A Screenable In Vivo Assay for Mitochondrial Modulators Using Transgenic Bioluminescent Caenorhabditis elegans
Summary
A protocol is described for in vivo detection of effects of mitochondrial inhibitors in the model organism Caenorhabditis elegans and for identification of potential enhancing compounds. This protocol can be used to screen drug libraries for compounds modulating mitochondrial function.
Abstract
The multicellular model organism Caenorhabditis elegans is a small nematode of approximately 1 mm in size in adulthood that is genetically and experimentally tractable. It is economical and easy to culture and dispense in liquid medium which makes it well suited for medium-throughput screening. We have previously validated the use of transgenic luciferase expressing C. elegans strains to provide rapid in vivo assessment of the nematode’s ATP levels.1-3 Here we present the required materials and procedure to carry out bioassays with the bioluminescent C. elegans strains PE254 or PE255 (or any of their derivative strains). The protocol allows for in vivo detection of sublethal effects of drugs that may identify mitochondrial toxicity, as well as for in vivo detection of potential beneficial drug effects. Representative results are provided for the chemicals paraquat, rotenone, oxaloacetate and for four firefly luciferase inhibitory compounds. The methodology can be scaled up to provide a platform for screening drug libraries for compounds capable of modulating mitochondrial function. Pre-clinical evaluation of drug toxicity is often carried out on immortalized cancerous human cell lines which derive ATP mostly from glycolysis and are often tolerant of mitochondrial toxicants.4,5 In contrast, C. elegans depends on oxidative phosphorylation to sustain development into adulthood, drawing a parallel with humans and providing a unique opportunity for compound evaluation in the physiological context of a whole live multicellular organism.
Introduction
The overall purpose of this procedure is the rapid quantification of the energy status of C. elegans in vivo with a view to using this as an endpoint in compound screening. The rationale is based on the transgenic expression of firefly luciferase in the nematode C. elegans1-3 via the plasmid pSLGCV (1https://www.addgene.org/49862). The luciferase enzyme is fused to the green fluorescent protein GFP and is expressed constitutively and ubiquitously in the cytoplasm (the luciferase peroxisome targeting signal was removed). This leads to light emission when the substrate luciferin is provided exogenously. As the worm is transparent, light can be measured in a luminometer as relative light units (RLU). Cellular ATP fuels the bioluminescence reaction and its availability determines the light levels produced. Consequently, luminometry offers a convenient means of assessing relative ATP levels and by extension mitochondrial function, since ATP production occurs mainly in mitochondria. A link between mitochondrial function and bioluminescence levels has been demonstrated previously through the silencing of mitochondrial electron transport chain genes and concomitant reduced light output.1
The bioluminescence strains generated were designated PE254(feIs4) and PE255(feIs5)1 (see materials list) and can be used interchangeably.3 A number of studies have utilized these sensor strains to report on cellular ATP levels in vivo following exposure to various xenobiotic chemicals such as sodium azide1, cadmium2, sewage sludge extract3, 5’-fluoro-2-deoxyuridine6, and a tobacco-specific nitrosamine7. The strains have also been useful to monitor effects of exposure to ultraviolet C radiation8 and effects of disrupting the mitochondrial respiratory chain function.1,9 An early version of the luminescence sensor expressing the luciferase gene without a GFP fusion (PE39) has also been used in the investigation of effects of heavy metals and of a respiratory uncoupler.10 Strains PE254 and PE255 carry the luciferase:GFP fusion and GFP fluorescence was shown to increase proportionally with nematode mass, offering a convenient means for normalization of luminescence values.6,9 The effects of differential amount of worms per well can also be taken into account by including multiple technical replicates in the assay (a minimum of 5 wells per condition).3
The protocol offers the possibility of in vivo monitoring of energy levels (as opposed to more technically laborious in vitro ATP determinations) enabling compound screening and repositioning in the physiological context of a whole multicellular organism. The procedure can be extended to a variety of genetic backgrounds by crossing the chromosomally integrated transgene into available mutant strains and/or by silencing genes by RNA interference; thus, taking full advantage of C. elegans as a model organism. The method should help reduce late phase failure of drug candidates due to mitochondrial toxicity and make a contribution towards reduction of higher animal testing.
Protocol
NOTE: Carry out all steps under sterile conditions (laminar flow cabinet) and with pre-sterilized materials (by autoclaving 126 °C, 11 min). An LB plate with streaked out E. coli OP50 kept at 4 °C is required, streak out onto fresh LB plates and restreak every month.
1. Bacterial Food (Escherichia coli OP50) Preparation
- Day 1. Inoculate 2 x 5 ml LB in two universal bottles with a single colony of E. coli OP50 and place in shaking incubator at 37 °C (220 rpm) for 8 hr.
- After 8 hr incubation, use 2 ml of E. coli OP50 LB culture to inoculate each of 3 x 200 ml LB. Place flasks in shaking incubator (220 rpm) O/N (17 hr) at 37 °C.
- Weigh 20 x 50 ml centrifuge tubes and write weight on the tube.
- Day 2. Using a serological pipette, aliquot 30 ml of O/N E. coli OP50 culture into the pre weighed centrifuge tubes. Centrifuge at 7,741 x g, 10 °C, 8 min.
- Carefully decant the supernatant, keep the tube inverted with lid on and leave to stand for several minutes. Using a pipette remove any excess supernatant that may have collected in the lid. Weigh the tube and calculate the weight of the pellet.
- Calculate the volume needed to provide a suspension of 30 g/L and mark this volume on the tube. Date, label and place tubes at -20 °C for use within 1-3 months or at – 80 °C if storing for longer than 3 months.
- Prepare bacterial suspension for growing nematodes; allow bacterial pellet to thaw out and add required volume of S complete medium11,12 to each tube, vortex gently to resuspend pellet, pool contents of different tubes to obtain the volume required. Work under sterile conditions.
2. Preparation of Drug Standards in a 96-well Plate Format
NOTE: If using a drug library, drug plates are provided at a single concentration of compound in DMSO. Primary screening will test compounds at a single concentration. Instructions follow for preparation of drug plate for confirmatory compound testing at a range of concentrations between 0-160 μM, selected after statistical significance at 10 μM. The steps below can be adapted to test other concentrations.
CAUTION! Follow necessary precautions for handling drugs (generally face mask, safety goggles, gloves are required).
- Prepare drug plate with working standards for confirmatory screening: weigh the required amount of compound into sterile 1.7 ml microcentrifuge tube and prepare 16 mM compound in DMSO (i.e., 100x concentrated relative to desired top concentration for exposure).
- Serially dilute 16 mM stock 1:2 in DMSO to obtain 8, 4, 2, 1, 0.5 and 0.25 mM standards (sterile conditions). These are 100x concentrated and will be diluted down to final concentrations of 0 (vehicle only), 2.5, 5, 10, 20, 40, 80 and 160 μM (in 1% DMSO) during drug exposure.
- In a laminar flow cabinet, place 20-50 μl per well of compound standards within a column of a 96-well plate, for example plate position A1: 16 mM, B1: 8 mM, C1: 4 mM, D1: 2 mM, E1: 1 mM, F1: 0.5 mM, G1: 0.25 mM drug and H1: DMSO. Use different columns for dilution series of different drugs.
- Aliquot vehicle to the wells of column 12 in the drug plate.
NOTE: This will facilitate testing of vehicle control down columns in addition to the testing along row H, ensuring that vehicle is tested in positions representative of the whole plate.
NOTE: Each drug plate for confirmatory screening holds dilution series for a maximum of 11 different drugs to be tested plus the vehicle control. - Label and seal plate, cover with foil and place at -20 °C until required.
3. Nematode Experiments
NOTE: Carry out all steps under sterile conditions, aseptically and with pre sterilized materials and reagents. Maintain C. elegans strains routinely on NGM plates with E. coli OP50.12 Suggested timings for the different protocol steps are provided in Table 1.
- Day 1 task: set up liquid culture of biosensor strain for subsequent synchronization.
- Take up nematodes from one 6 cm NGM plate (with plenty of young larvae where food has just run out) in 2 ml S complete and transfer to flask with 30 g/L E. coli OP50 in S complete (total volume 30 ml in a 250 ml capacity conical glass flask). Incubate 3 days at 20 °C, 160 rpm.
- Day 4 task (allow approximately 30-35 min): bleach culture to harvest eggs and synchronize worm population. Carry out all centrifugation steps for 1 min, 600 x g.
- Prepare bleach solution just before use: to each 16 ml of 0.156 M KOH/NaOH add 4 ml bleach.
- Pour 2 x 14 ml worm culture into 15 ml conical centrifugation tubes. Allow worms to settle by gravity (3 min). Remove and discard liquid above settled nematodes. Add 5 ml of bleach solution to each tube and start timer. Combine volumes into one tube.
- Invert tube gently for 2 min, check under stereoscope for the breaking of worms and the release of eggs into suspension. When most eggs are released or at a maximum time of 2 min in the bleach solution, centrifuge.
- Remove supernatant carefully. Wash pellet 1x with 14 ml S complete and centrifuge.
- Discard supernatant, add 10 ml bleach solution and start timer (this second bleach step ensures that most of the worm carcasses disintegrate and are no longer seen under stereoscope). After a maximum time in bleach solution of 2 min, centrifuge.
- Wash pellet 3x in S complete, gently decant supernatant and resuspend pellet in 14 ml S complete after each centrifugation. After final wash, resuspend egg pellet in 14 ml S complete.
- Transfer the volume to a conical glass flask. Incubate 18-24 hr at 20 °C, 160 rpm.
NOTE: Harvested eggs will hatch O/N and arrest development at the first larval instar (L1) due to lack of food, hence they will all be at the same stage of growth.
- Day 5 task: set up synchronized worm cultures for drug assays.
NOTE: Worms suspended in starvation medium tend to adhere to hydrophobic plastic surfaces such as pipette tips and tubes. 0.01% tween-20 is a surfactant used here to render plastic surfaces more hydrophilic and allow greater accuracy in counting (0.01% triton-X100 can also be used). When nematodes are in the bacterial supplemented medium, they don’t tend to stick to plastic.- Determine numbers of hatched L1’s in flask, keep flask on shaking platform (160 rpm).
- Take 3x 100 μl samples from flask (use a clean tip each time) into separate 1.7 ml microtubes with 900 μl liquid medium 0.01% Tween-20.
NOTE: Aspire 100 μl sample into the clean tip only once. Then when dispensing, pipette up and down a few times into the tween supplemented medium to release any worms adhering to the tip. - Count the nematodes in 4x 10 μl droplets from each triplicate dilution tube. Calculate average value and estimate the number of worms present in the conical glass flask.
- Take 3x 100 μl samples from flask (use a clean tip each time) into separate 1.7 ml microtubes with 900 μl liquid medium 0.01% Tween-20.
- Calculate the volume of hatched nematode suspension required to set up a 2 x 20 ml culture with 10 nematodes per 10 μl. Increase calculated volume by 10% to account for some subsequent nematode loss during centrifugation steps.
- Decant volume required into 2 x 15 ml centrifugation tubes (pre-weighed). Weigh tubes to obtain actual volume dispensed, vortex gently and adjust to target volume by removing excess volume with a 5 ml pipette (only the sterile tip should enter the tube).
- Make up volume to 14 ml with fresh S complete to wash nematodes. Centrifuge (1 min, 600 g). Remove and discard supernatant with care not to disturb the nematode pellet.
- Add 5 ml of S complete with 30 g/L E. coli OP50 to pelleted nematodes. Transfer the 5 ml in each tube to a conical glass flask containing 15 ml S complete with 30 g/L E. coli OP50. Note down time nematodes were first provided with food.
- Shake flask gently (160 rpm), take 9 x 10 μl droplets onto microscopic slides (use fresh tips each time) to count nematodes and confirm average of approximately 10 (±2) per 10 μl.
- Place nematode flasks in shaking incubator at 20 °C, 160 rpm for 42-44 hr.
- Determine numbers of hatched L1’s in flask, keep flask on shaking platform (160 rpm).
- Day 7 task: transfer nematodes to 96 well plates and initiate drug exposure.
- Combine nematode cultures into a single flask, keep swirling flask gently and place 3 x 4 ml of nematode culture in a 60 ml sterile trough. Place trough on a shaking platform (160 rpm).
- Use 8 channel pipette to aliquot 25 μl suspended nematodes per well of 96 well black microtiter plates with a flat transparent bottom (for taking both luminescence and fluorescence readings; or white plates for luminescence only). Cover with plate lid and set aside.
- After set up of every 2 plates, place another 2 x 2.5 ml nematodes in trough to replace lost volume (keep a ‘dead volume’ in trough of approximately 7 ml).
- Set up 13/14 plates with nematodes. 11 nematode plates will be required to test two 96-well drug plates. The 12th nematode plate will be loaded exclusively with vehicle as a control. The 13th plate will be used to establish background fluorescence on day 8 (see below). An extra plate can be prepared for use should mistakes occur during set up.
- Aliquot 74 μl of S complete to each well and place plate in a damp chamber (see materials list) in a shaking incubator (20 °C, 160 rpm) until ready to aliquot test compounds at the preselected developmental time (e.g., 45 hr 30 min after food provided). [Volume per well is now 99 μl].
- Thaw out drug plate(s) to test.
- Use multichannel pipette to set up nematode plates with drug, changing tips each time. Allow 5 min per 96 well plate for aliquoting drug.
- Take 1 μl from 1st column of drug plate and add to columns 1-5 of a plate containing nematodes; repeat from 2nd column in the drug plate into columns 8-12 of the plate containing nematodes. Add 1 μl of vehicle to columns 6 and 7 of the nematode plate.
NOTE: The total volume per well in the nematode plate will be 100 μl resulting in a 1:100 dilution of compound/vehicle. - Repeat process by adding 1 μl of the 3rd/ 4th column of the drug plate to columns 1-5 / 8-12 of the second nematode plate; add vehicle to columns 6 and 7 of the nematode plate.
- Continue to test remaining drug plate columns. Set up a minimum of one nematode plate with vehicle in all wells by sampling from column 12 of drug plate. Place plates back in damp chambers in shaking incubator (20 °C, 160 rpm) for 20-22 hr (see note for day 8).
- Take 1 μl from 1st column of drug plate and add to columns 1-5 of a plate containing nematodes; repeat from 2nd column in the drug plate into columns 8-12 of the plate containing nematodes. Add 1 μl of vehicle to columns 6 and 7 of the nematode plate.
- Part-prepare the luminescence buffer for next day: add DMSO and 10% Triton-X-100 to the required volume of S complete to obtain 1% DMSO and 0.15% Triton X-100. Allow 5 ml per plate read for luminescence, plus 1 ml for priming luminometer injector.
- Day 8 task: Read experimental endpoints — fluorescence and bioluminescence. (GFP fluorescence readings are recommended as a means to normalize bioluminescence data.)
NOTE: Aim to read endpoints by 66-67 hr after food provided to nematodes in order to prevent significant progeny production under the experimental conditions described.- Set up plate to use as background control for GFP readings. Combine well contents from 13th plate into a 15 ml tube. Allow nematodes to settle (2-3 min). Use supernatant to load a microplate (black with transparent bottom) with 100 μl bacterial suspension per well.
- Observe background control plate under microscope noting down wells where nematodes can be seen. Exclude these from the background estimate used in subsequent data analysis.
- Read fluorescence of each 96-well plate, including the background control. (Settings: optics from bottom of plate; filters 485/20 excitation; 528/20 emission.)
- Prepare buffer for luminescence readings: add luciferin (20 mM) to part-prepared buffer (from the previous day), to obtain luminescence buffer with 1% DMSO (1x), 0.15% Triton-X100 (3x) and 0.3 mM luciferin (3x). Prime luminometer injector 6x with 150 μl luminescence buffer.
- Previous step assumes 1% DMSO as vehicle during drug exposure. When vehicle is water, adjust the concentration of DMSO in luminescence buffer to 3% (i.e., 3x).
- Work with one plate at a time. Dispense 50 μl luminescence buffer per well. (150 μl final volume in well; 3x dilution of luminescence buffer: 1% DMSO, 0.05% Triton-X100 and 0.1 mM luciferin final concentrations.) Place immediately on shaking platform and start timer.
- After 3 min on shaking platform (160 rpm), read luminescence (1 sec per measurement).
- Set up plate to use as background control for GFP readings. Combine well contents from 13th plate into a 15 ml tube. Allow nematodes to settle (2-3 min). Use supernatant to load a microplate (black with transparent bottom) with 100 μl bacterial suspension per well.
4. Data Analysis
- Subtract average GFP background reading from fluorescence results. Divide bioluminescence by respective GFP reading.
- When a high GFP value is detected for nematodes exposed to a compound, measure fluorescence of the compound on its own to account for any compound fluorescence. If higher than average, use this as the background instead.
- Express bioluminescence, GFP normalized bioluminescence and GFP fluorescence data as a percentage of the average value for vehicle control in each plate.
- Plot average values and error bars for each concentration.
- Test for statistical significance using 2-way analysis of variance (ANOVA) with effects of ‘Concentration’, ‘Experiment’ and their interaction (each data point refers to a single well), and use the Dunnett test as post-hoc test (vs. vehicle control).
NOTE: If the “experiment” or the “interaction” terms are significant, further experiments may be required to confirm response. Where no variability between experiments is apparent from observation of data plots, a One-way ANOVA can be performed on pooled data from independent experiments.- For the statistical analysis of plate(s) with vehicle only, select all wells in columns 6 -7 and all wells in row H as control group (these are the positions routinely assigned to vehicle during compound testing).
Representative Results
Representative results to illustrate the method were obtained for rotenone (Figure 1), paraquat (Figure 2), oxaloacetate (Figure 3), and 4 compounds that were picked up as firefly luciferase inhibitors during screening of a drug library13 (Figure 4). The drug exposure was started at the L4 stage in all cases, but the paraquat exposure experiments were started at 41 hr after food first provided to nematodes compared to 45-46 hr for the other compounds. The protocol allows for a degree of flexibility in the selection of time for the initiation of the drug exposure and the exposure length. However, for screening purposes, set times should be adhered to once selected, for reproducibility between experiments. Endpoints should be measured by 66-67 hr developmental time in order to prevent extensive egg laying and hatching of progeny in wells. Guidelines for timings are provided in Table 1.
Rotenone, a mitochondrial complex I inhibitor, reduced bioluminescence after both relatively short (2 hr) and longer exposures (24 hr) to a range of concentrations (Figure 1). In this instance, no GFP measurements were taken, but the number of technical replicates used (n=6) is sufficient to even out any differences in worm numbers between wells3. The 24 hr exposure is a window of time over which nematodes grow, and slower development as a result of complex I inhibition was expected to contribute to the decline in bioluminescence. This was confirmed by visual observation under stereoscope with delayed development seen, particularly at concentrations of 20 μM and above; in addition some lethality was noticed from 40 μM (qualitative observations not quantified). No lethality was observed after 2 hr exposure but effects on worm movement were seen. A sharp decline in bioluminescence relative to controls occurred at the lowest concentration of rotenone tested 2.5 μM, for which effects were not easily detected by quick visual observation at 2 hr exposure. This decline in bioluminescence is consistent with reduced cellular ATP. Maximal inhibition was achieved with the lowest concentration of rotenone used (2.5 μM) indicating that any subsequent experiments specifically targeting rotenone should be carried out between 0 and 5 μM [the concentration range 0-160 μM was selected as part of a comparison with other drugs initially identified as having significant effects at 10 μM (to be published elsewhere)].
The herbicide paraquat affects mitochondrial function through increase of reactive oxygen species. Here, we demonstrate its effect on bioluminescence of C. elegans strain PE254 after exposure to a range of concentrations for 24 hr (Figure 2). As the strain carries a luc::GFP fusion, GFP fluorescence was also measured as a means of normalization.6,9 Paraquat decreased bioluminescence, GFP fluorescence and normalized bioluminescence significantly. The Dunnet post-hoc test indicated that the concentrations of paraquat with significant differences in relation to vehicle control were 4 and 8 mM for bioluminescence and normalized bioluminescence, as well as 2 mM for normalized bioluminescence. The only concentration significantly reducing GFP fluorescence was 8 mM, a concentration at which growth effects and the occasional dead worm were seen (qualitative observations). The decline in bioluminescence (and normalized bioluminescence) was greater than that of fluorescence, consistent with decreased mitochondrial function and effects on ATP production.
The citric acid cycle intermediate oxaloacetate tested on C. elegans strain PE254 at a single concentration of 8 mM, led to an increase in bioluminescence (Figure 3). No GFP fluorescence measurements were obtained or additional visual observation carried out; however such response to a single concentration would merit further confirmatory exposure to a range of concentrations, and more detailed observations of effects. An enhancement of bioluminescence by this compound is not surprising as it can be postulated to lead to greater activity of the citric acid cycle, with ultimately greater production of ATP (nevertheless it would be advisable to control for any effects on luciferase levels by assessing GFP fluorescence in subsequent experiments). The oxaloacetate data sets shown reveal the extent of variability in response seen in luciferase based experiments. In our experience this variability is a feature of the test system particularly for less detrimental exposure conditions.
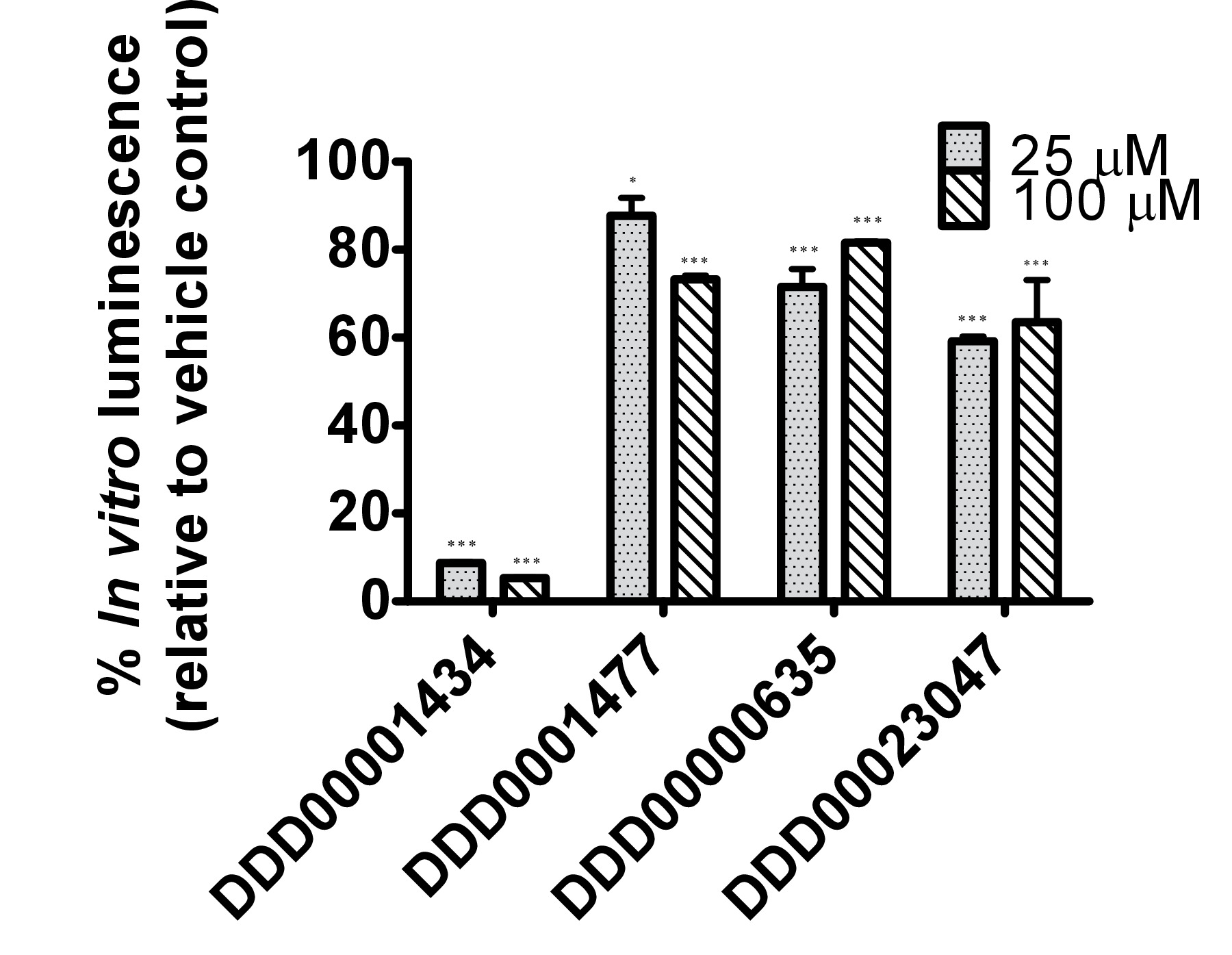
The firefly luciferase inhibitory compounds DDD00001434, DDD0001477, DDD00000635 and DDD00023047 were tested both in vitro by looking at effects on the purified enzyme and in vivo using the C. elegans luc:GFP expressing strain. There was no evidence that any of the compounds tested caused nematode death under the exposure conditions. All 4 compounds affected the luciferase activity in vitro at the 2 concentrations tested 25 and 100 μM (Figure 4A). DDD00001434 killed the activity of the purified luciferase almost completely, which provided a credible justification for the large decline in in vivo bioluminescence (Figure 4B). Although, the in vitro and in vivo assays are not directly comparable, the response to DDD00023047 was similar in both assays. DDD0001477 and DDD00000635 caused a greater decline in vivo than in in vitro. These compounds were provided to the live worms 22 hr prior to the luciferin substrate, and results may reflect that they were not easily displaced from the luciferase active site when luciferin became available. Alternatively, DDD0001477 and DDD00000635 may have an additional effect on cellular ATP levels. This would have to be assessed by means that do not involve firefly luciferase. Of note was the strong enhancement of GFP signal in live worms exposed to compound DDD00000635. This will be discussed below.
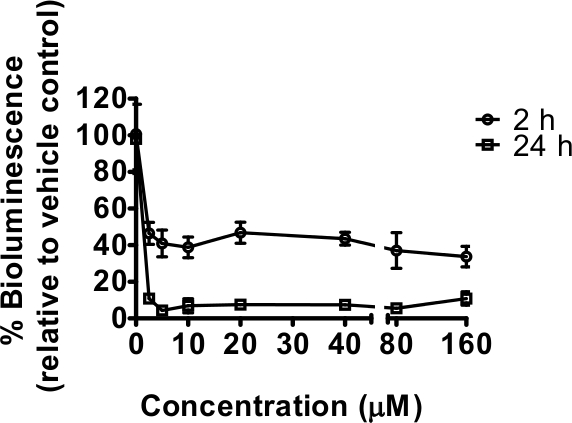
Figure 1. The mitochondrial complex I inhibitor rotenone decreased the energy status of C. elegans as measured by bioluminescence of strain PE254. Synchronized L4 stage worms (45-46 hr after food provided to L1 larvae) exposed to 0, 2.5, 5, 10, 20, 40, 80 and 160 μM rotenone in 1% DMSO for 2 hr (short) or 24 hr (long exposure) prior readings of bioluminescence respectively at 48 and 69 hr of worm development after food provided. Bioluminescence (unit Relative Light Units, RLU) was expressed as a percentage of the vehicle (1% DMSO) values. Error bars depict SEM of technical replicates at each rotenone concentration (n=6). All concentrations tested resulted in a highly statistically significant difference in relation to vehicle control (p<0.001).
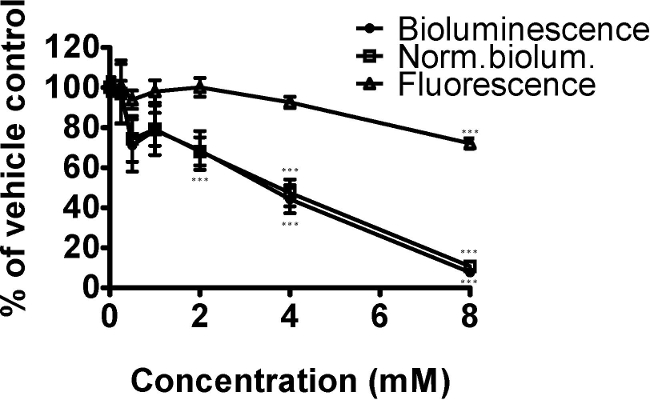
Figure 2. The oxidative stressor paraquat decreased the energy status of C. elegans strain PE254 in a concentration dependent manner (measured by bioluminescence). Synchronized L4 stage worms (for convenience in this instance at 41 hr after food provided to L1 larvae) were exposed to 0, 0.25, 0.5, 1, 2, 4 or 8 mM paraquat for 24 hr. GFP fluorescence (unit Relative Fluorescence Units, RFU) was used to normalize bioluminescence data (unit RLU), both endpoints obtained at 65 hr development. Data are expressed as a percentage of controls (1% DMSO). Error bars depict SEM of technical replicates (n=5 for different concentrations; n=18 for controls). One-way ANOVA: *** p<0.001 relative to vehicle control.
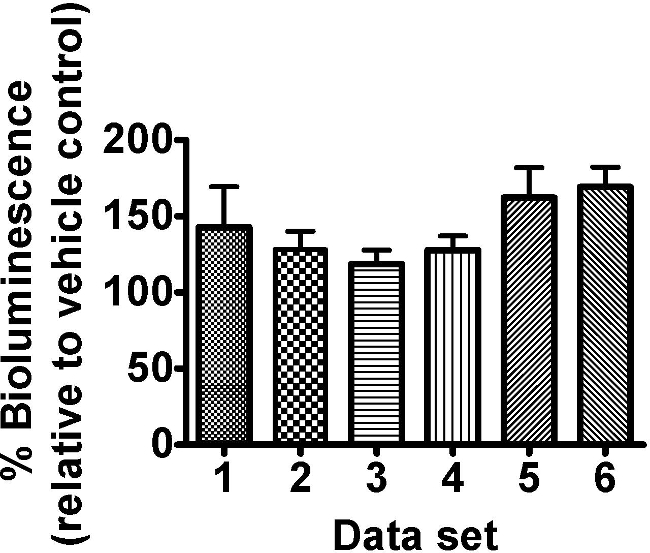
Figure 3. The citric acid cycle intermediate oxaloacetate (8 mM) enhanced the bioluminescence of C. elegans strain PE254. Synchronized L4 stage worms (45-46 hr after food provided to L1 larvae) were exposed to freshly prepared 8 mM oxaloacetate for 18 hr ± 30 min. Bioluminescence (unit RLU) was read at a developmental time of 63.5 hr ± 1 hr and was expressed as a percentage of the vehicle control (ddH2O). Different columns represent different data sets of 8 technical replicates allocated to different 96 well plates within the same experiment. Error bars depict SEM of technical replicates (n=8). 2 way-ANOVA with ‘set’ and ‘concentration’ as factors: ‘Set’ p>0.05, ‘concentration’ p<0.001 and interaction term p>0.05.
A
B
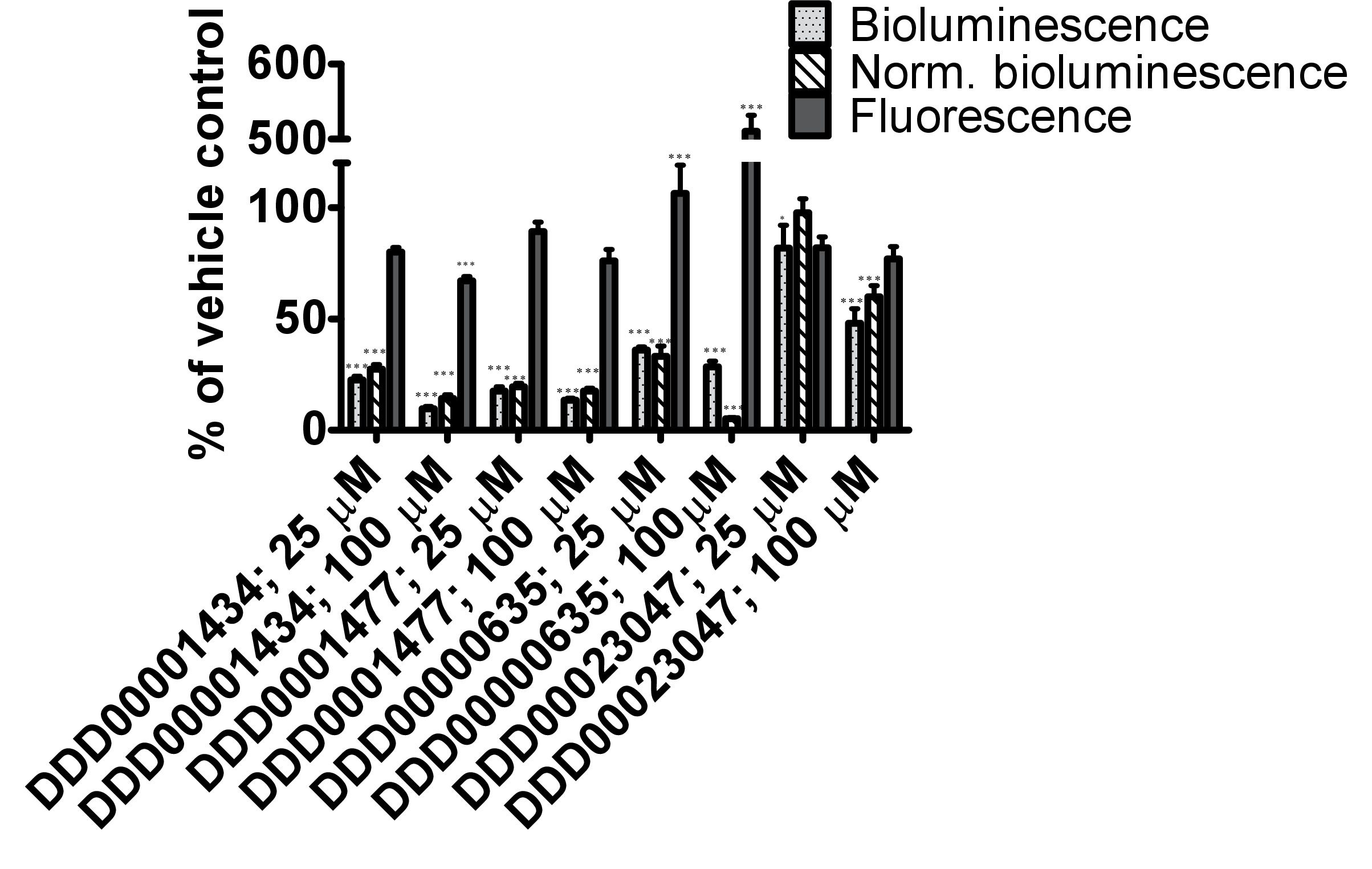
Figure 4. Testing of responses to 4 compounds from the Dundee drug discovery compound library13 (C1:DDD00001434, C2:DDD0001477, C3:DDD00000635 and C4:DDD00023047) with known inhibitory activity on firefly luciferase luminescence. (A) Effect of compounds on the activity of purified firefly luciferase (measured as light units, RLU), expressed as a percentage of vehicle control. The ATP bioluminescence CLSII kit (Roche, Manheim, Germany) was used according to instructions with the same amount of luciferase enzyme aliquoted to wells (white 96-well microtiter plates). ATP was provided at a final concentration of 10 μM and 1 μl of compound (final concentration indicated) or DMSO (1%) was added. Luminescence signal was integrated for 10 sec. Error bars depict SEM of technical replicates (n=4). Analysis: One-way ANOVA; *p<0.05 or *** p<0.001 relative to vehicle control. Differences between 25 and 100 μM highly significant for C1 (DDD00001434; p<0.001), and significant for C2 and C3 (respectively DDD0001477 and DDD00000635; p<0.05). (B) In vivo measurements of bioluminescence, bioluminescence normalized to GFP fluorescence and GFP fluorescence of C. elegans strain PE254 at 67 hr developmental time following exposure to test compounds for 22 hr. Error bars depict SEM of technical replicates (n=8). Statistical analysis (One way-ANOVA) carried out on pooled data from 3 data sets (3 x n=8). *p<0.05 or *** p<0.001 relative to respective vehicle control. Differences between 25 and 100 μM only statistically significant (p<0.05) for normalized bioluminescence following exposure to C4 (DDD00023047).
| Time of day | Day 1 | Day 2 | Day 3 | Day 4 | Day 5 | Day 6 | Day 7 | Day 8 |
| 9-10 | Step 5 | |||||||
| 10-11 | Step 5 | |||||||
| 11-12 | Step 4 | Step 5 | ||||||
| 12-13 | Step 4 | |||||||
| 13-14 | ||||||||
| 14-15 | ||||||||
| 15-16 | ||||||||
| 16-17 | Step 1 | Step 3 | ||||||
| 17-18 | ||||||||
| 18-19 | Step 2 |
Table 1. Overview of protocol steps to be carried out on each day of nematode experiments (Section 3) and suggested timings.
Discussion
The model organism C. elegans provides a powerful experimental system. It is easy to culture and produces plentiful genetically identical progeny with a 3.5 day life cycle from egg to reproducing adult (via juvenile larval stages L1 through to L4). Due to its small size, it can be conveniently grown in 96-well plates, facilitating compound screening. We present a phenotypic screening protocol based on strains of C. elegans that act as in vivo sensors of energy levels.14 The protocol is applicable to any laboratory, although a 96-well plate luminometer and a sterile cabinet is required. It is important to prevent contamination in assays by using good laboratory technique. If working manually, the maximum number of nematode plates achievable is 13 per experiment allowing the testing of 2x 96-well drug plates and including two vehicle plates. Representative results are presented for the mitochondrial complex I inhibitor rotenone, the superoxide generator paraquat, the citric acid cycle intermediate oxaloacetate and four compounds with known firefly luciferase inhibitory activity. The first two compounds led to a decrease in bioluminescence as predicted whereas oxaloacetate led to an enhancement, likely to result from greater generation of ATP. The oxaloacetate data sets also demonstrate the intrinsic variability in experiments based on firefly luciferase as a reporter. A minimum of three independent experiments are advisable to ascertain robustness of responses to unknown compounds.
Inhibition of firefly luciferase activity was identifiable with an in vitro assay using a commercial kit.3 One of the compounds resulted in a substantial increase in GFP fluorescence consistent with the inhibitor increasing the levels of GFP-tagged firefly luciferase by binding to the luciferase enzyme’s active site, and rendering it more stable.15 In some instances (not in this particular case) this can lead to increased levels of bioluminescence when the inhibitor is displaced by excess substrate.15 It is, therefore, important not to interpret results erroneously from compounds that interact with the firefly enzyme as decreased or increased ATP levels/mitochondrial function. Changes in luminescence signal quite often correlate with additional measurements of health such as movement, development and growth. As an example, a compound is a suspect luciferase inhibitor if a dramatic decline in bioluminescence signal is detected but worms are indistinguishable from controls by microscopic observation. An increase in GFP fluorescence, not explained by growth or compound autofluorescence, may also point to an inhibitor. A number of luciferase inhibitors15 are known and have been entered into PubChem. Therefore in the first instance, it is good practice to consult information on luciferase inhibitors held by PubChem; followed by testing of the compound effects in in vitro assays with the purified enzyme3 and/or confirmation of effects on mitochondrial function by additional means.16,17
GFP fluorescence measurements enable the detection of firefly luciferase levels (and the potential detection of some luciferase enzyme inhibitors as discussed above) making a strong case for measuring this endpoint alongside bioluminescence where possible. Normalization of bioluminescence readings to GFP is also a means of accounting for variations in worm numbers between wells (although this can also be achieved by inclusion of multiple technical replicates). For greater sensitivity, it is important to subtract background fluorescence from readings. The protocol assesses contribution of the bacterial suspension to the background fluorescence, however nematode autofluorescence is not accounted for (a limitation of the current assay, particularly critical for any aging studies given that nematode autofluorescence increases with age). Autofluorescence of test compounds should also be assessed in parallel. GFP normalization would seem indispensable where researchers wish to adapt protocol to compare cellular ATP pools in different genetic mutants or after silencing of different genes, in order to control for any strain differences at the level of expression of the bioluminescence transgene.
Critical parameters within the protocol include the developmental stage at which exposure is initiated, the length of exposure, and obtaining similar numbers of nematodes in different wells. Exposure can start at any stage of development of the nematodes, however L3 stage or older is preferable. From this stage on, nematodes depend on oxidative phosphorylation for development into adulthood, as evidenced by a very significant increase in mtDNA content, oxygen consumption and ATP levels.18,19,20 The present protocol initiates exposure at the L4 stage. The reason for aliquoting nematodes to 96-well plates only at this stage (rather than when they were first provided with food at the L1 stage), is that although plates are placed in damp chambers to minimize evaporation, a degree of evaporation still occurs in our shaking incubator, seen as very small droplets forming on the lids (this may not be an issue with other shaking incubators). By aliquoting nematodes to plates just before addition of drug standards, the actual drug concentration is not affected by any small variation in volume due to evaporation. Loading a nematode plate with vehicle only is useful to check that nematodes were evenly distributed between wells. Any significant differences found for the vehicle only plate would be indicative of poor technique in dispensing the nematodes to wells.
The length of exposure is an important factor to consider. If effects on energy status independently of growth are of interest, the protocol can be modified to accommodate shorter exposures (from almost immediate responses to, for instance, 2 hr). On the other hand, entry of compounds into C. elegans tissues may take some time. For example 12-24 hr are required to achieve maximum internal concentrations of resveratrol and 5-fluoro-2'-deoxyuridine (FuDR).21 Therefore, a longer exposure (18 to 24 hr) may be justifiable to maximize exposure levels. The exposure time should be limited to times that do not allow significant levels of reproduction to take place in the well. We recommend that the total developmental time of nematodes is kept under 66-67 hr. Although convenient, nematodes are gravid by then and have initiated egglaying. Nevertheless we found bioluminescence readings from egg preparations to be negligible (not shown). The sur-5 promoter driven transgene expression is first detected only two to two and a half hours after fertilization at the 100 cell stage22 and the chitinous eggshell is likely to limit entry of luciferin. Others have found that embryonated eggs did not contribute significantly to the metabolism of gravid adults.23 However, conditions allowing for significant offspring production are to be avoided. If preferred, the experimental timescale can be shifted back: we have previously initiated exposure to an environmental toxin at the late L3 larval stage (36 hr) and carried out bioluminescence and GFP measurements at 55 hr.2 Alternatively, genetic backgrounds that are conditionally sterile can be used to prevent progeny production, as for example the fer-15(b16) II; fem-1(hc17) IV double mutant which can be maintained at 15 °C, but is sterile at 25 °C.24 The use of FuDR to induce sterility is not recommended as this can in itself affect nematode metabolism.25 The assay could also be performed in strains with more permeable cuticles such as partial loss-of-function bus-8 mutants which are multidrug-sensitive due to increase permeability of drugs.26 This will facilitate shorter exposures and lower compound concentrations. The conditions for adding luciferin to these nematodes may also need adjusting i.e., 1% DMSO and 0.05% Triton-X100 in the luminescence buffer may not be required to enhance luciferin availability.
The protocol uses 1% DMSO as vehicle for compound delivery. Although not lethal, this concentration of DMSO has some biological effects as we have discussed in a previous publication.3 Many of the available drug libraries are prepared in 100% DMSO, at concentrations that will be suitable for cell-based assays after dilution of vehicle to 0.1%. Higher compound concentrations are required to elicit responses in C. elegans when compared with human cells, such that it is often not possible to conduct assays at lower than 1% DMSO. Again, the use of strains with more permeable cuticles may lead to testing with lower concentrations of vehicle. Concentrations of DMSO higher than 1% are not recommended.
A set incubation time with luciferin prior to readings should be adhered to. As previously shown, luminescence reaches its maximum levels within the second minute after adding luciferin, remaining relatively stable for the first 5 min, followed by a slow gradual decrease in luminescence over the next 30 min1. Kinetic responses to a particular chemical can be carried out during this initial stage of 30 min after initial dispensing of luciferin.
The protocol involves a starvation step following collection of eggs to achieve synchronization of the nematode population. We recommend synchronization of nematode test populations since different developmental stages differ in cellular ATP levels and may respond differently to the test compounds. By carrying out the starvation in S complete medium as opposed to M9, the hatching nematodes are provided with a carbon source (ethanol), so that larvae do not develop but are not completely starved.27,28 It has also been shown that arrested L1’s are primed for rapid response to food and normal L1 growth rate is achieved within 3 hr after food becomes available.29 However, it the possibility that the starvation step may alter the metabolism of C. elegans cannot be excluded.30 For this reason the length of starvation should not exceed 24 hr (18 hr is sufficient for synchronization). Certain laboratories may have the possibility of using a Copas Biosorter for age synchronization bypassing the starvation step altogether. Other laboratories may have a preference for expanding the nematode population on solid media (NGM plates with OP50) prior to bleaching (step 3.1) and this will work well too. We use S medium during compound exposure as a standard medium for nematode culturing, however other media can be selected, for instance K medium or EPA moderately hard water are often used for ecotoxicological studies.31,32
The protocol provides a means to screen and to identify candidates for further testing in terms of the effects on mitochondrial function. At the stage of primary screening it is likely that some compounds will have been missed due to only one concentration being tested, therefore drug screens should not be considered exhaustive. Hits can be validated by the use of techniques for assessing different aspects of mitochondrial function for example oxygen consumption, C. elegans strains with GFP expressing mitochondria, stains for mitochondrial membrane potential and/or ROS measurements.16,33,34 The greatest significance of the methodology is to have a means for screening large number of compounds and/or conditions at the multicellular organismal level, and importantly to be able to take advantage of the genetic tractability of C. elegans to investigate mechanisms of action. The technique may help to accelerate and find new pathways to the discovery of interesting compounds and targets. It is envisaged that the combination of the sensor with genetic backgrounds associated with human disease for screening of compound libraries in a disease relevant context will have much to offer.
Divulgations
The authors have nothing to disclose.
Acknowledgements
The authors would like to thank David Gray from the Dundee Drug discovery unit for kindly donating firefly luciferase inhibitory compounds DDD00001434, DDD0001477, DDD00000635 and DDD00023047; Tibor Harkani (Medical University of Vienna) for the suggestion of oxaloacetate as a test compound; and Charlie Dear (University of Aberdeen) for illustrations. This work was funded by a BBSRC Pathfinder award (BB/FOF/PF/4/11) and the University of Aberdeen.
Materials
| Orion II Microplate Luminometer | Berthold Detection Systems | with injector and the Simplicity 4.2 Software; transfer data to Excel at the end of plate measurement | |
| FLx800 fluorimeter | Biotek | with Gen5 Software; transfer data to Excel at the end of plate measurement | |
| Ks 130 basic shaking platform | Ika | #0002980000 | |
| Innova 4349 | New Brunswick Scientific | Refrigerated incubator shaker | |
| Eppendorf centrifuge 5804R | Eppendorf | with eppendorf rotor A-4-44 (for 50/15ml tubes) | |
| Stripette, Costar | Corning | #4489 | Serological pipette |
| Cellstar 50 ml tubes | Greiner bio-one | #227661 | |
| Cellstar 15 ml tubes | Greiner bio-one | #188261 | |
| Cellstar 60 mm tissue culture plates | Greiner bio-one | #628160 | |
| Superfrost Microscope slides | VWR | #631-0909 | |
| Sterile spatula | Corning | #3007; #3004 | for weighing out chemicals |
| 0.22 mM Millex GP filters | Millipore | #SLGP033RS | |
| Axygen eppendorf tubes 1.5 ml | Fisher Scientific | #MCT-150-R | |
| Corning 96 well assay plate | VWR | #3603 | Black plate clear bottom with lid |
| Nunc 96 well assay plate | Fisher Scientific | #236105 | White plate with lid |
| Reservoir reagent 60 mL | Thermo Scientific Finnpipette | #9510027 | used as trough for nematodes in protocol section 4.4.1) |
| Storage box/damp chamber | Roche Diagnostics | #10 800 058 001 | 174 x101 x 56.6 mm, used as a damp chamber with wet paper towels; 2X 96-well plates with lids can fit into one, the lower sitting on top of a microplate lid |
| Bleach: Sodium hypochlorite solution 4-4.99% Chlorine content | Sigma Aldrich | #239305 | Store in aliquots at 4°C, seal with parafilm to prevent loss of chlorine and cover in foil |
| D-Luciferin, potassium salt | Biotium Inc | # 10101-2 | Molecular Probes can also be used as supplier; prepare a 20 mM stock in ddH2O, keep at -20 °C in aliquots |
| Tween 20 | Sigma Aldrich | # P9416 | |
| DMSO | CalBiochem | #317275 | Purity 99.99% |
| Triton X100 | Alfa Aesar | #A16046 | Diltute to 10% in ddH20 |
| Nystatin | CalBiochem | #475914 | |
| C. elegans bioluminescent strains | Author's own laboratory | PE254, PE255 | contain integrated arrays feIs4 and feIs5 [Psur-5::luc+::gfp; rol-6(su1006)] respectively on chromossome V and X; select for homogeneous and strong expression of luc::GFP by fluorescence microscopy (e.g. pick 15 worms) every now and then. |
| E. coli OP50 | CGC | ||
| General chemicals | Sigma Aldrich/Fisher Scientific |
References
- Lagido, C., Pettitt, J., Flett, A., Glover, L. A. Bridging the phenotypic gap: real-time assessment of mitochondrial function and metabolism of the nematode Caenorhabditis elegans. BMC Physiol. 8, 7 (2008).
- Lagido, C., McLaggan, D., Flett, A., Pettitt, J., Glover, L. A. Rapid sublethal toxicity assessment using bioluminescent Caenorhabditis elegans, a novel whole-animal metabolic biosensor. Toxicol Sci. 109, 88-95 (2009).
- McLaggan, D., et al. Impact of sublethal levels of environmental pollutants found in sewage sludge on a novel Caenorhabditis elegans model biosensor. PloS one. 7, e46503 (2012).
- Rodriguez-Enriquez, S., Juarez, O., Rodriguez-Zavala, J. S., Moreno-Sanchez, R. Multisite control of the Crabtree effect in ascites hepatoma cells. Eur J Biochem. 268, 2512-2519 (2001).
- Marroquin, L. D., Hynes, J., Dykens, J. A., Jamieson, J. D., Will, Y. Circumventing the Crabtree effect: replacing media glucose with galactose increases susceptibility of HepG2 cells to mitochondrial toxicants. Toxicol Sci. 97, 539-547 (2007).
- Rooney, J. P., et al. Effects of 5′-fluoro-2-deoxyuridine on mitochondrial biology in Caenorhabditis elegans. Exp Gerontol. 56, 69-76 (2014).
- Bodhicharla, R., Ryde, I. T., Prasad, G. L., Meyer, J. N. The tobacco-specific nitrosamine 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK) induces mitochondrial and nuclear DNA damage in Caenorhabditis elegans. Environ Mol Mutagen. 55, 43-50 (2014).
- Leung, M. C., et al. Effects of early life exposure to ultraviolet C radiation on mitochondrial DNA content, transcription, ATP production, and oxygen consumption in developing Caenorhabditis elegans. BMC Pharmacol Toxicol. 14, 9 (2013).
- Kuang, J. J., Ebert, P. R. The failure to extend lifespan via disruption of complex II is linked to preservation of dynamic control of energy metabolism. Mitochondrion. 12, 280-287 (2012).
- Lagido, C., Pettitt, J., Porter, A. J., Paton, G. I., Glover, L. A. Development and application of bioluminescent Caenorhabditis elegans as multicellular eukaryotic biosensors. FEBS Lett. 493, 36-39 (2001).
- Lewis, J. A., Fleming, J. T. Basic culture methods. Methods Cell Biol. 48, 3-29 (1995).
- Stiernagle, T. Maintenance of C. elegans. WormBook. , 1-11 (2006).
- Brenk, R., et al. Lessons learnt from assembling screening libraries for drug discovery for neglected diseases. ChemMedChem. 3, 435-444 (2008).
- Lagido, C., Wilson, M. J., Kakouli-Duarte, T. . Nematodes as environmental indicators. , 225-251 (2009).
- Thorne, N., et al. Firefly luciferase in chemical biology: a compendium of inhibitors, mechanistic evaluation of chemotypes, and suggested use as a reporter. Chem Biol. 19, 1060-1072 (2012).
- Houtkooper, R. H., et al. Mitonuclear protein imbalance as a conserved longevity mechanism. Nature. 497, 451-457 (2013).
- Andreux, P. A., Houtkooper, R. H., Auwerx, J. Pharmacological approaches to restore mitochondrial function. Nat Rev Drug Discov. 12, 465-483 (2013).
- Tsang, W. Y., Lemire, B. D. Mitochondrial genome content is regulated during nematode development. Biochem Bioph Res Co. 291, 8-16 (2002).
- Decuyper, C., Vanfleteren, J. R. Oxygen-Consumption during Development and Aging of the Nematode Caenorhabditis-Elegans. Comp Biochem Phys A. 73, 283-289 (1982).
- Dillin, A., et al. Rates of behavior and aging specified by mitochondrial function during development. Science. 298, 2398-2401 (2002).
- Zheng, S. Q., Ding, A. J., Li, G. P., Wu, G. S., Luo, H. R. Drug absorption efficiency in Caenorhbditis elegans delivered by different methods. PloS one. 8, e56877 (2013).
- Yochem, J., Gu, T., Han, M. A new marker for mosaic analysis in Caenorhabditis elegans indicates a fusion between hyp6 and hyp7, two major components of the hypodermis. Génétique. 149, 1323-1334 (1998).
- Vanfleteren, J. R., DeVreese, A. Rate of aerobic metabolism and superoxide production rate potential in the nematode Caenorhabditis elegans. J Exp Zool. 274, 93-100 (1996).
- Garigan, D., et al. Genetic analysis of tissue aging in Caenorhabditis elegans: a role for heat-shock factor and bacterial proliferation. Génétique. 161, 1101-1112 (2002).
- Davies, S. K., Leroi, A. M., Bundy, J. G. Fluorodeoxyuridine affects the identification of metabolic responses to daf-2 status in Caenorhabditis elegans. Mech Ageing Dev. 133, 46-49 (2012).
- Partridge, F. A., Tearle, A. W., Gravato-Nobre, M. J., Schafer, W. R., Hodgkin, J. The C. elegans glycosyltransferase BUS-8 has two distinct and essential roles in epidermal morphogenesis. Dev Biol. 317, 549-559 (2008).
- Castro, P. V., Khare, S., Young, B. D., Clarke, S. G. Caenorhabditis elegans Battling Starvation Stress: Low Levels of Ethanol Prolong Lifespan in L1 Larvae. PloS one. 7, (2012).
- Baugh, L. R. To Grow or Not to Grow: Nutritional Control of Development During Caenorhabditis elegans L1 Arrest. Génétique. 194, 539-555 (2013).
- Baugh, L. R., DeModena, J., Sternberg, P. W. RNA Pol II Accumulates at Promoters of Growth Genes During Developmental Arrest. Science. 324, 92-94 (2009).
- Maxwell, C. S., Antoshechkin, I., Kurhanewicz, N., Belsky, J. A., Baugh, L. R. Nutritional control of mRNA isoform expression during developmental arrest and recovery in C. elegans. Genome Res. 22, 1920-1929 (2012).
- Cressman, C. P., Williams, P. L. Reference toxicants for toxicity testing using Caenorhabditis elegans in aquatic media. Am Soc Test Mater. 131, 518-532 (1997).
- Khanna, N., Cressman, C. P., Tatara, C. P., Williams, P. L. Tolerance of the nematode Caenorhabditis elegans to pH, salinity, and hardness in aquatic media. Arch Environ Con Tox. 32, 110-114 (1997).
- Benedetti, C., Haynes, C. M., Yang, Y., Harding, H. P., Ron, D. Ubiquitin-like protein 5 positively regulates chaperone gene expression in the mitochondrial unfolded protein response. Génétique. 174, 229-239 (2006).
- Yang, W., Hekimi, S. A Mitochondrial Superoxide Signal Triggers Increased Longevity in Caenorhabditis elegans. Plos Biol. 8, (2010).

