. Intraflagellar transport. Nat Rev Mol Cell Biol. 3 (11), 813-825 (2002).
- Ishikawa, H., Marshall, W. F. Ciliogenesis: building the cell’s antenna. Nat Rev Mol Cell Biol. 12 (4), 222-234 (2011).
- Niwa, S., et al. Autoinhibition of a Neuronal Kinesin UNC-104/KIF1A Regulates the Size and Density of Synapses. Cell Rep. 16 (8), 2129-2141 (2016).
- Snow, J. J., et al. Two anterograde intraflagellar transport motors cooperate to build sensory cilia on C. elegans neurons. Nat Cell Biol. 6 (11), 1109-1113 (2004).
- Ou, G., Blacque, O. E., Snow, J. J., Leroux, M. R., Scholey, J. M. Functional coordination of intraflagellar transport motors. Nature. 436 (7050), 583-587 (2005).
- Zhou, R., Niwa, S., Homma, N., Takei, Y., Hirokawa, N. KIF26A is an unconventional kinesin and regulates GDNF-Ret signaling in enteric neuronal development. Cell. 139 (4), 802-813 (2009).
- Zhao, C., et al. Charcot-Marie-Tooth disease type 2A caused by mutation in a microtubule motor KIF1Bbeta. Cell. 105 (5), 587-597 (2001).
- Niwa, S., Tanaka, Y., Hirokawa, N. KIF1Bbeta- and KIF1A-mediated axonal transport of presynaptic regulator Rab3 occurs in a GTP-dependent manner through DENN/MADD. Nat Cell Biol. 10 (11), 1269-1279 (2008).
- Zhou, R., Niwa, S., Guillaud, L., Tong, Y., Hirokawa, N. A molecular motor, KIF13A, controls anxiety by transporting the serotonin type 1A receptor. Cell Rep. 3 (2), 509-519 (2013).
- Klassen, M. P., et al. An Arf-like small G protein, ARL-8, promotes the axonal transport of presynaptic cargoes by suppressing vesicle aggregation. Neuron. 66 (5), 710-723 (2010).
- Mondal, S., Ahlawat, S., Koushika, S. P. Simple microfluidic devices for in vivo imaging of C. elegans, Drosophila and zebrafish. J Vis Exp. (67), (2012).
- Chai, Y., et al. Live imaging of cellular dynamics during Caenorhabditis elegans postembryonic development. Nat Protoc. 7 (12), 2090-2102 (2012).
- Kim, E., Sun, L., Gabel, C. V., Fang-Yen, C. Long-term imaging of Caenorhabditis elegans using nanoparticle-mediated immobilization. PLoS One. 8 (1), 53419 (2013).
- Berkowitz, L. A., Knight, A. L., Caldwell, G. A., Caldwell, K. A. Generation of stable transgenic C. elegans using microinjection. J Vis Exp. (18), (2008).
- Collet, J., Spike, C. A., Lundquist, E. A., Shaw, J. E., Herman, R. K. Analysis of osm-6, a gene that affects sensory cilium structure and sensory neuron function in Caenorhabditis elegans. Génétique. 148 (1), 187-200 (1998).
- Brenner, S. The genetics of Caenorhabditis elegans. Génétique. 77 (1), 71-94 (1974).
- Niwa, S. The nephronophthisis-related gene ift-139 is required for ciliogenesis in Caenorhabditis elegans. Sci Rep. 6, 31544 (2016).
- Maeder, C. I., San-Miguel, A., Wu, E. Y., Lu, H., Shen, K. In vivo neuron-wide analysis of synaptic vesicle precursor trafficking. Traffic. 15 (3), 273-291 (2014).
- Wu, Y. E., Huo, L., Maeder, C. I., Feng, W., Shen, K. The balance between capture and dissociation of presynaptic proteins controls the spatial distribution of synapses. Neuron. 78 (6), 994-1011 (2013).
- Dwyer, N. D., Adler, C. E., Crump, J. G., L’Etoile, N. D., Bargmann, C. I. Polarized dendritic transport and the AP-1 mu1 clathrin adaptor UNC-101 localize odorant receptors to olfactory cilia. Neuron. 31 (2), 277-287 (2001).
- Fang-Yen, C., Gabel, C. V., Samuel, A. D., Bargmann, C. I., Avery, L. Laser microsurgery in Caenorhabditis elegans. Methods Cell Biol. 107, 177-206 (2012).
- Kumar, J., et al. The Caenorhabditis elegans Kinesin-3 motor UNC-104/KIF1A is degraded upon loss of specific binding to cargo. PLoS Genet. 6 (11), 1001200 (2010).
- Zheng, Q., et al. The vesicle protein SAM-4 regulates the processivity of synaptic vesicle transport. PLoS Genet. 10 (10), 1004644 (2014).
- Blacque, O. E., et al. The WD repeat-containing protein IFTA-1 is required for retrograde intraflagellar transport. Mol Biol Cell. 17 (12), 5053-5062 (2006).
- Evans, J. E., et al. Functional modulation of IFT kinesins extends the sensory repertoire of ciliated neurons in Caenorhabditis elegans. J Cell Biol. 172 (5), 663-669 (2006).
- Shaner, N. C., Steinbach, P. A., Tsien, R. Y. A guide to choosing fluorescent proteins. Nat Methods. 2 (12), 905-909 (2005).
- Bindels, D. S., et al. mScarlet: a bright monomeric red fluorescent protein for cellular imaging. Nat Methods. 14 (1), 53-56 (2017).
- Prevo, B., Mangeol, P., Oswald, F., Scholey, J. M., Peterman, E. J. Functional differentiation of cooperating kinesin-2 motors orchestrates cargo import and transport in C. elegans cilia. Nat Cell Biol. 17 (12), 1536-1545 (2015).
- Robison, P., et al. Detyrosinated microtubules buckle and bear load in contracting cardiomyocytes. Science. 352 (6284), 0659 (2016).
- Hayashi, S., Okada, Y. Ultrafast superresolution fluorescence imaging with spinning disk confocal microscope optics. Mol Biol Cell. 26 (9), 1743-1751 (2015).

