µTongue: A Microfluidics-Based Functional Imaging Platform for the Tongue In Vivo
Summary
The article introduces the µTongue (microfluidics-on-a-tongue) device for functional taste cell imaging in vivo by integrating microfluidics into an intravital imaging window on the tongue.
Abstract
Intravital fluorescence microscopy is a tool used widely to study multicellular dynamics in a live animal. However, it has not been successfully used in the taste sensory organ. By integrating microfluidics into the intravital tongue imaging window, the µTongue provides reliable functional images of taste cells in vivo under controlled exposure to multiple tastants. In this paper, a detailed step-by-step procedure to utilize the µTongue system is presented. There are five subsections: preparing of tastant solutions, setting up of a microfluidic module, sample mounting, acquiring functional image data, and data analysis. Some tips and techniques to solve the practical issues that may arise when using the µTongue are also presented.
Introduction
The intravital fluorescence microscope is used widely to study the spatiotemporal dynamics on living tissues. Researchers are rapidly developing genetically encoded sensors that provide specific and sensitive transformations of the biological processes into fluorescence signals – which can be recorded readily using fluorescence microscopes that are widely available1,2. Although most internal organs in rodents have been investigated using the microscope, its successful application to the tongue has not yet been successful3.
Previous studies on the calcium imaging of taste cells were conducted ex vivo by thin-sectioning a tongue tissue to obtain circumvallate taste buds4,5,6 or by peeling off the taste epithelium to obtain fungiform taste buds7,8. The preparation of these samples was inevitably invasive, thus the natural microenvironments such as nerves innervation, permeability barriers, and blood circulation, were largely perturbed. The first intravital tongue imaging window was reported in 2015 by Choi et al., but reliable functional recording was not achievable because of the movement and optical artifacts caused by fluidic tastant stimuli9.
Recently, microfluidics-on-a-tongue (µTongue) was introduced10. This device integrates a microfluidic system with an imaging window on the mouse tongue. By attaining a quasi-steady-state flow of tastant stimuli throughout the imaging period, artifacts from fluidic motion could be minimized (Figure 1). The input port is fed by a series of multichannel pressure controllers, whereas the output port is connected to a syringe pump, which maintains 0.3 mL/min. Additionally, optical artifacts caused by the difference in refractive indices of tastant solutions were minimized by ratiometric analysis introducing a calcium-insensitive indicator (tdTomato) as well as the calcium indicator (GCaMP6)11. This design provided microscopic stability of taste cells in vivo even with abrupt switching between fluidic channels. Consequently, the µTongue implement a reliable functional screening of multiple tastants to the mouse taste buds in vivo.
In this protocol, the experimental procedures are explained in detail for calcium imaging of the mouse fungiform taste buds in vivo using µTongue. First, the preparation of artificial saliva and tastant solutions is described. Second, the setting up of the microfluidic system to achieve the quasi-steady-state flow is introduced. Third, the procedures used to mount the mouse tongue on the µTongue to permit image acquisition are delineated. Lastly, each step for image analysis, including correction of lateral motion artifacts and ratiometry, is specified. This protocol can be adapted readily to any research laboratory with a mouse facility and a two-photon microscope or equivalent equipment.
Protocol
All surgical procedures were approved by the Institutional Animal Care and Use Committee (IACUC) of Sungkyunkwan University and Seoul National University.
1. Preparation of solutions: artificial saliva and tastants
- Prepare artificial saliva by dissolving 2 mM NaCl, 5 mM KCl, 3 mM NaHCO3, 3 mM KHCO3, 0.25 mM CaCl2, 0.25 mM MgCl2, 0.12 mM K2HPO4, 0.12 mM KH2PO4, and 1.8 mM HCl in distilled water (>1 L), and adjust the pH of the solution to 7 (see Table of Materials)12.
- Prepare tastants, such as sour: 10 mM citric acid; salty: 400 mM NaCl, optionally with 50 µM amiloride; sweet: 40 mM acesulfame K; bitter: a mixture of 5 mM quinine, 5 mM denatonium and 20 µM cycloheximide, by dissolving the tasting chemical in the artificial saliva prepared in step 1.1.
2. Preparation of the microfluidic system
NOTE: Tastants were delivered to the mouse tongue using a pressurized multichannel fluidic delivery system (refer Figure 1 and Table of Materials).
- Fill the reservoirs of the pressurized flow perfusion system with the artificial saliva and tastants.
- Connect the compressed air line to the regulator input and set the air pressure between 30 and 50 psi in the fluidic delivery system.
- Set the output pressure of the regulator to 0.4 psi and check if liquid comes out of the tube under this pressure.
- Connect the manifold from the reservoirs to the input port of µTongue.
- Connect the output port of µTongue to a syringe pump and withdraw liquid with ~300 µL∙min-1 to establish the steady-state condition. Observe the constant volume of a hanging droplet under the µTongue. Adjust the value of the setting parameter depending on the sample height.
- Disconnect the compressed air line and stop the syringe pump until protocol step 3 is completed.
3. Mouse preparation for in vivo imaging (Figure 2).
NOTE: All animal preparations were carried out during the daytime under aseptic conditions on a laboratory workbench.
- Mouse anesthesia
- Prepare a 7-week-old or older mouse of either sex. Use a genetically modified mouse line that expresses calcium-sensing fluorescence proteins in the taste cells.
- The mouse is restrained for anesthesia. A mixture of 100 mg/kg ketamine and 10 mg/kg xylazine is injected intraperitoneally into the mouse13.
- TRITC-dextran (500 kDa) in 2.5% W/V phosphate buffer saline is administrated intravenously into the mouse through a retro-orbital route to observe blood circulation during an imaging session.
- Attach a head fixer on the mouse skull to minimize movement artifacts.
- The mouse head is sprayed with 70% ETOH while the mouse is placed in a supine position. Lift the head skin lightly with forceps and snip off approximately 7 mm2 with scissors.
- Clean the hair around the scalp, remove the periosteum under the skin, apply an instant adhesive to the skull, and attach a customized head fixer.
- After the instant adhesive is hardened, apply dental glue around the head fixer and illuminate with a blue light to solidify dental glue.
- Place the mouse tongue on the bottom unit of µTongue.
- Attach the lower lip of the mouse to the bottom unit of µTongue with an instant adhesive.
- Place the mouse on the board (mouse preparation board in Figure 1B) and put the bottom unit of the µTongue to the posts (µTongue hold post in Figure 1B). Make sure that the holes at the edge of the bottom unit are aligned to the post.
- Tighten the mouse head fixer to the head fixer holder at the board. Then, adjust the distance between the mouse head and the device. Rotate the mouse head smoothly approximately 45° using the head fixer holder. This process prevents physical contact of the mouse nose with microscope objective.
- Draw the mouse tongue gently using plastic tweezers and attach the ventral side of the tongue to the upper side of the bottom unit of the µTongue. Then, wipe the surface of the mouse tongue with a wet cotton swab.
- Soak a piece of paper in the artificial saliva and place it on the exposed surface of the mouse tongue to maintain a wet condition.
- Place the curved washers on the posts that hold both ends of the bottom part of µTongue.
- Place the mouse preparation on the microscope stage. Position the exposed mouse tongue under the approximate center of the microscope objective area. Be sure not to deviate from the dynamic range of the stage. Then, tighten the mouse board on the stage with screws.
- Place the heating pad under the mouse body and maintain the temperature at 36.5 °C-37.5 °C. Monitor the mouse body temperature with a temperature sensor and control the temperature of the heating pad using a feedback signal from the temperature sensor.
- Twist a thin piece of paper and place it at the mouth of the mouse to prevent liquid from entering the mouse trachea.
- Remove the wet tissue from the mouse tongue and place the prepared µTongue on the mouse tongue. Put a microfluidic channel on the tongue and adjust its position to observe the surface of the tongue through the imaging window.
- Secure the µTongue by gently screwing on both ends with minimal compressive pressure.
4. Imaging acquisition
- Turn on the 920 nm two-photon laser and the microscope in advance of use.
- Mount the water-immersion objective (16x, NA 0.80 or 25x, NA 1.1) on the microscope. Drop the distilled water on the imaging window of the µTongue and immerse the objective.
- In camera mode, turn on the light using mercury lamp and illuminate the surface of the tongue.
- By adjusting the Z-axis, search the autofluorescent signal from the filiform papillae to find the approximate focal plane. Then, using the X and Y adjustment knob, locate a taste bud.
- Switch to the multiphoton mode. Set the image acquisition conditions as follows: excitation wavelength: 920 nm; emission filter set: 447/60 nm, 525/50 nm, and 607/70 nm; bidirectional raster scan mode, frame size: 512 x 512.
- Adjust the X and Y positions to place the taste bud on the center of the image window.
- Search the blood vessels surrounding the taste bud at about two-third height of the taste bud. Visualize the blood circulation by TRITC-dextran (500 kDa) injection from protocol step 3.2. If the blood flow clogs, loosen the fixing screws slightly to allow blood flow.
- Adjust Z-axis and find the Z-plane of taste bud that contains an adequate number of taste cells.
- Proceed calcium imaging with 2-6 Hz for 80 s. Provide a taste solution of 20 s by switching on the reservoir of the fluidic system after imaging starts. After 20 s of taste stimulation, switch the reservoir back to artificial saliva.
- After sequential imaging is finished, wait for about 3-4 min in advance to the next imaging session. Keep the artificial saliva flowing to the mouse tongue to wash away the tastant remanent from the previous imaging session. Depending on the design of the experiment, repeat the session as required.
- When in vivo calcium imaging is complete, euthanize the mouse according to the IACUC procedure. The mouse under anesthesia is sacrificed in the CO2 chamber.
NOTE: Check the depth of the anesthesia using a toe-pinch reflex. During an imaging session, artificial saliva from the reservoirs should be provided consistently. If bubbles appear at the imaging window of the µTongue, remove the bubbles by pushing them through the input or the output of the µTongue using strong liquid pressure.
5. Image analysis (Figure 3)
- Image conversion
- Open the raw image files using Fiji14 or a similar image analysis software.
- Convert the image file to a RGB stack file to use the NPL Bud Analyzer code.
- Image > Color > Split Channels
- Image > Color > Merge Channels and select the image from step 5.2.1.
- Image > Color > Stack to RGB
- Image registration
NOTE: Use the custom-written code for data analysis. Please refer to https://github.com/neurophotonic/Tastebud-analyzer.- Run the code named Taste_GUI.m; a GUI window named NPL Bud Analyzer will pop up. Click on the New Analysis button on the upper-right corner, then load the converted image from step 5.1. Set the frame rate above the loaded image.
- Draw the region of interest (ROI) over the loaded image for registration. Double click on the selected ROI and an auto-calculated registration will start.
- Obtain the relative fluorescence intensity changes (ΔF/F)
- Go back to the NPL Bud Analyzer window to show the registered image from step 5.2 automatically. If the user already has a reg file, click on the Load Data button and select the _reg.tif file.
- Click on the CIRCLE or POLYGON button and position the ROI of taste cell over the taste bud image.
- This presents the raw fluorescence intensity and the calcium trace (ΔF/F) of the selected taste cell automatically under the taste bud image.
- Click on Save Trace to present the calcium trace (ΔF/F) on the right side of the GUI, while ROI is shown over the taste bud image. Repeat steps 5.3.2-5.3.4 until the ROI selection is finished.
NOTE: Click on the Delete Trace button if the ROI is mis-selected to eliminate the last selected ROI and calcium trace. - After the ROI selection is finished, write the file name on the bottom-right corner, and click on the Finish button to export the ΔF/F calcium trace as an .xls format, and the tase bud image with ROIs in a .bmp format.
- Analysis of the calcium trace
- Analyze the calcium trace obtained from step 5.3. Consider that taste cells have reacted to tastant when fluorescence intensity rises more than two standard deviations of the baseline after the tastant is delivered4, and p-values are less than 0.01, using paired or unpaired t-tests10.
- Consider the taste cell as a responder cell, if it responds to a certain tastant more than two times out of three trials(~60%)15.
- Obtain the representative calcium traces by averaging individual calcium traces acquired from step 5.4.1.
Representative Results
The Pirt-GCaMP6f-tdTomato mouse was used to obtain a taste bud image. The surface of the mouse tongue was covered with autofluorescent filiform papillae. Taste buds are spread sparsely over the surface of the tongue (Figure 4A). The images of the taste bud and its structure were acquired using three different filter detectors. Using the 607/70 nm filter set, the tdTomato signal from the taste cells was obtained for ratiometric analysis (Figure 4B). Using the 525/50 nm filter set, the GCaMP signal from the taste cells and blood vessels that surround the taste bud were acquired (Figure 4B). Using the 447/60 nm filter set, the collagen connective tissue, which structurally supports the taste bud, was acquired (Figure 4B).
After acquiring the images of the taste bud and relative structures, in vivo calcium imaging was carried out using the protocol. The Pirt-GCaMP6f-tdTomato mouse was used to screen on taste cells (Figure 5A)16. Taste cells responded repeatedly to their respective taste stimuli (Figure 5B). Taste cells were considered to have reacted to the tastant when they met the conditions presented in protocol 5.1.4. In this trial, cell 2 responded to both sweet and umami tastants. The result is consistent with previous research observing cellular activity using electrophysiology17. Cell 3 responded to both 400 mM NaCl and 400 mM NaCl under amiloride. It implicates that cell 3 have used an ENaC independent pathway for the response to salty taste. The taste bud in this experiment did not include a cell responding to sour tastes. The screening of taste cells was carried out under stable imaging conditions, and each taste cell showed a repeatable response to a distinct type of taste.
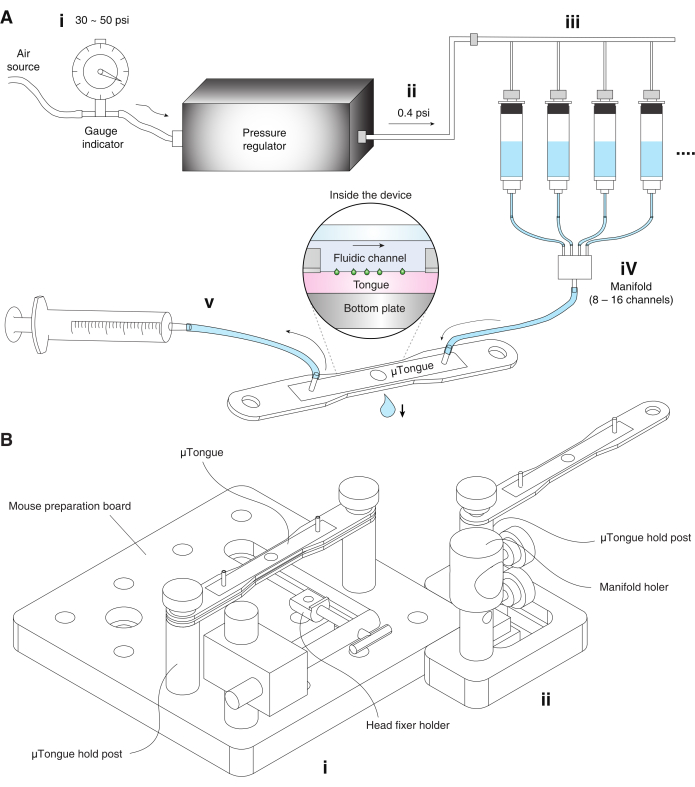
Figure 1: The µTongue, a microfluidics-based functional imaging platform. (A) Pressurized fluidic delivery system. (i) The pressure regulator of the fluidic system is connected to the external air source. The pressure of the air source is adjusted between 30-50 psi before entering the pressure regulator. (ii) Air pressure from the regulator is approximately 0.4 psi. (iii) Reservoirs containing artificial saliva and different taste solutions are connected to the output of the air pressure regulator. (iv) Each reservoir converges to a manifold that is connected to the input port of the µTongue. (v) A syringe pump is connected to the output of the µTongue and controls the flow. (B) The µTongue, a microfluidics-based functional imaging platform. The name of each part is specified in the figure. (i) Mouse preparation board. (ii) Fluidic system setup board. Please click here to view a larger version of this figure.
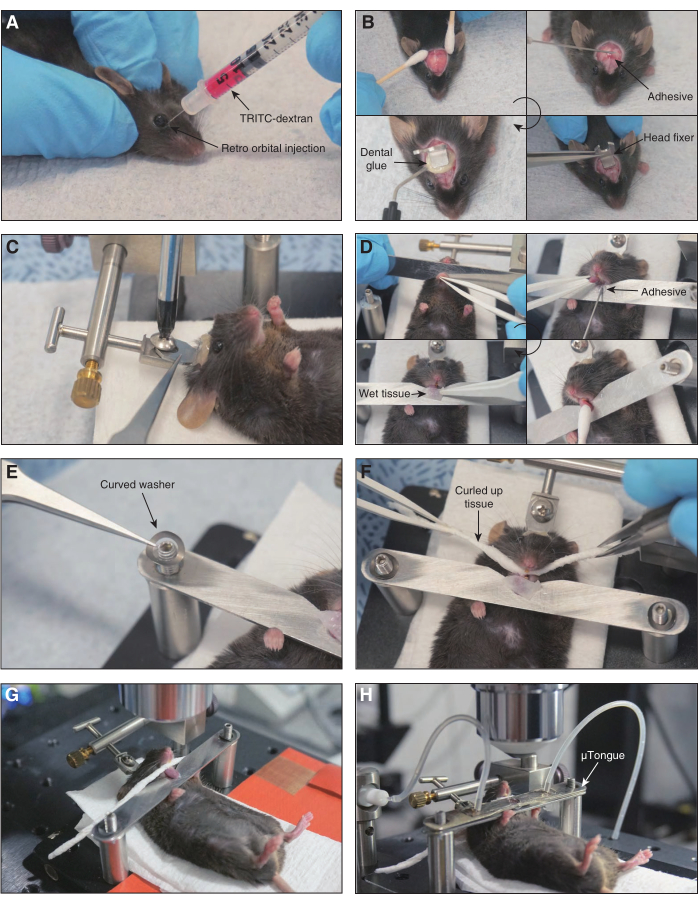
Figure 2: Sequential description of mouse preparation. Important steps in the mouse preparation are shown. (A) Retro-orbital injection of TRITC-dextran. (B) Process of attachment of a head fixer to the mouse skull is shown. The head skin and periosteum are cleared. Adhesive glue and dental glue are used for attachment. (C) The head fixer on the mouse skull is screwed onto the mouse preparation board. (D) Procedure of mounting a tongue on the bottom unit of the µTongue. An instant adhesive is used for tongue fixation. The tongue is cleaned using a wet cotton swab and covered with wet paper tissues to prevent dryness. (E) Curved washers are applied to both ends of the bottom unit of the µTongue. (F) A piece of twisted paper tissue is placed in the mouse oral cavity. (G) Mouse preparation board is mounted on the microscope stage and screwed tightly to ensure stable imaging conditions. (H) The µTongue is placed on the tongue. An objective lens is adjusted over the imaging window. Please click here to view a larger version of this figure.
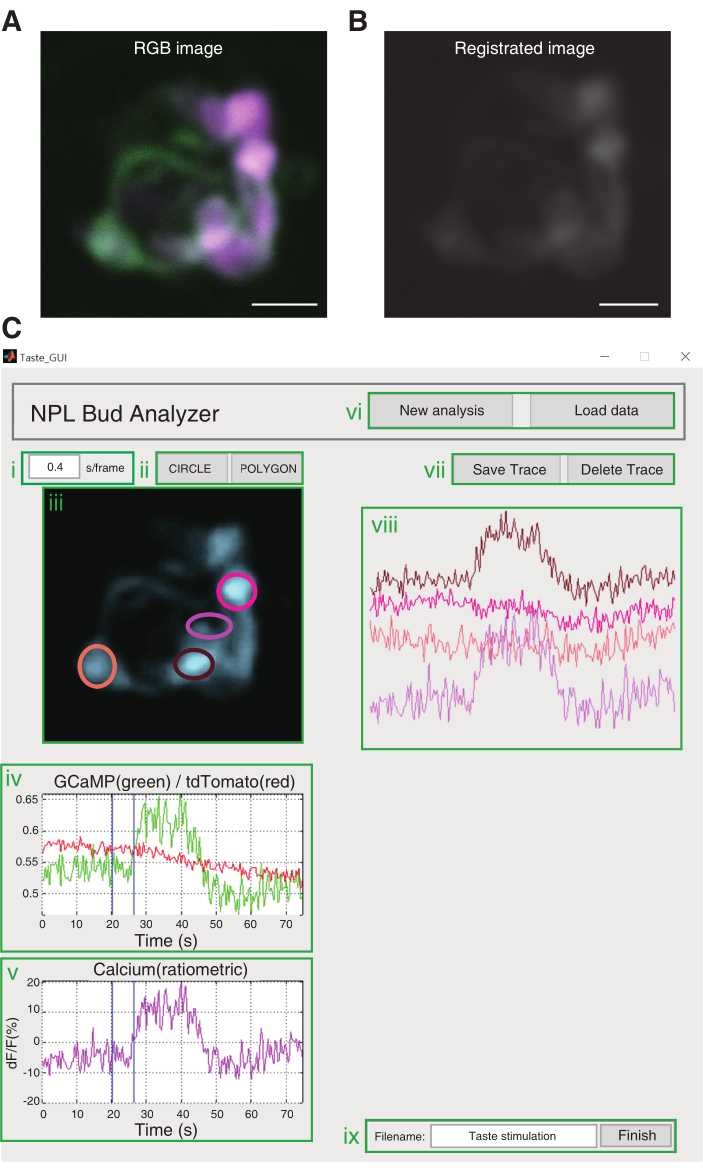
Figure 3: Image analysis. (A) An RGB image is converted from each single-color image. Scale bar, 10 µm. (B) Image registration using a conducted custom code. (C) GUI of the custom code. (i) Input location for the frame rate. The default frame rate is 0.16 s/frame. (ii) Buttons to draw ROIs. (iii) The area in which the loaded image is shown. (iv) The calcium signal of the ROI selected is shown as a green trace, whereas the calcium-insensitive signal of the ROI selected is shown as a red trace. (v) Ratiometric analysis and ΔF/F are calculated automatically. The ΔF/F graph is presented in magenta. (vi) Buttons for image loading. New Analysis is for loading an RGB converted image. Load Data is for loading an image that has already undergone registration. (vii) The Save Trace button is to keep the ΔF/F graph and the ROI selected at viii. The Delete Trace button is to remove ΔF/F graph from viii. (viii) Saved calcium traces are shown. (ix) Area to fill in the file name. The Finish button is to extract data and save them in the same directory of the code. Please click here to view a larger version of this figure.
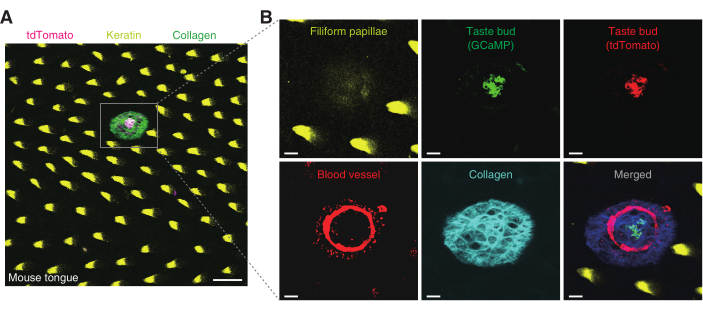
Figure 4: The surface of the mouse tongue and a taste bud in the fungiform papillae. (A) The surface of the mouse tongue is captured in a large field. A taste bud keratinized filiform papillae, and the collagen structure are shown. Each structure is indicated using different colors: magenta, yellow, and green, respectively. Scale bar, 100 µm. (B) A taste bud from (A) is magnified in and captured using three different emission filter detectors. The filiform papillae, in yellow, are captured using a 525/50 nm detector. This structure is observed from the very surface of the tongue up to ~25 µm in depth. GCaMP signals in green and tdTomato signals in red represent the taste cells. These signals are detected by 525/50 and 607/70 nm detectors, respectively. Rhodamine dextran representing blood circulation is captured at both 525/50 and 607/70 nm detectors. The collagen structure shown in cyan blue is acquired by 447/60 nm detectors. The last picture shows all the previous images merged. Scale bar, 20 µm. Please click here to view a larger version of this figure.
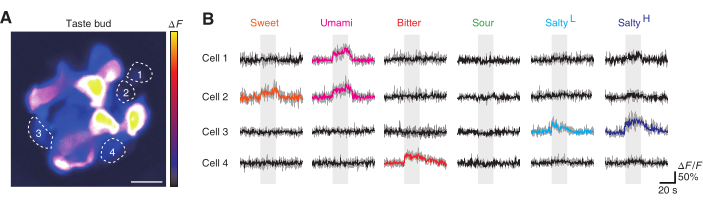
Figure 5: Taste screeningof a Pirt-GCaMP6f-tdTomato mouse in vivo. (A) A representative taste bud of the Pirt-GCaMP6f-tdTomato mouse. The image is shown as an intensity-based pseudo-color. Dashed lines demarcate each taste cell. The brightness of each taste cell depends on the expression of the fluorescent protein and the depth of the taste cell location. Scale bar, 10 µm. (B) The calcium trace of each taste cell for the five basic taste stimuli. Every repeated trial is shown in gray on the back and the averaged trace is presented above each trial. Colored traces are defined as responsive whereas black traces are defined as non-responsive. Each color represents a different taste. Salty(L) represents low salty, with a mixture of 400 mM NaCl and 50 µM amiloride used for taste stimulation. Salty(H) represents high salty, with 400 mM NaCl used for stimulation. Taste stimulation is shown as a gray box on the back of each calcium trace. Please click here to view a larger version of this figure.
Discussion
Described here is a detailed protocol to apply µTongue to the investigation of functional activities of taste cells in vivo. In this protocol, the functional imaging on the taste cells using genetically encoded calcium indicators is performed. In addition to the use of transgenic mice, the electrophoretic loading of calcium dyes (or voltage sensing dyes) onto the taste cells can be an alternative option.
All the taste solutions less than 1.336 of refractive index were used in this experiment. Although µTongue provides a stable fluidic delivery and the ratiometric analysis ameliorates imaging artifacts, it will be challenging for the researchers to use a higher concentration of tastant (e.g., >100 mM sucrose with refractive index in 1.338). The large difference in refractive index between artificial saliva and taste solution shifts the image focal plane more than the compensation range by the post-image process. Empirically, a certain range of refractive index of taste solution (less than 1.336) that allows stable cellular imaging in real-time is obtained.
For researchers experienced in fluorescence imaging and animal handling, this protocol can be learned readily over repeated practice. However, it contains critical steps, which often impede successful data acquisition. First, once externalized from the oral civility, the tongue should be kept moist with artificial saliva to preserve the natural mucosal microenvironment. Second, blood circulation around the taste bud should be intact, to maintain a physiologic supply of oxygen, nutrients, and blood-borne factors.
Divulgaciones
The authors have nothing to disclose.
Acknowledgements
This work was supported by the Institute of Basic Science (IBS-R015-D1), the National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIT) (No. 2019M3A9E2061789), and by the National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIT) (No. 2019M3E5D2A01058329). We are grateful to Eunsoo Kim and Eugene Lee for their technical assistance.
Materials
| acesulfame K | Sigma Aldrich | 04054-25G | Artificial saliva / tastant |
| calcium chloride solution | Sigma Aldrich | 21115-100ML | Artificial saliva / tastant |
| citric acid | Sigma Aldrich | C0759-100G | Artificial saliva / tastant |
| cycloheximide | Sigma Aldrich | 01810-5G | Artificial saliva / tastant |
| denatonium | Sigma Aldrich | D5765-5G | Artificial saliva / tastant |
| Dental glue | Denkist | P0000CJT-A2 | Animal preparation |
| Image J | NIH | ImageJ | Data analysis |
| IMP | Sigma Aldrich | 57510-5G | Artificial saliva / tastant |
| Instant adhesive | Loctite | Loctite 4161, Henkel | Animal preparation |
| K2HPO4 | Sigma Aldrich | P3786-100G | Artificial saliva / tastant |
| KCl | Sigma Aldrich | P9541-500G | Artificial saliva / tastant |
| Ketamine | Yuhan | Ketamine 50 | Animal preparation |
| KH2PO4 | Sigma Aldrich | P0662-25G | Artificial saliva / tastant |
| KHCO3 | Sigma Aldrich | 237205-500G | Artificial saliva / tastant |
| MATLAB | Mathwork | MATLAB | Data analysis |
| MgCl2 | Sigma Aldrich | M8266-100G | Artificial saliva / tastant |
| MPG | Sigma Aldrich | 49601-100G | Artificial saliva / tastant |
| Mutiphoton microscope | Thorlab | Bergamo II | Microscope |
| NaCl | Sigma Aldrich | S3014-500G | Artificial saliva / tastant |
| NaHCO3 | Sigma Aldrich | 792519-500G | Artificial saliva / tastant |
| Objective | Nikon | N16XLWD-PF | Microscope |
| Octaflow | ALA Scientific Instruments | OCTAFLOW II | Fluidic control |
| PC | LG | Lg15N54 | Fluidic control |
| PH meter | Thermoscientific | ORION STAR AZ11 | Artificial saliva / tastant |
| Phosphate-buffered saline | Sigma Aldrich | 806562 | Artificial saliva / tastant |
| quinine | Sigma Aldrich | Q1125-5G | Artificial saliva / tastant |
| Syringe pump | Havard Apparatus | PHD ULTRA 4400 | Fluidic control |
| TRITC-dextran | Sigma Aldrich | 52194-1G | Animal preparation |
| Ultrafast fiber laser | Toptica | FFultra920 01042 | Microscope |
| Xylazine | Bayer Korea | Rompun | Animal preparation |
Referencias
- Mao, T., O’Connor, D. H., Scheuss, V., Nakai, J., Svoboda, K. Characterization and subcellular targeting of GCaMP-type genetically-encoded calcium indicators. PLoS One. 3 (3), 1-10 (2008).
- Shih, A. Y., et al. Two-photon microscopy as a tool to study blood flow and neurovascular coupling in the rodent brain. Journal of Cerebral Blood Flow & Metabolism. 32 (7), 1277-1309 (2012).
- Choi, M., Kwok, S. J. J., Yun, S. H. In vivo fluorescence microscopy: Lessons from observing cell behavior in their native environment. Physiology. 30 (1), 40-49 (2015).
- Caicedo, A., Samir Jafri, M., Roper, S. D. In situ Ca2+ imaging reveals neurotransmitter receptors for glutamate in taste receptor cells. Journal of Neuroscience. 20 (21), 7978-7985 (2000).
- Tomchik, S. M., Berg, S., Kim, J. W., Chaudhari, N., Roper, S. D. Breadth of tuning and taste coding in mammalian taste buds. Journal of Neuroscience. 27 (40), 10840-10848 (2007).
- Dando, R., Roper, S. D. Cell-to-cell communication in intact taste buds through ATP signalling from pannexin 1 gap junction hemichannels. The Journal of Physiology. 587 (24), 5899-5906 (2009).
- Chandrashekar, J., et al. The cells and peripheral representation of sodium taste in mice. Nature. 464 (7286), 297-301 (2010).
- Oka, Y., Butnaru, M., von Buchholtz, L., Ryba, N. J. P., Zuker, C. S. High salt recruits aversive taste pathways. Nature. 494 (7438), 472-475 (2013).
- Choi, M., Lee, W. M., Yun, S. H. Intravital microscopic interrogation of peripheral taste sensation. Scientific Reports. 5 (8661), 1-6 (2015).
- Han, J., Choi, M. Comprehensive functional screening of taste sensation in vivo. bioRxiv. 16419 (371682), 1-22 (2018).
- Thestrup, T., et al. Optimized ratiometric calcium sensors for functional in vivo imaging of neurons and T lymphocytes. Nature Methods. 11 (2), 175-182 (2014).
- Danilova, V. Glossopharyngeal nerves to taste stimuli in C57BL / 6J mice. BME Neuroscience. 15, 1-15 (2003).
- Wu, A., Dvoryanchikov, G., Pereira, E., Chaudhari, N., Roper, S. D. Breadth of tuning in taste afferent neurons varies with stimulus strength. Nature Communications. 6 (8171), 1-11 (2015).
- Schindelin, J., et al. Fiji: An open-source platform for biological-image analysis. Nature Methods. 9 (7), 676-682 (2012).
- Tan, H. E., et al. The gut-brain axis mediates sugar preference. Nature. 580 (7804), 511-516 (2020).
- Roebber, J. K., Roper, S. D., Chaudhari, N. The role of the anion in salt (NaCl) detection by mouse taste buds. The Journal of Neuroscience. 39 (32), 6224-6232 (2019).
- Kusuhara, Y., et al. Taste responses in mice lacking taste receptor subunit T1R1. Journal of Physiology. 591 (7), 1967-1985 (2013).

