Genetic Manipulation of the Plant Pathogen Ustilago maydis to Study Fungal Biology and Plant Microbe Interactions
Summary
We describe a robust gene replacement strategy to genetically manipulate the smut fungus Ustilago maydis. This protocol explains how to generate deletion mutants to investigate infection phenotypes. It can be extended to modify genes in any desired way, e.g., by adding a sequence encoding a fluorescent protein tag.
Abstract
Gene deletion plays an important role in the analysis of gene function. One of the most efficient methods to disrupt genes in a targeted manner is the replacement of the entire gene with a selectable marker via homologous recombination. During homologous recombination, exchange of DNA takes place between sequences with high similarity. Therefore, linear genomic sequences flanking a target gene can be used to specifically direct a selectable marker to the desired integration site. Blunt ends of the deletion construct activate the cell's DNA repair systems and thereby promote integration of the construct either via homologous recombination or by non-homologous-end-joining. In organisms with efficient homologous recombination, the rate of successful gene deletion can reach more than 50% making this strategy a valuable gene disruption system. The smut fungus Ustilago maydis is a eukaryotic model microorganism showing such efficient homologous recombination. Out of its about 6,900 genes, many have been functionally characterized with the help of deletion mutants, and repeated failure of gene replacement attempts points at essential function of the gene. Subsequent characterization of the gene function by tagging with fluorescent markers or mutations of predicted domains also relies on DNA exchange via homologous recombination. Here, we present the U. maydis strain generation strategy in detail using the simplest example, the gene deletion.
Introduction
Ustilago maydis is a phytopathogenic model fungus that has been studied extensively for decades 1,2. It exists in two morphologies, a yeast-like, non-pathogenic stage and a filamentous, infectious form 3. Universal breakthrough discoveries such as homologous recombination and DNA repair mechanisms were made in the yeast-like growth stage of this fungus 4. Furthermore, the morphological switch to the infectious filament and virulence factors important for infection are well-characterized 5,6. The increasing molecular knowledge about biology and virulence of this smut fungus relies on a straightforward gene replacement strategy 7-9 supported by an excellent genome annotation 10 and the ease of reverse genetics using, e.g., the well-organized plasmid collection at our institute (http://www.mikrobiologie.hhu.de/ustilago-community.html). Standardized, rapid infection assays in maize seedlings allow detailed studies of pathogenicity factors 11.
The genome of U. maydis contains about 6,900 genes 10. To study their function, they can be deleted individually or in combination due to an efficient homologous recombination system. Flanking regions of about 1 kb containing perfectly homologous ends are ideal for rates of homologous recombination greater than 50%, but already 250 bp with non-homologous ends allow some degree of correct integration of the construct 9. Currently, five different resistance cassettes, hygR, cbxR, natR, G418R, and phleoR mediating resistance against hygromycin, carboxin, nourseothricin, G418 and phleomycin, are employed to select for transformants 7,9. In addition, the hygromycin resistance has been developed into a recyclable cassette (FRT-hygR) that can be removed by the transient expression of a heterologous FLP recombinase 12. This allows removal of the resistance cassette and thereby in theory unlimited genetic modifications. Phleomycin is mutagenic 13, so that with the new cassettes, in particular the recyclable hygR cassette, the use of phleoR is decreasing. Quadruple mutants can thus be generated using the other four cassettes, but for quintuple mutants, the FRT-hygR system is recommended 14.
This general gene deletion strategy has been successfully transferred to other smut fungi such as Sporisorium reilianum 15, U. hordei 16, or U. esculenta 17 and therefore offers the potential for further applications in yet genetically intractable organisms with an efficient homologous recombination system. Moreover, organisms lacking homologous recombination can be modified to improve genetic engineering as exemplified by the deletion of genes involved in non-homologous-end-joining in Neurospora crassa 18,19.
Here we describe the published gene deletion strategy for U. maydis 7,9 in experimental detail with a focus on the rapid and accurate verification of the candidates. As an example, we use the fungal chitinases and depict the generation of single mutants as well as multiple deletion strains 20,21. Chitinases are interesting examples, because they act on chitin in the rigid cell wall. Cell wall remodeling is required for morphological changes during cell division, switch to filamentous growth, and spore formation. Hence, in deletion mutants phenotypes throughout the lifecycle can be expected.
Protocol
1. Generation of Deletion Constructs
- Generate a plasmid containing the deletion construct (Figure 1) comprising a respective 1 kb upstream flank (UF) and downstream flank (DF) each and the appropriate resistance cassette flanked by blunt cutting restriction sites.
NOTE: Any cloning strategy can be used (for cloning see reference 22) we recommend Golden Gate cloning for such plasmids 9. - Excise the deletion construct from the plasmid using a blunt cutter to obtain 1 µg DNA of the deletion construct 9. Purify the deletion construct to eliminate enzyme and buffer 22 e.g. by using commercial reagents and follow the manufacturer's instructions.
NOTE: The vector backbone can remain in the transformation mixture.
2. Preparation of Protoplasts
NOTE: Keep sterile conditions during all times of the experiment. The cell wall is a strong protective barrier that limits access of molecules to the plasma membrane. To allow uptake of the DNA containing the deletion construct, the cell wall needs to be removed by cell wall degrading enzymes in a protoplasting reaction. Critical steps in protoplast preparation are 1) osmotic stabilization of the medium and 2) avoiding mechanical stress on the protoplasts.
- Mark 2 ml tubes with a round bottom to freeze the protoplasts and cool them down at -20 °C. Start a pre-culture in 3 ml YEPS-Light from a fresh plate of the strain with the desired genetic background. Incubate on a rotating wheel for 24 hr at 28 °C.
NOTE: The strain can be the solopathogenic strain SG200 10, wildtype strains, tester strains such as AB33 23, or any mutant background for generation of double mutants. Make sure the strain is free of the desired resistance. - Dilute the culture in 50 ml YEPS-Light in a baffled flask and incubate on an orbital shaker at 28 °C with 200 rpm.
NOTE: The doubling time is about 2 hr for wildtype strains. Allow three duplications minimum. - Grow until exponential phase. Ensure that the optical density, measured at a wavelength of 600 nm (OD600), is around 0.8 (0.6 to 1.0 is acceptable). An OD600 of 1.0 corresponds to about 1 to 2 x 107U. maydis cells per ml.
- Check the cells for contamination under a microscope. Use 40x magnification. Pellet the cells of the complete 50 ml culture for 5 min, 1,500 x g and discard the supernatant. Resuspend the pellet in 25 ml sodium citrate, sorbitol solution (SCS), centrifuge 5 min, 1,500 x g, and discard supernatant.
NOTE: Bacterial contaminations can be identified as small and often moving cells. Fungal contaminations can be identified based on a different cell shape or size compared to the cigar shaped cells of U. maydis. - During centrifugation prepare protoplasting solution (12.5 mg/ml Trichoderma lysing enzymes, in SCS; prepare 3 ml per cell pellet; filter sterilize through a 22 µm filter; solution has to be fresh for optimal enzymatic activity). SCS is an osmotic stabilizer to prevent rupture of protoplast upon removal of the cell wall.
- Resuspend the pellet in 2 ml protoplasting solution. Incubate for 5 to 20 min at RT and check under the microscope until 30 to 40% of the cells are round or like pinheads (Figure 2).
NOTE: Perform all steps on ice from now on and handle protoplasts with care! - Wash 3x in 10 ml cold (4 °C) SCS, centrifuge at 1,000 x g for 5 min.
- Resuspend the pellet in 10 ml cold sorbitol, TrisHCl, CaCl2 solution (STC), spin at 1,000 x g for 5 min, discard supernatant. Resuspend the pellet in 1 ml cold STC, make 100 µl aliquots in the cooled tubes. Freeze the aliquots at -80 °C until further use.
3. Transformation of U. maydis
NOTE: The transformation of U. maydis protoplasts relies on the polyethylene glycol (PEG)-mediated transformation method, which is technically simple and opposed to electroporation or biolistic transformation does not require specialized equipment.
- Prepare the two-layered selection plates (2 plates per transformation reaction). The bottom layer contains the antibiotic in a doubled concentration. Pipet 12 ml of RegLight containing 400 µg/ml hygromycin (or 300 µg/ ml nourseothricin or 4 µg/ml carboxin or 1 mg/ml G418) into a Petri dish. Avoid bubbles.
NOTE: Bottom layer plates may be stored a couple of days at 4 °C, but prepare the top layer freshly during transformation (see below). - Thaw protoplasts on ice (1 tube/transformation). Add 1 µl heparin (15 mg/ml). Add 1 µg DNA (linearized construct) or 10 µl H2O (water control). Incubate for 10 min on ice.
- During this time, for the upper layer spread 12 ml liquefied RegLight (boil and cool to 60 °C) onto the RegLight bottom plate.
NOTE: This layer does not contain antibiotics. - Add 500 µl STC/PEG (polyethylene glycol) to the transformation tube (3. 2) and mix carefully by inverting the tube. Incubate 15 min on ice.
- Distribute each transformation reaction on two of the two-layered RegLight plates. Spread carefully and slowly with a glass pipet. Incubate plates upright at 28 °C for 5 to 7 days until transformants grow as colonies. Obtain about 100-200 colonies per plate.
NOTE: Lower numbers might be caused by "bad" protoplasts that either contain too much cell wall and therefore cannot take up DNA or cannot regenerate. The former can be tested by transformation with a self-replicating plasmid, the latter by testing regeneration on plates without antibiotics. - Prepare YEPS-Light plates containing the appropriate antibiotic at 200 µg/ml hygromycin (or 150 µg/ ml nourseothricin or 2 µg/ml carboxin or 500 µg/ml G418; these concentrations equal half of the one used for bottom plates). Re-purify at least 24 putative transformant colonies on these plates. Single colonies should be obtained. At the same time streak out the original strain on non-selective media.
NOTE: Plates can be stored at 4 °C for 1-2 weeks.
4. Verification of Correct Deletion Events
NOTE: The mutations are verified in a three-step procedure (Figure 1). First, candidates that contain the wildtype copy of the gene are excluded by PCR. Second, the integration of the resistance cassette into the locus is verified by PCR. Third, the integration of the resistance cassette only into the desired locus is verified by Southern blot analysis. Careful strain verification is essential, so that mutant phenotypes truly correlate with the deletion.
- First Diagnostic PCR 20
- Design primers pairing in the gene to be deleted that result in a product of 250-600 bp (Figure 1). Such small products are easy to amplify in a colony PCR, and long enough for detection in standard gel electrophoresis.
- For each candidate and the original strain as a positive control, resuspend a little bit (amount of a pinhead) of cell material of a single colony with a toothpick in 20 µl 0.02 M NaOH. Incubate at RT for 30 min.
NOTE: Use fresh colonies. If the plates are older than 1 week re-streak to obtain a fresh colony. - Use 1 µl of the cell suspension for the PCR immediately. Run a standard PCR reaction and analyze resulting fragments.
NOTE: These samples cannot be stored.
NOTE: This PCR tests for presence of the wildtype gene and thereby allows rapid identification of negative candidates in which the gene of interest has not been replaced. Clones yielding bands in this PCR reaction will be discarded. One such negative clone should anyway be carried along as an additional negative control. Candidates without a PCR product have to be analyzed further. For genes without detrimental effects approximately half of the candidates should contain the wildtype gene, while the other half should not result in a band. This would correspond to a homologous recombination rate of 50%.
- Second Diagnostic PCR
- Preparation of genomic DNA.
NOTE: Genomic DNA (gDNA) can be prepared using two different protocols. The first involves toxic reagents like phenol and chloroform and hence, should be handled only by experienced researchers (described in 4.2.1.1 to 4.2.1.5) while the second can also be handled by less experienced students (described in 4.2.1.6 to 4.2.1.11). Both protocols are working reliably. Step 4.2.1.1 is identical for both protocols.- Inoculate 5 ml YEPS-Light with each candidate and incubate on a rotating wheel for 24 hr at 28 °C. Drop 2 µl of the culture on a CM plate. Keep the culture sterile.
NOTE: Following is a standard protocol (CAUTION: toxic steps). - Put 1 scoop (~200 µl) of HCl-washed glass beads to 2 ml tubes. Add 2 ml of the U. maydis culture. Spin 12,000 x g for 5 min. Discard supernatant (pellet can be frozen at this stage). Add 500 µl Phenol/Chloroform/Isoamylalcohol (25:24:1) to the pellet. CAUTION! PHENOL IS TOXIC!
- Add 500 µl Usti-lysis-buffer 1. Shake for 6 to 10 min on a vibrax at 1,000 rpm. Verify that the cell pellet is completely resuspended, but avoid extended shaking to prevent shearing of the gDNA. Spin at 12,000 x g for 15 min. Transfer 400 µl of the aqueous phase into the new reaction tube (do not touch the interphase).
- Add 1 ml 100 % (v/v) ethanol. Invert the tube 3x to mix, observe the cloud of DNA that shortly appears. Spin at 12,000 x g for 5 min. Remove supernatant and wash with 200 µl 70% EtOH. Do not resuspend the pellet in this stage.
- Remove supernatant completely (do not let the pellet dry completely). Add 50 µl TE/RNase. Dissolve gDNA by shaking at 400 rpm at 50°C for 10 min. Analyze 2 µl of the gDNA on a 0.8 % agarose gel 20.
NOTE: The steps below comprise an alternative non-toxic protocol.
- Inoculate 5 ml YEPS-Light with each candidate and incubate on a rotating wheel for 24 hr at 28 °C. Drop 2 µl of the culture on a CM plate. Keep the culture sterile.
- Alternate preparation of genomic DNA
- Put 1 scoop (~200 µl) of HCl washed glass beads to 2 ml tubes. Add 2 ml of the U. maydis culture and spin at 12,000 x g for 5 min. Discard supernatant (pellets can be frozen at this stage).
- Add 500 µl Usti Lysis buffer 2 to the pellet. Shake for 5 to 15 min on a vibrax at 1,000 rpm. Verify that the cell pellet is completely resuspended.
- Incubate the tubes at 65 °C for 15 to 20 min. Using appropriate incubators designed to host 2 ml tubes is critical. Then, place it on ice for 5 min.
- Add 100 µl 8 M potassium acetate, vortex or invert 8 to 10 times. Spin 12,000 x g for 15 min at RT.
- Transfer 500 µl of the supernatant to a fresh 1.5 ml tube and add 400 µl isopropanol. Vortex or invert 8 to 10 times. Spin 12,000 x g for 15 min. Remove supernatant and wash with 500 µl 70% EtOH. Spin 12,000 x g for 5 min.
- Soak off supernatant completely! Eventually, add a short spin (10 sec) to remove residual liquid. Let the DNA pellet dry for 3 to 5 min. Add 50 µl TE/RNAase and incubate at 50 °C for 10 to 15 min at 400 rpm.
- PCR
- Design two primer pairs with one of them binding outside the flanks and the corresponding one binding within the resistance cassette (UF-f, RC-r; RC-f, DF-r, Figure 1) 9.
- Use 1 µl of a 1:10 dilution of the gDNA of every candidate as a template in 2 PCR reactions verifying the correct insertion upstream and downstream 9.
- Use standard PCR reactions for each candidate and analyze resulting fragments 22.
NOTE: These PCRs test for the insertion of the resistance cassette in the correct genomic locus. If products are obtained for both sides, the candidate is likely a correct insertion. However, also candidates where only one side (upstream or downstream) could be confirmed should be carried along for further analysis in case that PCR reactions simply failed for one flank.
- Preparation of genomic DNA.
- Verification of Correct Genomic Insertion by Southern Blot Analysis 22
- Digest 15 µl of gDNA (4.2.1) of the mutant candidates and the corresponding wildtype/progenitor strain with an appropriate enzyme that cuts outside the locus/construct and in addition, cuts either in the gene or in the resistance cassette leading to clearly distinguishable patterns (Figure 1) 8.
NOTE: None of the expected bands should be larger than 10 kb, so that they are clearly distinguishable from undigested DNA. - Use the upstream and the downstream flank as probes. To label them for example use the PCR DIG labelling Kit or any similar standard method. Mix the labelled probes in a 1:1 molecular ratio and carry out the Southern blot. Keep at least two independent correct transformants.
NOTE: The wildtype pattern should be clearly detectable in the original strain and the correct clones should contain only the expected bands. Additional bands only present in the candidates indicate other insertions of the construct in a wrong locus. Such clones should be discarded. The negative candidate identified in the first diagnostic PCR should contain the wildtype bands and the bands (unpredictable in size) of the wrongly integrated deletion construct.
- Digest 15 µl of gDNA (4.2.1) of the mutant candidates and the corresponding wildtype/progenitor strain with an appropriate enzyme that cuts outside the locus/construct and in addition, cuts either in the gene or in the resistance cassette leading to clearly distinguishable patterns (Figure 1) 8.
- Glycerol Stocks
NOTE: Glycerol stocks serve to maintain the clones for years in strain collections. At least two independent transformants should be kept for each mutant. This allows comparing phenotypes that should be identical.- Grow 5 ml cultures of the confirmed mutant strains in YEPS-Light on a rotating wheel for 24 hr at 28 °C Mix 500 µl of each culture with 500 µl NSY-Glycerol-medium. Invert tube until culture and medium are well mixed. Glycerol in the medium prevents the formation of ice crystals.
- Freeze at -80 °C. Re-streak a part of the frozen culture to test if the stock is functional.
5. Microscopic Phenotypes
- Grow 5 ml cultures of wildtype and mutant strains in YEPS-Light on a rotating wheel for 12 hr at 28 °C.
- Next day, dilute the culture to an OD600 of about 0.1 and grow it till an OD600 between 0.5 and 0.7. An OD600 of 1.0 corresponds to about 1 to 2 x 107 U. maydis cells per ml.
- Inspect the cell morphology microscopically.
NOTE: Healthy cells appear cigar shaped (Figure 4, WT). Pronounced vacuoles or filament formation indicate that the cells are stressed.
NOTE: Depending on the expected mutant phenotype, the analysis can be adapted, e.g., if reporter strains are used the appropriate assay can be carried out at this stage. If cell morphology is changed statistical analysis should be conducted, e.g., to determine the mean cell length.
6. Infection Assay
NOTE: The seedling infection assay has been visualized previously in this journal 11. It can either be carried out using a haploid, solopathogenic strain background such as SG200 10 or by mixing compatible mating partners which both carry the mutation.
- Grow at least 40 maize seedlings per strain for 7 days at a light cycle of 16 hr, 200 µE, 28 °C/22 °C (day/night). They should be at the 3-leaf stage and 10-15 cm tall at the day of infection.
- Two days before infection, start a pre-culture of the strains in 5 ml YEPS-Light on a rotating wheel for 24 hr at 28 °C. Grow a main culture in 50 ml YEPS-Light up to OD600 = 1.0 (cell division rate = 2 hr, allow 3 divisions minimum, can be done O/N).
- Spin down the cells of 50 ml culture at 1,500 x g for 5 min and discard the supernatant. Wash 3x with sterile H2O. Resuspend the cells in sterile H2O to an OD600 of 3.0. If necessary, mix the mating partners.
- Inject 250 – 500 µl of the cell suspensions into the stem of 7 day old maize seedlings using a small syringe. Use sterile H2O as a control. Keep infected seedlings for an additional 7-14 days in the same light and temperature conditions. Score the phenotype of each plant according to healthy, chlorosis, anthocyanin formation, small tumors, large tumors, dead plants 10.
NOTE: Scoring can be done as early as 7 days and repeated at 14 days to follow infection over time.
Representative Results
The deletion constructs for all four chitinolytic genes encoded in the U. maydis genome were generated by Golden Gate cloning using the hygR cassette for deletion of cts1, the natR for deletion of cts2, the G418R cassette for deletion of cts3 and the cbxR for deletion of cts4 20. A general overview of the gene replacement strategy is exemplified by the deletion of cts3 (Figure 1). Single and double mutants of cts1 with a second chitinolytic gene were generated sequentially with the same constructs in two different genetic backgrounds for analysis of the role of chitinases in virulence. Two different strain backgrounds were employed: AB33 allows induction of filamentous growth, the first step towards pathogenicity 23, SG200 is a solo-pathogenic strain that enables rapid disease scoring 10. After transformation of the cts3-deletion construct into the appropriate strain background, single colonies of the second selection plate were tested for the correct deletion by diagnostic PCRs (Table 1) and Southern blot (Figure 3). Interestingly, for cts3 the homologous recombination rate was much higher in SG200 than in AB33, but this difference was not consistently observed in deletions of the other chitinases.
The stringent strain verification ensures a single insertion of the deletion construct at the desired position. However, additional modification such as point mutations can remain undetected, which might mislead the interpretation of phenotypes. Therefore, at least two independent transformants are phenotyped.
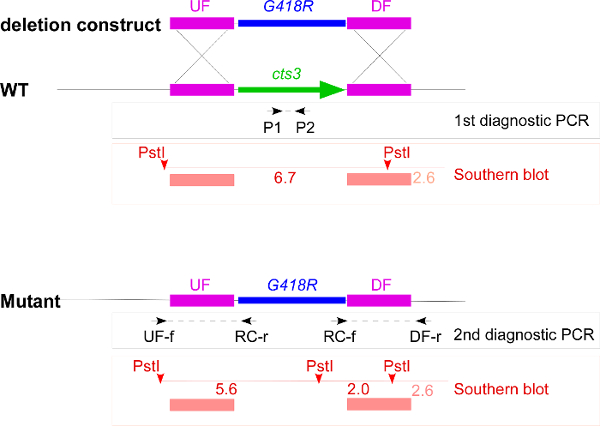
Figure 1. Overview of the deletion strategy exemplified by cts3. This schematic overview includes the deletion construct with upstream flank (UF), downstream flank (DF), and the geneticin resistance cassette (G418R), the genomic locus of wildtype (WT) and cts3 deletion mutant and all primers as well as the restriction sites used in the strain verification. The first diagnostic PCR results in a product only if the wildtype gene copy is present, the two second diagnostic PCRs result in products if the resistance cassette has been correctly integrated. For the Southern blot the genomic DNA is digested with PstI, the probes span the UF and DF (red bars) leading to detection of a 6.7 kb band in wildtype and two bands of 5.6 kb and 2.0 kb in the mutant. In addition, the DF probe weakly recognizes a 2.6 kb product. Please click here to view a larger version of this figure.
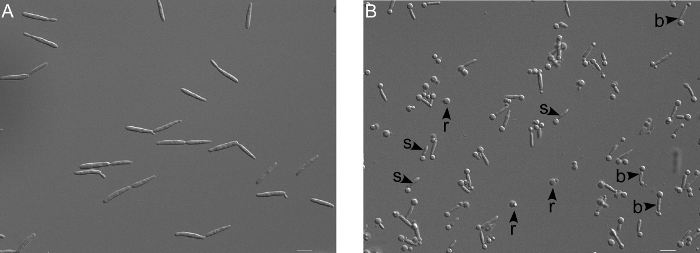
Figure 2. Microscopic verification of protoplasting reaction. (A) Untreated wildtype cells (FB2) appear cigar shaped. (B) Upon treatment with cell wall degrading enzymes (here for for 30 min), the appearance of round spheres at one end (s) and bar-bell like structures (b) indicates that the cell wall degradation has started. Finally the protoplasts are completely round (r), which is due to the lack of cell wall. Scale bar = 10 µm. Please click here to view a larger version of this figure.
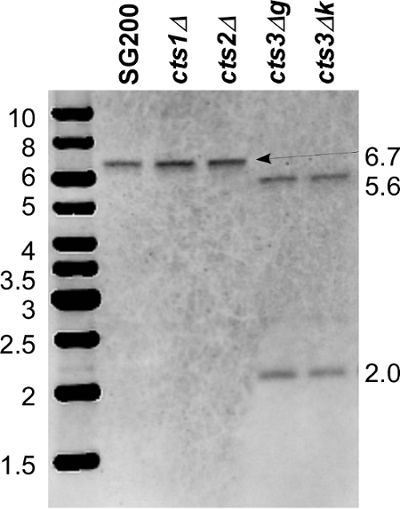
Figure 3. Southern blot for cts3 deletion. Genomic DNA was digested with PstI. Both flanks were labelled, mixed in equimolar ratios and used as a single probe. In the wildtype SG200 and deletions of the chitinases cts1 and cts2, a single band of 6.7 kb is detected. In cts3 deletions (g and k), two bands of 5.6 kb and 2.0 kb are visible indicative of the correct deletion of cts3. An additional band of 2.6 kb can be visualized by massive overexposure. This corresponds to a fragment that is recognized by an overlap of only 100 bp of the downstream flank probe. Please click here to view a larger version of this figure.
Some mutations lead to obvious growth defects that can be detected microscopically. In the chitinase deletions, single mutants did not display any obvious defect, while cts1/2 double mutants showed a pronounced cell separation defect 20. Staining of septa showed that cytokinesis was completed but the cells remained connected likely via residual chitin (Figure 4 from Langner et al. 2015). A similar microscopic growth phenotype is known for the deletion of khd4 24, a gene encoding an RNA-binding protein mediating post-transcriptional RNA turnover of genes involved in morphology and pathogenicity 25.
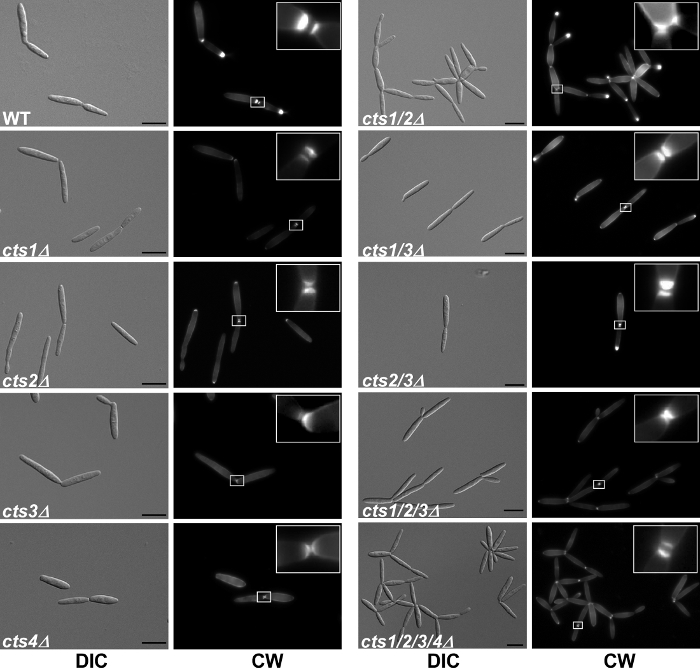
Figure 4. Microscopic analysis of deletion mutants. Cell morphology and septum formation of the chitinase deletion strains. cts1/2Δ strains exhibit a cell separation defect and form large aggregates, which is not due to a lack of septum formation. Primary and secondary septa (see 5x enlargement in insets) were stained with Calcofluor White (CW) prior to microscopy. Scale bars = 10 µm. This figure is reprinted with permission from Langner et al. 2015 Eukaryotic Cell 20. Please click here to view a larger version of this figure.
To test the contribution of chitinases to virulence, seedling infection assays were carried out using the mutants in the SG200 strain background. While mutants in khd4 were reduced in virulence, deletion of the chitinases did not affect infection of maize seedlings (Figure 5) 20,25.
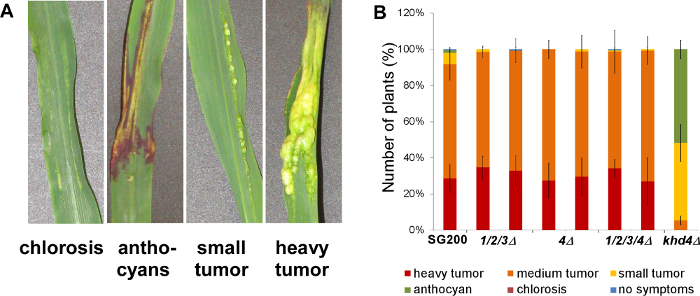
Figure 5. Infection assay of deletion mutants. Disease rating of maize seedlings 9 days after infection with the respective U. maydis strains. (A) Macroscopic symptoms that are used for disease scoring. (B) Two independent transformants were tested for each strain (N >100). All chitinase-deficient mutants (1/2/3Δ: triple mutant endochitinases, 4Δ: single mutant N-acetylglucosaminidase, 1/2/3/4Δ: quadruple mutant) infected the host plant leading to heavy tumor formation as observed in wildtype infections with the solopathogenic strain SG200. By contrast, a strain carrying a deletion of the gene encoding the RNA-binding protein Khd4 was reduced in virulence as reported previously 24. Error bars indicate standard deviation of three independent experiments. This figure is modified with permission from Langner et al. 2015 Eukaryotic Cell 20. Please click here to view a larger version of this figure.
| Genetic background | 1st diagn. PCR (excluded/tested) |
2nd diagn. PCR (confirmed/tested) |
Southern (confirmed/tested) |
% recomb. |
| AB33 | 7/22 | – | 7/15 | 32 |
| AB33 cts1Δ | 19/24 | – | 5/5 | 21 |
| SG200 | 0/10 | 8/10 | 7/8 | 70 |
| SG200 cts1Δ | 0/20 | 19/20 | 14/19 | 70 |
Table 1: Strain verification for cts3 deletions. The chitinase gene cts3 was deleted in four different genetic backgrounds to allow studies of filamentous growth (AB33) and infection (SG200) in single and multiple deletions.
Discussion
This protocol describes how to generate deletion mutants for reverse genetic studies in U. maydis. The starting point is a deletion construct that contains flanking sequences of the gene-of-interest containing sequences of about 1 kb upstream of the start and downstream of the stop-codon as well as an appropriate resistance cassette as it was previously optimized 7,9. The constructs have to be individually generated for each gene and carefully verified for sequence errors prior to deleting the gene. Point mutations in the flanks can cause unwanted alterations in the genomic sequence, in particular if the flanks reach into the neighboring gene or the mutation modifies regulatory elements. This would affect all transformants and can cause phenotypes and side-effects unrelated to the desired gene deletion.
When planning the deletion strategy, the expected phenotype has to be considered to choose the genetic background. In the present case, AB33 23 and SG200 10 were chosen to allow for testing phenotypes in the morphological switch and plant infection. Similarly, many other read-outs may be of interest, e.g. fusions with cts1 are employed in export of heterologous proteins 14,26, and a strain background with fluorescently tagged rrm4 allows analysis of RNA transport on endosomes 27,28.
During strain verification, the high-throughput diagnostic PCRs allow a rapid reduction of the number of candidates to be tested in the rather laborious Southern blot. In particular, the first diagnostic PCR allows exclusion of candidates still containing the wildtype copy of the gene and thereby prevents the massive use of toxic phenol in gDNA preparations. In a standard case with a non-essential gene, approximately half of the colonies may contain the wildtype gene. This pre-screening can be of great interest for genes with potentially detrimental growth phenotypes. Hundreds of candidates can rapidly be screened.
If all candidates contain the wildtype copy, the gene deletion is most likely lethal. This can be confirmed by deleting one copy in a diploid strain background (e.g. d132) and analysis of the segregation after teliospore germination 29. Alternatively, a conditional deletion strategy using an inducible promoter can be chosen to follow the mutant phenotype. However, also contamination resulting from the crude NaOH-based method for gDNA preparation can interfere with the PCR and lead to false-negatives i.e. candidates do not show a band and therefore are kept even though they contain the gene. This can be excluded by testing a control gene with a comparable size product in an independent PCR reaction or by multiplex-PCR with the two gene-specific primers and primers for a control gene that result in a bigger fragment.
If a mutant is already available, e.g., cts1Δ30 or rrm4Δ, strain generation of tagged versions can be accelerated by replacing the gene deletion resistance cassette (e.g. hygR) in the genome with a novel construct containing, e.g., the fluorescently labelled rrm4 and a different resistance marker (e.g. natR). In this case, transformants can be screened by loss of the initial resistance marker 9. Correct candidates are resistant only against the new antibiotic (nourseothricin) and have lost their initial resistance (hygromycin). Additionally, complementation of the mutant phenotype in trans can be carried out to verify functionality of tagged constructs 30,31.
The here described gene deletion and modification strategy offers a reliable and exact tool in genetic analysis of U. maydis. For multiple deletions however, the strain generation can get time-consuming, since the alterations have to be carried out sequentially. Therefore, as a complementary tool, last year the CRISPR-Cas system has been established for U. maydis 32. It offers the possibility to disrupt several genes simultaneously and is an excellent addition to the genetic toolbox of U. maydis.
In summary, the protocol for genetic manipulation and U. maydis strain generation described here is a robust method that has been widely used in the community for years. Together with the comprehensive collection of respective resistance cassette modules it allows all sorts of genetic engineering in this fungus, addressing diverse questions ranging from basic to applied research.
Divulgaciones
The authors have nothing to disclose.
Acknowledgements
Special thanks to Dr. Benedikt Steuten for critical reading of the manuscript. The original work on the chitinases was carried out by Dr. Thorsten Langner. The laboratory of VG is supported by the Cluster of Excellence in Plant Sciences (CEPLAS, DFG EXC 1028) and BioSC, the laboratory of KS is supported by BioSC. KB is supported by BioSC. The scientific activities of the Bioeconomy Science Center (BioSC) were financially supported by the Ministry of Innovation, Science and Research within the framework of the NRW Strategieprojekt BioSC (No. 313/323-400-00213). LF is supported by a doctoral fellowship of the DFG International Research Training Group 1525 iGRADplant.
Materials
| Aminobenzoeic acid (Free Acid) | Sigma Aldrich | A-9878 | |
| Bacto Agar | BD | 214010 | alternatively use local supplier |
| Bacto Peptone | BD | 211677 | alternatively use local supplier |
| Bacto Yeast Extract | BD | 212750 | alternatively use local supplier |
| CaCl2*2H2O | Grüssing GmbH | 10234 | alternatively use local supplier |
| Ca-pantothenat (Hemi-Ca. salt) | Sigma Aldrich | P-2250 | |
| Carboxin | Sigma Aldrich | 45371 | |
| Casamino acids | BD | 223050 | |
| Cholinchlorid | Sigma Aldrich | C-1879 | |
| Citric acid | ChemSolute | 24,321,000 | alternatively use local supplier |
| CuSO4*5H2O | Fluka | 61240 | alternatively use local supplier |
| D(+)Sucrose | Roth | 4621.1 | alternatively use local supplier |
| DNA degr. free acid | Sigma-Aldrich | D-3159 | |
| EDTA | Sigma Aldrich | E4378 | |
| FeCl3*6H2O | Grüssing GmbH | 10288 | alternatively use local supplier |
| Geneticin (G418) disulfate salt | Sigma Aldrich | A1720 | |
| Trichoderma lysing enzymes | Sigma Aldrich | L1412 | |
| Glucose | Caelo | 2580 | alternatively use local supplier |
| Glycerin | Fisher Chemical | G065015 | alternatively use local supplier |
| H3BO3 | AppliChem | A2940 | Dangerous substance. Please check manufacturer's safety instructions. |
| Heparin sodium salt | Sigma Aldrich | H3393-50KU | |
| Hygromycin B-solution | Roth | 1287.2 | Dangerous substance. |
| KCl | VWR | 26764298 | alternatively use local supplier |
| KH2PO4 | AppliChem | A3620 | alternatively use local supplier |
| MgSO4 waterfree | Merck | 7487-88-9 | Water free is critical. Alternatively use local supplier |
| MnCl2*4H2O | AppliChem | A2087 | alternatively use local supplier |
| myo-Inositol | Sigma Aldrich | I-5125 | |
| Na2-EDTA*2H2O | AppliChem | A2937 | alternatively use local supplier |
| Na2MoO4*2H2O | Roth | 0274.2 | alternatively use local supplier |
| Na2SO4 | Grüssing GmbH | 12174 | alternatively use local supplier |
| NaCl | Fisher Chemical | S316060 | alternatively use local supplier |
| NaOH | ChemSolute | 13,751,000 | alternatively use local supplier |
| NH4NO3 | Roth | K299.1 | alternatively use local supplier |
| Nicotinic acid (Free Acid) | Sigma Aldrich | N-4126 | |
| Nourseothricin dihydrogen sulfate | Werner BioAgents | 5,001,000 | |
| Nutrient broth | Difco | local suppliers | |
| Phenol:Chloroform:Isoamyl Alcohol (25:24:1) pH 6.7 | Sigma Aldrich | P3803 | Dangerous substance. Please check manufacturer's safety instructions. |
| polyethylene glycol (PEG) | Sigma Aldrich | P-3640 | |
| Potassium acetate | AppliChem | 121479 | alternatively use local supplier |
| Pyridoxin (Monohydrochlorid) | Sigma Aldrich | P-9755 | |
| Riboflavin | Sigma Aldrich | R4500 | |
| RNaseA | Sigma Aldrich | R5503 | |
| SDS | Roth | Cn30.3 | alternatively use local supplier |
| small syringe | BD | 300300 | alternatively use local supplier |
| sterile filter, 22 µm | VWR | 28145-477 | alternatively use local supplier |
| Sorbitol | Roth | 6213.1 | alternatively use local supplier |
| Thiamin-Hydrochloride | Serva | 36020.02 | alternatively use local supplier |
| tri-Na-Citrate | Fisher Chemical | S332060 | alternatively use local supplier |
| Tris- (hydroxymethyl) aminomethane | VWR | 103156X | alternatively use local supplier |
| Tris hydrochloride | Roth | 9090.4 | alternatively use local supplier |
| Triton X-100 | Serva | 37240 | alternatively use local supplier |
| ZnCl2 | Fluka | 96470 | alternatively use local supplier |
| Name | Company | Catalog Number | Comments |
| Composition of solutions/preparation of material | Composition of solutions | ||
| Carboxin | Stock: 5 mg/ml in methanol, final concentration: 2 µg/ml | ||
| CM plates | 0.25 % (w/v) Casamino acids, 0.1 % (w/v) Yeast Extract, 1.0 % (v/v) Holliday vitamin solution, 6.25 % (v/v); Holliday salt solution, 0.05 % (w/v) DNA degr. free acid, 0.15 % (w/v) NH4NO3, 2.0 % (w/v) Bacto Agar; adjust to pH 7.0 using 5 M NaOH; after autoclaving add 1 % glucose | ||
| Geneticin (G418) | Stock: 50 mg/ml in H2O, final concentration: 500 µg/ml | ||
| HCl-washed glass beads (0,35-0,45 mm) | Cover glass beads with concentrated HCl (25 %, 7.8 M) and incubate for 60 min. Sway several times. Decant HCl (keep decanted liquid) and wash glass beads with 3 M HCl (keep decanted liquid). Wash glass beads several times with double distilled H2O until the pH is 7 (the liquid should not be yellow-green anymore). Aliquot the glass beads and dry them at 180 °C. The decanted HCl has to be neutralized before disposal. | ||
| Heparin | Stock: 15 mg/ml | ||
| Holliday salt solution | 16.0 ‰ (w/v) KH2PO4, 4.0 ‰ (w/v) Na2SO4, 8.0 ‰ (w/v) KCl, 1.32 ‰ (w/v) CaCl2*2H2O, 8.0 ‰ (v/v) trace elements, 2.0 ‰ (w/v) MgSO4; sterile filtrate | ||
| Holliday vitamin solution | 0.1‰ (w/v) Thiamin, 0.05‰ (w/v) Riboflavin, 0.05‰ (w/v) Pyridoxin, 0.2‰ (w/v) Ca-Pantothenat, (0.05‰ (w/v) Aminobenzoeic acid, 0.2‰ (w/v) Nicotinic acid, 0.2‰ (w/v) Cholinchlorid, 1.0‰ (w/v) myo-Inositol; may be stored at -20 °C | ||
| Hygromycin | Stock: 50 mg/ml in PBS, final concentration: 200 µg/ml | ||
| Nourseothricin | Stock: 200 mg/ml in H2O, final concentration: 150 µg/ml | ||
| NSY-glycerol-medium | 0.8 % (w/v) Nutrient Broth, 0.1 % (w/v) Yeast Extract, 0.5 % (w/v) Sucrose, 80.0 % (v/v) 87% Glycerin (f.c. 69.6%) | ||
| RegLight | 1.0% (w/v) Yeast Extract 0.4 % (w/v) Bacto Peptone, 0.4 % (w/v) Sucrose, 18.22 % (w/v) Sorbitol, 1.5 % (w/v) Agar | ||
| SCS, pH 5.8 | Solution 1: 20 mM tri-Na-citrate, 1 M Sorbitol; colution 2: 20 mM Citric acid, 1 M Sorbitol, add solution 2 into solution 1 until pH 5.8 is reached; autoclave | ||
| STC, pH 8 | 1 M Sorbitol, 10 mM Tris-HCl pH 7.5, 100 mM CaCl2; filter sterile | ||
| STC/PEG | 40 % (v/v) PEG in STC-buffer | ||
| TE buffer, pH 8 | 1.31 mM Tris-Base, 8.69 mM Tris-HCl, 10 mM Na2-EDTA*2H2O | ||
| TE/RNase | 10 µg/ml RNaseA in TE buffer | ||
| Trace elements | 0.06‰ (w/v) H3BO3, 0.14‰ (w/v) MnCl*4H2O, 0.4 ‰ (w/v) ZnCl2, 0.4 ‰ (w/v) Na2MoO4*2H2O, 0.1 ‰ (w/v) FeCl3*6H2O, 0.04‰ (w/v) CuSO4*5H2O | ||
| Trichoderma lysing enzymes solution | 12.5 mg/ml SCS; filter sterile; prepare shortly before use | ||
| Tris-HCl pH 7.5 | 806 mM Tris-HCl, 194 mM Tris-Base; check the pH and if necessary adjust with HCl; autoclave | ||
| Usti-lysis buffer 1, pH 8 | 10 mM Tris-HCl (pH 8.0), 10 mM NaCl, 1 % (w/v) SDS, 2 % (v/v) TritonX-100, 1 mM EDTA. Do not measure pH using pH meter. | ||
| Usti-lysis buffer 2 | mix Usti lysis buffer 1 with 1 x TE in a 1:1 ratio | ||
| YEPS-Light medium | 1.0% (w/v) Yeast Extract, 0.4% (w/v) Bacto Peptone, 0.4% (w/v) Sucrose, for plates: 1.5% (w/v) Bacto Agar |
Referencias
- Dean, R., et al. The Top 10 fungal pathogens in molecular plant pathology. Molecular Plant Pathology. 13, 414-430 (2012).
- Vollmeister, E., et al. Fungal development of the plant pathogen Ustilago maydis. Fems Microbiol Rev. 36, 59-77 (2012).
- Banuett, F. Ustilago maydis, the delightful blight. Trends in Genetics. 8, 174-180 (1992).
- Holloman, W. K., Schirawski, J., Holliday, R. The homologous recombination system of Ustilago maydis. Fungal Genetics and Biology. 45, S31-S39 (2008).
- Feldbrügge, M., Kämper, J., Steinberg, G., Kahmann, R. Regulation of mating and pathogenic development in Ustilago maydis. Current Opinion in Microbiology. 7, 666-672 (2004).
- Ökmen, B., Doehlemann, G. Inside plant: Biotrophic strategies to modulate host immunity and metabolism. Current Opinion in Plant Biology. 20, 19-25 (2014).
- Brachmann, A., König, J., Julius, C., Feldbrügge, M. A reverse genetic approach for generating gene replacement mutants in Ustilago maydis. Molecular Genetics and Genomics. 272, 216-226 (2004).
- Kämper, J. A PCR-based system for highly efficient generation of gene replacement mutants in Ustilago maydis. Molecular Genetics and Genomics. 271, 103-110 (2004).
- Terfrüchte, M., et al. Establishing a versatile Golden Gate cloning system for genetic engineering in fungi. Fungal Genetics and Biology. 62, 1-10 (2014).
- Kämper, J., et al. Insights from the genome of the biotrophic fungal plant pathogen Ustilago maydis. Nature. 444, 97-101 (2006).
- Chavan, S., Smith, S. M. A Rapid and Efficient Method for Assessing Pathogenicity of Ustilago maydis on Maize and Teosinte Lines. Journal of Visualized Experiments. (83), e50712 (2014).
- Khrunyk, Y., Münch, K., Schipper, K., Lupas, A. N., Kahmann, R. The use of FLP-mediated recombination for the functional analysis of an effector gene family in the biotrophic smut fungus Ustilago maydis. New Phytologist. 187, 957-968 (2010).
- van Peer, A. F., de Bekker, C., Vinck, A., Wösten, H. A. B., Lugones, L. G. Phleomycin Increases Transformation Efficiency and Promotes Single Integrations in Schizophyllum commune. Appl Environ Microb. 75, 1243-1247 (2009).
- Sarkari, P., et al. Improved expression of single-chain antibodies in Ustilago maydis. Journal of Biotechnology. 191, 165-175 (2014).
- Schirawski, J., Heinze, B., Wagenknecht, M., Kahmann, R. Mating type loci of Sporisorium reilianum: Novel pattern with three a and multiple b specificities. Eukaryotic Cell. 4, 1317-1327 (2005).
- Cervantes-Chavez, J. A., Ali, S., Bakkeren, G. Response to Environmental Stresses, Cell-wall Integrity, and Virulence Are Orchestrated Through the Calcineurin Pathway in Ustilago hordei. Mol Plant Microbe In. 24, 219-232 (2011).
- Yu, J. J., et al. An efficient genetic manipulation protocol for Ustilago esculenta. FEMS Microbiology Letters. 362, (2015).
- Ninomiya, Y., Suzuki, K., Ishii, C., Inoue, H. Highly efficient gene replacements in Neurospora strains deficient for nonhomologous end-joining. P Natl Acad Sci USA. 101, 12248-12253 (2004).
- Colot, H. V., et al. A high-throughput gene knockout procedure for Neurospora reveals functions for multiple transcription factors. P Natl Acad Sci USA. 103, 10352-10357 (2006).
- Langner, T., et al. Chitinases Are Essential for Cell Separation in Ustilago maydis. Eukaryot Cell. 14, 846-857 (2015).
- Langner, T., Göhre, V. Fungal chitinases: function, regulation, and potential roles in plant/pathogen interactions. Current Genetics. , (2015).
- Sambrook, J., Fritsch, E. F., Maniatis, T. . Molecular Cloining: a laboratry manual. , (1989).
- Brachmann, A., Weinzierl, G., Kämper, J., Kahmann, R. Identification of genes in the bW/bE regulatory cascade in Ustilago maydis. Molecular Microbiology. 42, 1047-1063 (2001).
- Becht, P., Vollmeister, E., Feldbrügge, M. Role for RNA-binding proteins implicated in pathogenic development of Ustilago maydis. Eukaryotic Cell. 4, 121-133 (2005).
- Vollmeister, E., et al. Tandem KH domains of Khd4 recognize AUACCC and are essential for regulation of morphology as well as pathogenicity in Ustilago maydis. RNA. 15, 2206-2218 (2009).
- Stock, J., et al. Applying unconventional secretion of the endochitinase Cts1 to export heterologous proteins in Ustilago maydis. Journal of Biotechnology. 161, 80-91 (2012).
- Baumann, S., Pohlmann, T., Jungbluth, M., Brachmann, A., Feldbrügge, M. Kinesin-3 and dynein mediate microtubule-dependent co-transport of mRNPs and endosomes. Journal of Cell Science. 125, 2740-2752 (2012).
- Pohlmann, T., Baumann, S., Haag, C., Albrecht, M., Feldbrügge, M. A FYVE zinc finger domain protein specifically links mRNA transport to endosome trafficking. eLife. 4, (2015).
- Kronstad, J. W., Leong, S. A. Isolation of two Alleles of the b-Locus of Ustilago. P Natl Acad Sci USA. 86, 978-982 (1989).
- Koepke, J., et al. The RNA-binding protein Rrm4 is essential for efficient secretion of endochitinase Cts1. Molecular & cellular proteomics. 10, (2011).
- Schuler, D., et al. Hxt1, a monosaccharide transporter and sensor required for virulence of the maize pathogen Ustilago maydis. New Phytologist 206. , 1086-1100 (2015).
- Schuster, M., Schweizer, G., Reissmann, S., Kahmann, R. Genome editing in Ustilago maydis using the CRISPR-Cas system. Fungal Genetics and Biology. , (2015).

