How to Study Basement Membrane Stiffness as a Biophysical Trigger in Prostate Cancer and Other Age-related Pathologies or Metabolic Diseases
Summary
Here we explain a protocol for modelling the biophysical microenvironment where crosslinking and increased stiffness of the basement membrane (BM) induced by advanced glycation endproducts (AGEs) has pathological relevance.
Abstract
Here we describe a protocol that can be used to study the biophysical microenvironment related to increased thickness and stiffness of the basement membrane (BM) during age-related pathologies and metabolic disorders (e.g. cancer, diabetes, microvascular disease, retinopathy, nephropathy and neuropathy). The premise of the model is non-enzymatic crosslinking of reconstituted BM (rBM) matrix by treatment with glycolaldehyde (GLA) to promote advanced glycation endproduct (AGE) generation via the Maillard reaction. Examples of laboratory techniques that can be used to confirm AGE generation, non-enzymatic crosslinking and increased stiffness in GLA treated rBM are outlined. These include preparation of native rBM (treated with phosphate-buffered saline, PBS) and stiff rBM (treated with GLA) for determination of: its AGE content by photometric analysis and immunofluorescent microscopy, its non-enzymatic crosslinking by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS PAGE) as well as confocal microscopy, and its increased stiffness using rheometry. The procedure described here can be used to increase the rigidity (elastic moduli, E) of rBM up to 3.2-fold, consistent with measurements made in healthy versus diseased human prostate tissue. To recreate the biophysical microenvironment associated with the aging and diseased prostate gland three prostate cell types were introduced on to native rBM and stiff rBM: RWPE-1, prostate epithelial cells (PECs) derived from a normal prostate gland; BPH-1, PECs derived from a prostate gland affected by benign prostatic hyperplasia (BPH); and PC3, metastatic cells derived from a secondary bone tumor originating from prostate cancer. Multiple parameters can be measured, including the size, shape and invasive characteristics of the 3D glandular acini formed by RWPE-1 and BPH-1 on native versus stiff rBM, and average cell length, migratory velocity and persistence of cell movement of 3D spheroids formed by PC3 cells under the same conditions. Cell signaling pathways and the subcellular localization of proteins can also be assessed.
Introduction
The basement membrane (BM) is a sheet of specialized extracellular matrix (ECM) that maintains stable tissue borders by separating layers of epithelial cells from the stroma1. Covalent crosslinking between adjacent triple helices of collagen IV in the BM stabilizes their lateral association by establishing an irregular network of super-twisted helices2. These collagen IV lattices act as a scaffold for its interaction with laminin and other BM components1. The structural arrangement of the BM provides it with the mechanical strength and rigidity necessary for the normal development of glandular epithelia3.
During aging and disease the BM progressively thickens and stiffens3,4. For example, a 3-fold increase in the elastic modulus (E) of the ocular BM occurs between the ages of 50 and 80 in the normal population, and this stiffening is further exacerbated in metabolic disorders like diabetes5. The structural and biomechanical changes in the BM that result in its increased stiffness occur when its ECM components, collagen IV and laminin, become non-enzymatically crosslinked following their exposure to advanced glycation endproducts (AGEs).
The purpose of the method described here was to establish a model for the investigation of how BM stiffness, due to AGE exposure, promotes prostate epithelial cell (PEC) and prostate tumour cell (PTC) invasiveness in the context of the switch to metastatic prostate cancer (PCa). To do this a previous method used for generating 3D glandular acini from mammary epithelial cells (MECs) in reconstituted rBM gels6 was adapted to include an additional step where the rBM gels are pre-treated with glycolaldehyde (GLA). Several techniques for assessing GLA induced crosslinking and stiffening of pre-treated rBM gels are described, including photometric analysis, sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS PAGE), confocal microscopy and rheometric analysis. The prostate cell types selected for culture on the pre-stiffened rBM include: RWPE-1, PECs derived from a normal prostate gland7; BPH-1, PECs derived from a prostate gland affected by BPH8; and PC3, metastatic PTCs derived from a secondary tumor located in the vertebral bone of a prostate cancer (PCa) patient9.
In addition to advancing the study of prostate gland pathology, the protocol for stiffening of rBM gels by their treatment with GLA can be adapted to investigate how BM stiffness contributes to other age-related pathologies and metabolic disorders. For example, the model can be directly applied to investigate how metastatic cancer is induced by BM stiffness in organs such as the breast, colon, ovary and pancreas by the incorporation of appropriate cell types. Furthermore, the protocol can be adapted to investigate how stiff BM promotes biomechanical mechanisms of disease progression in diabetes-related microvascular disease, retinopathy, nephropathy and neuropathy.
Protocol
1. Induction of BM Stiffness Induced by GLA Treatment (Non-enzymatic Crosslinking)
- Thaw a frozen vial of BM matrix (10 ml) by incubating at 4 °C (standing on ice in a cold room or refrigerator) until the contents of the vial have become liquid (8-16 hr).
Caution: If a cold room/refrigerator is not used, cover the entire bottle with ice. This will prevent the stock solution of BM from solidifying. - For future experiments and to avoid repeated freeze thaw cycles, prepare 25 x 0.4 ml aliquots from each new 10 ml vial of BM matrix. Store vials at -80 °C until the expiry date indicated by the manufacturer. When needed, thaw vials at 4 °C standing on ice for 2 hr.
- Prepare an even surface of ice. Place an 8-well chamber glass slide on top of the ice to maintain a temperature of 4 °C during the coating procedure. Thaw a vial of BM matrix at 4 °C.
Note: One 0.4 ml vial of BM matrix is sufficient to coat an entire 1 x 8-well chamber slide. Keep the vial covered in ice while handling to prevent the BM matrix from solidifying. - Cut off the dispensing end of a 200 µl pipette tip using scissors. Cool the blunt-ended 200 µl pipette tip to 4 °C and place it on to a 200 µl capacity pipetting aid. Take up 40 µl of the cold BM matrix solution into the pipette tip and transfer it into a well on the chilled 8-well chamber glass slide.
Note: 40 µl of BM solution is enough to cover a surface area of 0.8 cm2. Keep pipette tips chilled during the coating procedure to avoid solidification of the BM solution. Do not introduce air bubbles into the BM matrix solution and ensure the well is evenly coated without the formation of a visible meniscus at the edges. - Repeat step 1.4 according to the number of wells and chambers required.
- After coating, place the 8-well chamber slide at 37 °C for 30 min to promote polymerization of the BM. Close the incubator door very carefully to avoid unwanted disturbance of the liquid rBM. Do not exceed the 30 min incubation time to avoid dehydration of the rBM gel.
Note: The resulting gel is the native reconstituted BM (rBM). The 37 °C incubation step does not require 5 % CO2. However, for convenience perform this step in a tissue culture incubator set at 37 °C and 5 % CO2 (with humidification). - Prepare 50 mM glycolaldehyde (GLA) diluted in 0.2 M phosphate buffer (pH 7.8). Sterilize the solution by passing it through a 0.22 micron syringe filter using a 50 ml syringe.
- For a crosslinking reaction in a final volume of 250 µl of 50 mM GLA, add 25 µl of 0.5 M sodium cyanoborohydride or 2.5 M aminoguanidine to 125 µl of 100 mM GLA (2x stock) and 100 µl of 0.2 M phosphate buffer (pH 7.8). Sterilize the stock solutions by passing them through a 0.22 micron syringe filter using a 50 ml syringe.
Caution: Handle sodium cyanoborohydride wearing a lab coat, gloves, faceshield and respirator while working in a fume hood.
- For a crosslinking reaction in a final volume of 250 µl of 50 mM GLA, add 25 µl of 0.5 M sodium cyanoborohydride or 2.5 M aminoguanidine to 125 µl of 100 mM GLA (2x stock) and 100 µl of 0.2 M phosphate buffer (pH 7.8). Sterilize the stock solutions by passing them through a 0.22 micron syringe filter using a 50 ml syringe.
- Add 250 µl of GLA solution to cover the polymerized rBM gel and incubate at 37 °C for 6 hr to produce a semi-stiff rBM gel or 14 hr to produce a stiff rBM gel.
Note: The volume of GLA added must cover the polymerized rBM gel and should be adjusted accordingly. If different GLA incubation times are used, the rBM gels should be analyzed to determine the fold-increase in rBM stiffness (see Step 2.4).- Prepare a negative control by incubating a native rBM gel in 250 µl of sterile phosphate buffered saline (PBS) for 14 hr at 37 °C.
- Prepare two additional controls where the formation of Schiff base or Amadori adduct rearrangement during the crosslinking reaction are inhibited, by the addition of 50 mM sodium cyanoborohydride or 250 mM aminoguanidine.
- Prepare 1 M glycine ethyl ester (GEE) diluted in PBS. Sterilize the solution by passing it through a 0.22 micron syringe filter using a 50 ml syringe.
- After the indicated incubation time, carefully remove the GLA solution from the crosslinked rBM gels, GLA solution containing inhibitors from the control rBM gels and PBS from the control native rBM gels. Add 250 µl of GEE solution to all of the rBM gels and incubate at 37 °C for 1 hr.
Note: This step quenches the crosslinking reaction. - Wash all rBM gels 10 times in 500 µl PBS to remove all traces of GLA and GEE. Incubate the rBM gels overnight at 37 °C in 400 µl of PBS to prevent their dehydration.
- Analyze the rBM gels for AGE accumulation, non-enzymatic crosslinking and viscoelastic properties (Steps 2.1-2.4). For rheometric analysis of their viscoelastic properties prepare the rBM gels in cloning rings (Step 2.4).
- For cell culture, rinse rBM gels 2 times with 500 µl culture media before seeding the cells (Steps 5 and 6). Perform washes gently without the pipette tip touching the gel surface.
2. Quantification of Non-Enzymatic Crosslinking and Stiffness of rBM Treated with GLA
- Photometric Analysis
- Measure AGE accumulation in GLA-treated and control rBM gels using photometric analysis to determine the extent of the Maillard reaction.
- After Step 1.11, remove the PBS from rBM gels in the 8-wells chamber slides and add 250 µl ice-cold double distilled water. Incubate at 4 °C for 16-24 hr to ensure that the matrix is completely liquefied.
Note: rBM peptides in this solution contain AGEs with auto-fluorescent properties. - Transfer the liquefied BM solution to a 1.5 ml tube and measure the fluorescent emission of the solution using a spectrophotometer (excitation wavelength = 370 nm; emission wavelength = 440 nm).
- After Step 1.11, remove the PBS from rBM gels in the 8-wells chamber slides and add 250 µl ice-cold double distilled water. Incubate at 4 °C for 16-24 hr to ensure that the matrix is completely liquefied.
- Measure AGE accumulation in GLA-treated and control rBM gels using photometric analysis to determine the extent of the Maillard reaction.
- SDS-PAGE Analysis of Cyanogen Bromide Peptides
- Resolve the GLA-treated and control rBM gels on a polyacrylamide gel to confirm that GLA has induced crosslinking and the formation of macro-fibres.
- Centrifuge the liquefied BM solution collected at Step 2.1.1.2 at 10,000 x g for 5 min at room temperature.
- Prepare a stock solution containing 2 g/ml of cyanogen bromide diluted in acetonitrile.
Caution: Always handle cyanogen bromide in a fume hood while wearing a lab coat, gloves, faceshield and respirator. - Remove the supernatant, re-suspend the BM gel pellet in 500 µl of 20 mg/ml cyanogen bromide + 70 % v/v formic acid and incubate overnight at RT.
- Use a 1 ml disposable syringe to transfer the resuspended BM gel pellet into a dialysis cassette with a molecular weight cut off 3.5 kDa.
- Submerge the cassette into a 500 ml glass beaker containing 500 ml of double distilled water and a magnetic stir bar. Place this onto a magnetic stirrer and dialyze overnight (16 hr) at 4 °C (in a cold room) to remove all traces of cyanogen bromide and formic acid.
- Use a 1 ml disposable syringe to transfer the dialyzed BM solution from the cassette into a 1.5 ml tube.
- Analyse 25 µl of each BM sample on a 12% v/v polyacrylamide gel10,11. Following SDS-PAGE, carry out silver staining of the polyacrylamide gel12 to visualize the electrophoretic pattern of cyanogen bromide-matrix peptides13.
- Resolve the GLA-treated and control rBM gels on a polyacrylamide gel to confirm that GLA has induced crosslinking and the formation of macro-fibres.
- Immunofluorescent Microscopy Analysis
- Perform immunofluorescent staining of GLA treated and control rBM gels with anti-AGE/pentosidine, anti-collagen IV and anti-laminin antibodies followed by confocal microscopy to visualize accumulated AGEs and collagen IV/laminin fibre structural rearrangements in the crosslinked rBM gels13.
Note: Always use a sufficient volume to cover the entire rBM gel during incubations and washes without touching the rBM surface with the pipette tip. For details of analyzing 3D acini cultures by immunofluorescence see reference6 and for confocal microscopy of 3D acini see reference14.- Wash GLA treated and control rBM gels in 8-wells chamber slides 2 times with 300 µl of PBS+ (PBS containing 0.1 mM CaCl2 and 0.5 mM MgCl2) for 5 min at RT.
- Remove the PBS+ then add 300 µl of 4 % w/v paraformaldehyde (PFA) diluted in PBS+ to cover each rBM gel. Incubate for 30 min at room temperature to fix the rBM components.
- Remove the 4 % w/v PFA solution. Add add 300 µl of 75 mM NH4Cl + 0.5 mM MgCl2 solution and incubate for 5 min at RT (repeat 5x) to quench the fixation.
- Prepare Immunofluorescence buffer (IF buffer) by making the following solution in sterile water: 130 mM NaCl, 7 mM Na2HPO4, 3.5 mM NaH2PO4, 7.7 mM NaN3, 0.1% w/v bovine serum albumin, 0.5% v/v polyethylene glycol tert-octylphenyl ether and 0.05% v/v polyethylene glycol sorbitan monolaurate.
- Prepare IF blocking buffer by supplementing IF buffer with 20 % v/v goat serum.
- Remove the quenching solution and add 300 µl of IF blocking buffer to the rBM gels to prevent nonspecific reactions. Incubate 2 hr at RT on a shaking platform.
- Remove the IF blocking buffer and incubate the rBM gels for 16 hr at 4 °C with 300 µl of primary antibody diluted in IF blocking buffer (1: 500 mouse anti-pentosidine mAb; 1/250 rabbit anti-collagen IV pAb; 1/250 rabbit anti-laminin A/C pAb).
Note: Incubations for longer than 20 hr at 4 °C can liquidize the rBM. - Remove the primary antibody and wash 3 times (10 min each) with 300 µl of IF buffer at room temperature on a shaking platform.
- Remove the IF buffer and add 300 µl of the secondary antibody (goat anti-rabbit or anti-mouse IgG [H+L]) conjugated with a fluorochrome diluted 1: 500 in IF blocking buffer. Incubate for 2 hr at RT on a shaking platform.
- Remove the secondary antibody and incubate in 300 µl of IF buffer for 10 min at room temperature. Remove the IF buffer and wash 3 x 10 min in 300 µl of PBS+ at room temperature.
- Fix and quench a second time, as described above (Steps 2.3.1.2 and 2.3.1.3).
- Mount stained rBM gels in mounting media and analyze the formation of dense bundles of major components using epifluorescent or confocal microscopy.
Note: For details of analyzing 3D acini cultures by immunofluorescence see reference6 and for epifluorescent and confocal microscopy of 3D acini see reference14.
- Perform immunofluorescent staining of GLA treated and control rBM gels with anti-AGE/pentosidine, anti-collagen IV and anti-laminin antibodies followed by confocal microscopy to visualize accumulated AGEs and collagen IV/laminin fibre structural rearrangements in the crosslinked rBM gels13.
- Rheological Analysis
- Perform rheometric analysis of GLA treated and control rBM gels to measure their viscoelasticity (stiffness).
- Set up rBM gels that are 1 mm thick in a circular mold with a diameter of 8 mm. To do this, place a cloning ring (8 mm diameter) inside a well of a 24-well culture plate and add BM matrix solution prepared as described in Steps 1.3-1.6.
Note: For accurate recapitulation of the rBM gels used for experiments, the rBM gels prepared for rheometric analysis need to have the same surface area and thickness as the rBM gels set up in the 8-well chambers. The rBM gels analyzed in Figure 3 were 1 mm thick and 8 mm in diameter. - Treat the rBM gels set up in the cloning rings with PBS, GLA for 6 hr and GLA for 14 hr as described above (Steps 1.8 to 1.11).
- Measure the elastic modulus (E) of the 8 mm diameter rBM gels on a rheometer with an 8 mm parallel plate serrated geometry, over a range of 1-3% strain, at a fixed frequency oscillation of 1Hz and temperature of 21 °C. For additional details about the rheometric analysis of ECM gels see references see reference13,15,16.
Note: E is determined from the resulting shear storage modulus (G') through the use of the following equation E = 2 * G' * (1+v) where v is the Poisson's ratio of 0.5, as described in reference13,15,16.
- Set up rBM gels that are 1 mm thick in a circular mold with a diameter of 8 mm. To do this, place a cloning ring (8 mm diameter) inside a well of a 24-well culture plate and add BM matrix solution prepared as described in Steps 1.3-1.6.
- Perform rheometric analysis of GLA treated and control rBM gels to measure their viscoelasticity (stiffness).
3. Culture and Handling of the Normal PEC line, RWPE-1
- Grow RWPE-1 cells in keratinocyte serum-free media (KSFM) supplemented with 5 ng/ml epidermal growth factor (EGF), 50 µg/ml bovine pituitary extract (BPE) and 50 U/ml penicillin with 50 µg/ml streptomycin (complete KSFM).
Note: To avoid induction of epithelial-to-mesenchymal (EMT)-like transition do not expose RWPE-1 cells to serum. Allow complete KSFM to reach RT for 30 min after removing from storage at 4 °C and do not warm in a 37 °C water bath as this will inactivate the EGF and BPE. - Aspirate the complete KSFM from a confluent 10 cm2 plate of RWPE-1 cells, rinse with 5 ml of pre-warmed PBS and add 5 ml of 0.05 % v/v trypsin ensuring that all cells are covered with the solution.
- Place the cells in a tissue culture incubator set at standard conditions of 37 °C and 5% CO2 (with humidification) for 5 to 10 min. Check the extent of trypsinization after 5 min and gently tap the culture plate to detach the cells.
Note: RWPE-1 cells do not tolerate long periods of trypsinization so it is advised not to handle more than two plates at the same time. It is also important to dissociate all cells from the plate to avoid clonal selection.
- Place the cells in a tissue culture incubator set at standard conditions of 37 °C and 5% CO2 (with humidification) for 5 to 10 min. Check the extent of trypsinization after 5 min and gently tap the culture plate to detach the cells.
- When all RWPE-1 cells have disassociated, add 5 ml of warm PBS containing 2 % v/v fetal calf serum (FCS) to quench the trypsin. Gently pipette up and down to break up the cell aggregates before transferring the cells to a centrifuge tube.
- Centrifuge the disassociated cells at 125-150 x g for 5 min at 25 °C, discard the supernatant and re-suspend the pellet of cells in 5 ml of complete KSFM until a suspension of single cells is obtained.
- Transfer 1 ml of the re-suspended cells into a new tube and add 9 ml of complete KSFM to propagate the cells at a 1:5 passage dilution for subsequent experimental use. Count the rest of the cells using a hemocytometer for setting up acini (see Section 5.1).
Note: Do not culture RWPE-1 cells for more than 10 passages since after prolonged periods of culture they do not form acini with the correct architecture. - Change the culture media every 48 hr to ensure the EGF and BPE remain active.
Note: Include this medium change for any treatments that extend beyond 48 hr.
4. Culture and Handling of the BPH Cell Line, BPH-1
- Culture BPH-1 cells in RPMI 1640 media complemented with 5 % v/v FCS, 50 U/ml penicillin and 50 µg/ml streptomycin. Warm the culture media, PBS and 0.25% w/v trypsin-0.53 M EDTA solution to 37 °C before use.
Note: Cells can also be cultured in media with 2.5 % v/v FCS8. - Aspirate the culture media from a confluent 10 cm2 plate of BPH-1 cells and wash the cells 2 x with 3 ml of PBS to remove all the traces of culture media with serum that may quench the trypsin reaction.
- Aspirate the PBS and add 3 ml of trypsin-EDTA solution to cover the cells. Place the plate in an incubator set at 37 °C and 5% CO2 (with humidification) for 5 min. Remove the trypsin-EDTA solution when the cells are round but remain attached to the dish. Wash cells with 5 ml of PBS.
- After removal of the PBS, add 5 ml of culture media and gently pipette up and down to produce a suspension of single cells. Transfer the cells to a 15 ml tube.
- Take 2 ml of the cell suspension into a new centrifuge tube with 8 ml of complete media and plate the BPH-1 cells onto a 10 cm2 culture plate at a 1:5 passage dilution for subsequent experimental use. Count the rest of the cells using a hemocytometer for setting up acini (see Section 5.2).
Note: Keep a record of the passage number, as older BPH-1 cells do not form acini with a proper architecture. A passage number more than 10 is not desired. - Change the culture media every 72 hr.
5. 3D Culture of Prostate Gland Acini on Native and Stiff rBM
- If RWPE-1 cells are being used to form acini, dilute 5,000 cells prepared in step 3.5 in 300 µl of complete KSFM supplemented with 2 % v/v of BM solution.
- If BPH-1 cells are being used to form acini, dilute 2,500 cells prepared in step 4.5 in 300 µl of RPMI 1640 culture media supplemented with 2 % v/v of BM solution.
Note: BPH-1 cells are larger than RWPE-1 cells so lower numbers of BPH-1 cells are used to obtain a similar distribution of acini after 6 days of culture. - Gently seed the cells onto the native and AGE-stiffened rBM and carefully place the cultures in an incubator set at 37 °C and 5% CO2 (with humidification) to ensure an even distribution of growing acini in the well and that each cell divides to produce one acina.
- Every 2 days replace the culture media with fresh culture media containing 2 % v/v BM solution to ensure that cells have the growth factors required for normal acina homeostasis.
- Monitor acinar morphology in growing cultures using brightfield microscopy13.
Note: After 3 days in culture individual cells will form a cluster of >3 cells and after 1 week prostate gland acini with a diameter of ~50 µm will be observed. - Follow protocol described in 2.3 to perform immunofluorescence using antibodies specific for markers of cell-matrix adhesions, cell-cell adhesions, apico-basal polarity and invasiveness13.
- Use a mounting media with 4',6-diamidino-2-phenylindole (DAPI) or include an extra step (after 2.3.12) to stain cell nuclei by incubating with DAPI for 5 min and wash 2 x 5 min with PBS+.
6. 3D Culture of Prostate Tumor Cell Aggregates on Native and Stiff rBM
- Culture PC3 cells in RPMI 1640 medium containing 10 % v/v FCS and 50 U/ml penicillin with 50 µg/ml streptomycin. Warm the culture media, PBS and 0.25 % w/v trypsin-0.53 M EDTA solution to 37 °C before use.
- Aspirate the culture media from a confluent 10 cm2 culture dish of PC3 cells and wash the cells 2x with 3 ml of PBS to remove all traces of FCS that can quench the trypsin reaction.
- Aspirate the PBS and add 3 ml of trypsin-EDTA solution to cover the cells and incubate for 1 min.
- When the cells become rounded, but remain attached to the dish, carefully aspirate the trypsin-EDTA solution and wash with 3 ml of PBS to remove all traces of trypsin.
- After removal of the PBS, add 5 ml of culture media and gently pipette up and down to produce a suspension of single cells. Transfer the cells to a 15 ml tube.
- Take 1 ml of the PC3 cell suspension into a new centrifuge tube and add 9 ml of culture media. Plate the cells on a 10 cm2 culture dish (1:10 dilution) for subsequent experimental use. Count the remaining cells using a hemocytometer.
- Dilute 2,500 PC3 cells prepared in step 6.6 in 300 µl of RPMI 1640 culture media supplemented with 2 % v/v of BM solution to allow for the formation of a gradient gel in the culture.
- Gently seed the cells onto the native and AGE-stiffened rBM and carefully place the culture into the incubator set at 37 °C and 5% CO2 (with humidification) to ensure even distribution of growing spheroids in the well.
- Change the culture media every 72 hr.
- To study the effect of stiff (AGE-rich) rBM on prostate tumor cell migration, image PC3 cells using brightfield video time-lapse microscopy using temperature/CO2 control and a humidified chamber17.
Note: PC3 cells grow in strands on native rBM and do not form acini with a lumen, but if left to grow more than 72 hr on native rBM they will form 3D spheroids . - Following data acquisition, manually track PC3 cells and calculate their migration speed, shape (elongation ratio) and persistence of migration17-19.
Note: Persistence = ratio D/T, D = distance from start to end of cell trajectory, T = total length of cell trajectory.
Representative Results
3D Prostate Acini Cultured on Stiff rBM
After 6 days in culture, PECs derived from normal prostate tissue (RWPE-1) (Figure 1A) and BPH tissue (BPH-1) (Figure 1B) form acini on native (PBS treated) rBM that are organized into uniform spheroids of epithelial cells. These acini also have the characteristics of highly organized PECs with apical-to-basal polarity and a visible luminal space13,20.
The acini formed by PECs derived from normal prostate tissue (RWPE-1) (Figure 1A) and BPH tissue (BPH-1) (Figure 1B) on stiffened (AGE-rich) rBM (treated with GLA) have a disrupted architecture (shifting from spheroidal to polygonal in shape and cells protruding/migrating from the acini into the AGE-rich rBM) (Figure 1A). These acini are also characterized by highly disorganized PECs that have lost their apical-to-basal polarity with a small or non-existent luminal space13.
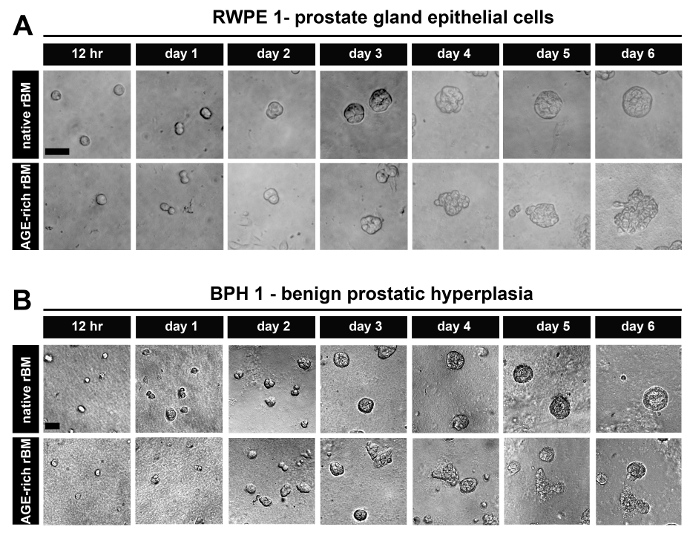
Figure 1: Prostate Epithelial Cells Grown as 3D Glandular Acini on Native and Stiff Reconstituted Basement Membrane (rBM). (A) Brightfield images of RWPE-1 cells grown for 12 hr up to 6 days on rBM gels treated with PBS (native) or 50 mM glycolaldehyde for 14 hr (AGE-rich; stiff); Scale bar = 50 µm. (B) BPH-1 cells, grown as described in panel A; Scale bars = 50 µm; data is representative of 3 independent experiments. Please click here to view a larger version of this figure.
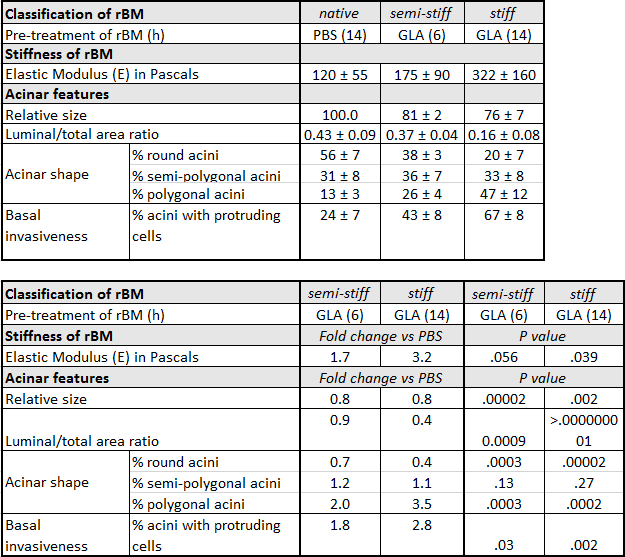
Table 1: Characteristics of Prostate Epithelial RWPE-1 acini Grown on Native, Semi-Stiff and Stiff Reconstituted Basement Membrane (rBM). RWPE-1 acini were grown on rBM pre-treated with PBS for 14 hr (native), glycolaldehyde (GLA) for 6 hr (semi-stiff) or GLA for 14 hr (stiff). For acinar shape, the percentage (%) ± standard deviation (SD) of round, semi-polygonal and polygonal acini were calculated from 5 independent experiments (50 acini quantified per condition). Relative acinar size was calculated (native rBM = 100 %) from 3 independent experiments. For invasiveness, % ± SD acini with one or more protruding cells were calculated from 3 independent experiments. Fold change is calculated by dividing the average value obtained under semi-stiff or stiff conditions by the corresponding value for native conditions. P values calculated using Student's t-test (α = 0.05).
AGE dependent increased rBM stiffness promotes PC3 prostate tumor cell migration
PC3 cells grown on native rBM migrate by maintaining continuous cell-cell contact, whereas PC3 cells grown on AGE-rich (stiff) rBM move independently from each other (Figure 2A). After 72 hr in culture PC3 cells form foci (spheroids) on native (PBS treated) rBM, whereas PC3 cells on stiff (AGE-rich) rBM do not from spheroids and migrate independently (Figure 2B). PC3 cells on stiff (AGE-rich) rBM are more elongated than PC3 cells grown on native rBM (Figure 2C). PC3 cells on stiff rBM migrate faster than PC3 cells grown on native rBM (Figure 2D). PC3 cells on stiff rBM display a decrease in persistence compared to PC3 cells grown on native rBM (Figure 2E).
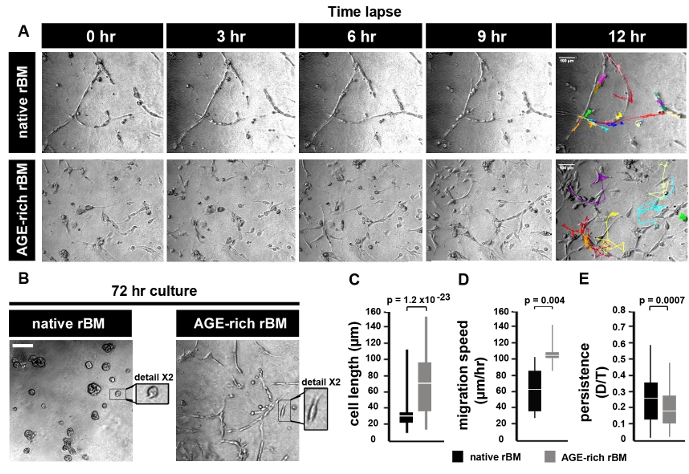
Figure 2: Prostate Tumor Cell Migration on Native and Stiff Reconstituted Basement Membrane (rBM). (A) Brightfield images of PC3 cells grown on rBM gels treated with PBS (native) or 50 mM glycolaldehyde for 14 hr (AGE-rich, stiff). Cells were imaged using a brightfield microscope (10X objective) and an acquisition rate of 1 image per hr for 12 hr followed by cell tracking to generate trajectories. Images shown correspond to the time points after 0, 3, 6, 9 and 12 hr. Trajectories of single cells are shown for the 12 hr time point. Scale bar = 100 µm. (B) PC3 cells cultured on native or stiff rBM for 72 hr, and imaged as described in panel (A). Scale bar = 100 µm. Detail shows selected area at 2X magnification. (C) Mean ± S.D. cell length (µm); significant difference between native rBM and stiff rBM (p = 1.2 x 10-23). (D) Mean ± S.D. velocity (µm/hr) calculated from cell trajectories; significant difference between native rBM and stiff rBM (p = 0.004). (E) Mean ± S.D. persistence of cell movement (ratio D/T, where D = distance from start to end of cell trajectory, T = total length of cell trajectory); significant difference between native rBM and stiff rBM (p = 0.0007). For panels C-E >10 cells were analyzed, data is representative of 3 independent experiments. Please click here to view a larger version of this figure.
Discussion
A protocol for the generation of 3D glandular acini from MECs in pure rBM gels6 was modified in a previous study by the addition of 4 mg/ml type I collagen to the rBM matrix. The addition of collagen resulted in the elastic modulus of the rBM gel increasing from 175 ± 37 to 1589 ± 380 Pascals. This 9.1-fold increase in stiffness modulated the growth, survival, migration and differentiation of MECs21. The protocol was modified again by including a treatment step with D-(-)-ribose to promote non-enzymatic crosslinking of the type I collagen that had been added to the rBM gel. The resultant 15-fold increase in stiffness was found to cooperate with oncogenic transformation of MECs to promote their invasive behavior22. The experimental approach of adding type I collagen to rBM gels facilitates the direct interaction of MECs with collagen fibres, which only occurs in human tissue after the physical barrier between the stroma and epithelium provided by the BM undergoes proteolytic degradation. By generating 3D glandular acini from PECs in pure rBM gels pre-treated with GLA, the current protocol opens the way to study how BM stiffness per se can trigger their invasive behaviour (Figure 3). The levels of BM stiffness induced in this protocol have physiological relevance. Incubation with 50 mM GLA for 6 hr and 14 hr respectively increased the elastic moduli of the pure rBM gel to 175 ± 90 and 322 ± 160 compared to 122 ± 55 Pascals in rBM gels treated with PBS (Table 1). This 1.7 to 3.2-fold increase in rBM stiffness recapitulates the 2.5- to 3.4-fold increase in stiffness observed in malignant compared to normal prostate or BPH tissue23-26. As outlined in a recent publication13 the morphological changes induced by the accumulation of AGE and rBM stiffness in PEC acini can be quantified for a statistically significant shift from a rounded to polygonal shape, decreased luminal/total acinar area, and protruding cells migrating from the acina into the AGE-rich rBM (Figure 3). Immunoblotting can also be used to assess markers of EMT (e.g. loss of E-cadherin13) and the contractile behavior (e.g. phosphorylated myosin light chain-2, pMLC213) in PECs grown in normal versus stiff rBM (Figure 3). Further evaluation using immunofluorescent staining and confocal microscopy can be applied to visualize the BM (e.g. laminin, collagen IV and AGE accumulation13), cellular apical-to-basal polarity (e.g. apical localization of EEA1: early endosomal antigen 1; and GM130: 130 kDa cis-Golgi marker13) and cellular patterns of adhesion molecules (e.g. E-cadherin localization to cell-cell junctions13) (Figure 3).
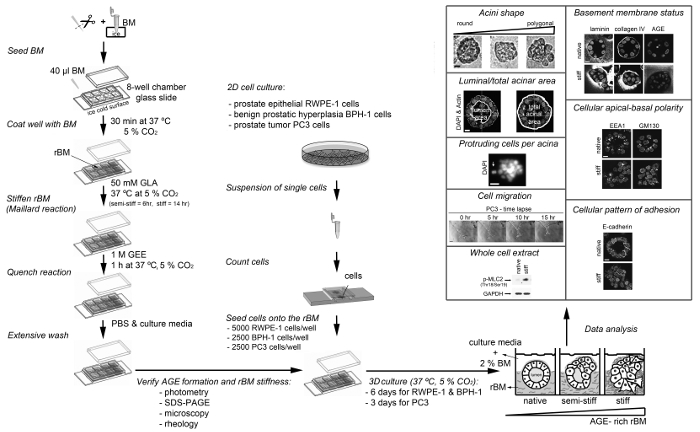
Figure 3: Overview of the Different Protocols Presented Here. The diagram depicts how to prepare and stiffen the reconstituted basement membrane (rBM) with glycolaldehyde (Maillard reaction), how to seed cells on to the stiff rBM, how to analyze the stiff rBM (extent of Maillard reaction) and procedures that can be used to analyze the cellular and molecular changes induced by AGE-rich rBM. AGE, advanced glycation endproducts; BM, basement membrane; DAPI, 4',6-diamidino-2-phenylindole; EEA1, early endosomal antigen 1; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; GLA, glycolaldehyde; GEE, glycine ethyl ester; GM130, 130 kDa cis-Golgi marker; p-MLC2 (Thr18/Ser19), myosin light chain-2 phosphorylated at sites threonine 18 and serine 19; rBM, reconstituted basement membrane; SDS-PAGE, sodium dodecyl sulfate polyacrylamide gel electrophoresis. For RWPE1 acini Scale bar = 10 µm; for PC3 tumor cell spheroids Scale bar = 100 µm. This figure has been modified from reference13. Please click here to view a larger version of this figure.
Troubleshooting steps will be necessary if D-(-)-ribose is chosen as the crosslinking agent for rBM. During protocol development it was found that treatment with 1 M D-(-)-ribose for 72 hr, as previously described for rBM/collagen gels22, resulted in the dehydration and shrinkage of rBM gels. The evaluation of lower concentrations of D-(-)-ribose and shorter treatment times may help to overcome this limitation.
A potential limitation in future applications of the protocol could be encountered where higher levels of rBM stiffness are desired. If longer incubation times and higher concentrations of GLA are used to induce higher levels of rBM gel stiffness it will be necessary to assess whether these treatment conditions have an impact on cell survival and proliferation, as previously described13. It should also be noted that incubation of RWPE-1 cells with serum induces a phenotypic EMT-like transition and exposure to serum or serum-containing materials should be avoided. For example, if experiments involve the transfection of short interfering RNA (siRNA) oligonucleotides, the procedure should be optimized using RWPE-1 cells grown in KSFM, without switching the cells to low serum transfection media. This drawback could compromise the level of gene silencing achieved when using transient siRNA approaches in the model. For some protein targets it would be advised to employ inducible shRNA vectors for tunable gene silencing and the desired decrease in protein levels. Adaptations that incorporate enzymatic crosslinking by stromal cell or tumor cell associated lysyl oxidase (LOX)17 could also be incorporated into future models.
This protocol will facilitate the future study of pro-invasive mechanisms triggered by AGE-dependent BM stiffness in PECs (RWPE-1, BPH-1) and evaluation of anti-metastatic targets in invasive PTCs (PC3). Given that BPH is considered to be a metabolic disorder27, this protocol also paves the way towards our improved understanding of the link between metabolic disorders and increased prostate cancer risk. Given that BM stiffness induced by its exposure to AGEs may be a trigger for invasiveness in other cancer types, it will be of interest to use the protocol to set up similar models that incorporate normal epithelial cells and tumor cells from other organs (e.g. breast, colon, ovaries, pancreas).
Critical steps within the protocol, together with their timings, are summarized in Figure 4. During the initial step it is essential to maintain the stock solution of rBM at 4 °C while it thaws to prevent its polymerization. Pipette tips should not be placed into the rM stock solution until they have been chilled to 4 °C. For the next step it is also important to ensure the chamber slides have equilibrated to 4 °C before they are coated with the rBM solution. As soon as the temperature of the rBM solution is increased above 4 °C it will undergo irreversible polymerization to form a gel. It essential that the rBM is not disturbed during the polymerization stage to ensure that it forms an even surface suitable for cell culture and microscopic analysis. The duration of incubation with GLA with or without inhibitors of the Maillard reaction (sodium cyanoborohydride and amingoguanidine) will determine how stiff the rBM gel becomes. It is recommended to use a 6 hr incubation with GLA if semi-stiff conditions are required, and 14 hr incubation if stiff conditions are required (Table 1). Alternate incubation times or concentrations of GLA can be used if different levels of stiffness are desired. In this case rheological analysis of the rBM gels need to be incorporated as an essential step. Following the step of quenching the Maillard reaction by incubation with GEE and the subsequent washing steps with PBS, the rBM gels can be used immediately or stored at 4 °C for up to 48 hr prior to their use for cell culture. Once cell cultures are set up it is important to change the culture medium (including any treatments) every two days. It is recommended to maintain the 3D cell cultures for 3-12 days according to the parameters under investigation. For 3D PEC acini it is recommended to analyse the cultures after 6 days, and for 3D PTC spheroids analysis is recommended after 3 days of culture in the first instance.
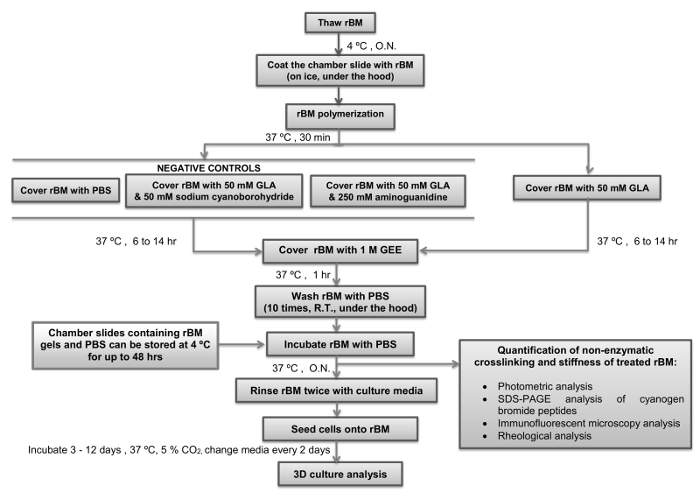
Figure 4: Simple Overview of the Protocol with Critical Steps and Timings Indicated. The flow diagram depicts how to prepare and stiffen the reconstituted basement membrane (rBM) with glycolaldehyde (Maillard reaction) with critical steps and timings indicated. Points where the protocol can be stopped, and rBM gels stored, are also indicated. rBM, reconstituted basement membrane; GLA, glycolaldehyde; GEE, glycine ethyl ester; O.N., overnight; PBS, phosphate buffered saline; R.T., room temperature. Please click here to view a larger version of this figure.
Divulgaciones
The authors have nothing to disclose.
Acknowledgements
We thank Simon Hayward (Vanderbilt University Medical Center) for the BPH-1 cells; and Thomas Cox and Janine Erler (Biotech Research & Innovation Centre, University of Copenhagen) for their assistance with rheological measurements. MR-T was funded by Worldwide Cancer Research, formerly The Association of International Cancer Research (Grant 08-0803 to JS), The British Embassy Montevideo and Agencia Nacional de Investigacion e Innovacion (UK_RH_2015_1_2 to MR-T). MC was supported by Prostate Cancer UK (Grant S14-017 to JS and GS). KW was funded by The China Scholarship Council. MAM was funded by The Saudi Arabian Cultural Bureau.
Materials
| I – Material for monolayer culture | |||
| BPH-1 | CaP Cell Line Database | PCaCL-132 | Contact: simon.hayward@mcmail.vanderbilt.edu |
| Complete keratinocyte serum-free media | ThermoFisher Scientific | 17005-075 | Do not warm at 37 ºC before use |
| Fetal calf serum | First Link UK Ltd | 02-00-850 | Store at -20 ºC in aliquots |
| PC3 | American Type Culture Collection | CRL-1435 | |
| Penicillin/streptomycin | ThermoFisher Scientific | 15070-063 | |
| Phosphate Buffer Saline (Dulbecco A) tablets | Oxoid | BR0014G | |
| RPMI 1640 medium | Sigma-Aldrich | R5886 | warm in 37 ºC water bath before use |
| RWPE-1 | American Type Culture Collection | CRL-11609 | |
| Trypsin-EDTA solution | Sigma-Aldrich | T4049 | |
| Name | Company | Catalog Number | Comments |
| II – Material for 3D culture | |||
| Acetonitrile | Sigma-Aldrich | 271004 | |
| Aminoguanidine hydrochloride | Sigma-Aldrich | 396494 | Irritating to eyes, respiratory system and skin |
| Chamber slides, 8-well | Thermo Scientific Nunc Lab-Tek | TKT-210-816M | |
| Culture Matrix reconstituted basement membrane (rBM) reduced growth factor extract | AMS Biotechnology | 3445-005-01 | Store Basement membrane (BM) at -80 ºC in aliquots |
| Cyanogen bromide | Sigma-Aldrich | C91492 | Toxic by contact skin and inhalation |
| Formic acid | Sigma-Aldrich | 695076 | |
| Glycine ethyl ester hydrochloride (GEE) | Sigma-Aldrich | 50060 | Irritating to eyes |
| Glycolaldehyde dimer (GLA) | Sigma-Aldrich | G6805 | |
| Sodium cyanoborohydride | Sigma-Aldrich | 71435 | Highly flammable; Toxic by contact skin and inhalation |
| Syringe filter 0.22 microns | Appleton Woods | BC680 | |
| Name | Company | Catalog Number | Comments |
| III – Material to quantify Maillard reaction | |||
| 4’,6-diamidino-2-phenylindole (DAPI) | ThemoFisher Scientific | D3571 | light sensitive and store at -20 ºC in aliquots |
| Cloning cylinders | Sigma-Aldrich | C1059 | |
| Goat anti-mouse IgG (H+L) Alexa Fluor 488 conjugate | ThemoFisher Scientific | A-11001 | light sensitive |
| Goat anti-rabbit IgG (H+L) Alexa Fluor 488 conjugate | ThemoFisher Scientific | A-11034 | light sensitive |
| Goat serum | Abcam | ab7481 | Store at -20 ºC in aliquots |
| Vectashield mounting media | Vector Laboratories | H-1000 | |
| Mouse anti-pentosidine clone PEN-12 mAb | TransGenic Inc | KH012 | |
| Paraformaldehyde (PFA) | Sigma-Aldrich | F8775 | Store at -20 ºC in aliquots |
| Rabbit anti-human collagen IV polyclonal antibody | Acris Antibodies | R1041 | Store at -20 ºC in aliquots |
| Rabbit anti-laminin A/C pAb | Santa Cruz Biotechnology Inc | sc-7292 | Store at -20 ºC in aliquots |
| Polyethylene glycol tert-octylphenyl ether (Triton-X100) | Sigma-Aldrich | T9284 | |
| Polyethylene glycol sorbitan monolaurate (Tween-20) | Sigma-Aldrich | P1379 | |
| Dialysis cassette Slide-A-Lyzer | ThemoFisher Scientific | 66333 | |
| Name | Company | Catalog Number | Comments |
| IV – Equipment | |||
| ARG2 controlled strain rotational rheometer | T.A. Instruments | ||
| Axiovert S100 (20x magnification) microscope | Zeiss | ||
| CO2 controlled humidified incubation chamber for Zeiss Axio S100 microscope | Solent Scientific | ||
| Confocal Axiovert 200M (40x, 63x magnification) microscope | Zeiss | ||
| Olympus LH50A microscope fitted with a digital camera using phase-contrast | Olympus | ||
| PHERAstar Plus plate reader spectrophotometer | BMG Labtech | ||
| Name | Company | Catalog Number | Comments |
| V – Software | |||
| Image J 1.47v | National Institute of Health, USA | ||
| MetaXpress | Molecular Divices |
Referencias
- Timpl, R., Brown, J. C. Supramolecular assembly of basement membranes. Bioessays. 18 (2), 123-132 (1996).
- Yurchenco, P. D., Ruben, G. C. Type IV collagen lateral associations in the EHS tumor matrix. Comparison with amniotic and in vitro networks. Am J Pathol. 132 (2), 278-291 (1988).
- Halfter, W., et al. Protein composition and biomechanical properties of in vivo-derived basement membranes. Cell Adh Migr. 7 (1), 64-71 (2013).
- Candiello, J., Cole, G. J., Halfter, W. Age-dependent changes in the structure, composition and biophysical properties of a human basement membrane. Matrix Biol. 29 (5), 402-410 (2010).
- To, M., et al. Diabetes-induced morphological, biomechanical, and compositional changes in ocular basement membranes. Exp Eye Res. 116, 298-307 (2013).
- Debnath, J., Muthuswamy, S. K., Brugge, J. S. Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods. 30 (3), 256-268 (2003).
- Bello, D., Webber, M. M., Kleinman, H. K., Wartinger, D. D., Rhim, J. S. Androgen responsive adult human prostatic epithelial cell lines immortalized by human papillomavirus 18. Carcinogenesis. 18 (6), 1215-1223 (1997).
- Hayward, S. W., et al. Establishment and characterization of an immortalized but non-transformed human prostate epithelial cell line: BPH-1. In Vitro Cell Dev Biol Anim. 31 (1), 14-24 (1995).
- Kaighn, M. E., Narayan, K. S., Ohnuki, Y., Lechner, J. F., Jones, L. W. Establishment and characterization of a human prostatic carcinoma cell line (PC-3). Invest Urol. 17 (1), 16-23 (1979).
- Laemmli, U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 227 (5259), 680-685 (1970).
- Towbin, H., Staehelin, T., Gordon, J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci U S A. 76 (9), 4350-4354 (1979).
- Rabilloud, T. Mechanisms of protein silver staining in polyacrylamide gels: a 10-year synthesis. Electrophoresis. 11 (10), 785-794 (1990).
- Rodriguez-Teja, M., et al. AGE-modified basement membrane cooperates with Endo180 to promote epithelial cell invasiveness and decrease prostate cancer survival. J Pathol. 235 (4), 581-592 (2015).
- Graf, B. W., Boppart, S. A. Imaging and analysis of three-dimensional cell culture models. Methods Mol Biol. 591, 211-227 (2010).
- Yao, N. Y., Larsen, R. J., Weitz, D. A. Probing nonlinear rheology with inertio-elastic oscillations. J Rheology. 52 (4), 1013-1025 (2008).
- Baker, A. M., Bird, D., Lang, G., Cox, T. R., Erler, J. T. Lysyl oxidase enzymatic function increases stiffness to drive colorectal cancer progression through FAK. Oncogene. 32 (14), 1863-1868 (2013).
- Caley, M. P., et al. Tumor-associated Endo180 requires stromal-derived LOX to promote metastatic prostate cancer cell migration on human ECM surfaces. Clin Exp Metastasis. 3 (2), 151-165 (2016).
- Sturge, J., Wienke, D., East, L., Jones, G. E., Isacke, C. M. GPI-anchored uPAR requires Endo180 for rapid directional sensing during chemotaxis. J Cell Biol. 162 (5), 789-794 (2003).
- Sturge, J., Wienke, D., Isacke, C. M. Endosomes generate localized Rho-ROCK-MLC2-based contractile signals via Endo180 to promote adhesion disassembly. J Cell Biol. 175 (2), 337-347 (2006).
- Rodriguez-Teja, M., et al. Survival Outcome and EMT Suppression Mediated by a Lectin Domain Interaction of Endo180 and CD147. Mol Cancer Res. 13 (3), 538-547 (2015).
- Paszek, M. J., et al. Tensional homeostasis and the malignant phenotype. Cancer Cell. 8 (3), 241-254 (2005).
- Levental, K. R., et al. Matrix crosslinking forces tumor progression by enhancing integrin signaling. Cell. 139 (5), 891-906 (2009).
- Carson, W. C., et al. Material characterization of ex vivo prostate tissue via spherical indentation in the clinic. Med Eng Phys. 33 (3), 302-309 (2011).
- Hoyt, K., et al. Tissue elasticity properties as biomarkers for prostate cancer. Cancer Biomark. 4 (4-5), 213-225 (2008).
- Krouskop, T. A., Wheeler, T. M., Kallel, F., Garra, B. S., Hall, T. Elastic moduli of breast and prostate tissues under compression. Ultrason Imaging. 20 (4), 260-274 (1998).
- Zhang, M., et al. Quantitative characterization of viscoelastic properties of human prostate correlated with histology. Ultrasound Med Biol. 34 (7), 1033-1042 (2008).
- Corona, G., et al. Benign prostatic hyperplasia: a new metabolic disease of the aging male and its correlation with sexual dysfunctions. Int J Endocrinol. , (2014).

