Super-resolution Imaging of the Cytokinetic Z Ring in Live Bacteria Using Fast 3D-Structured Illumination Microscopy (f3D-SIM)
Summary
Spatiotemporal information about dynamic proteins inside live cells is crucial for understanding biology. A type of super-resolution microscopy called fast 3D-structured illumination microscopy (f3D-SIM) reveals unique information about the cytokinetic Z ring in bacteria: both its bead-like appearance and the rapid dynamics of FtsZ within the ring.
Abstract
Imaging of biological samples using fluorescence microscopy has advanced substantially with new technologies to overcome the resolution barrier of the diffraction of light allowing super-resolution of live samples. There are currently three main types of super-resolution techniques – stimulated emission depletion (STED), single-molecule localization microscopy (including techniques such as PALM, STORM, and GDSIM), and structured illumination microscopy (SIM). While STED and single-molecule localization techniques show the largest increases in resolution, they have been slower to offer increased speeds of image acquisition. Three-dimensional SIM (3D-SIM) is a wide-field fluorescence microscopy technique that offers a number of advantages over both single-molecule localization and STED. Resolution is improved, with typical lateral and axial resolutions of 110 and 280 nm, respectively and depth of sampling of up to 30 µm from the coverslip, allowing for imaging of whole cells. Recent advancements (fast 3D-SIM) in the technology increasing the capture rate of raw images allows for fast capture of biological processes occurring in seconds, while significantly reducing photo-toxicity and photobleaching. Here we describe the use of one such method to image bacterial cells harboring the fluorescently-labelled cytokinetic FtsZ protein to show how cells are analyzed and the type of unique information that this technique can provide.
Introduction
A number of different imaging technologies have been used over the years to study bacteria including various electron and optical microscopy techniques. Whilst electron microscopy offers extremely high resolution, down to the nanometer, the need for a vacuum and use of contrast agents has limited this technique to fixed and embedded specimens. Artifacts can also be introduced due to lengthy sample preparation and a skilled practitioner is needed to prepare the samples, and to analyze and interpret the resulting images. Optical microscopy offers the benefits of simpler sample preparation and the application of multiple labels with relative ease compared with electron microscopy. Using fluorescent proteins and dye conjugates, the ability to study live cells with fluorescence microscopy is also possible. With improvements in fluorophore stability and fluorescent protein size/structure leading to decreased cell toxicity, as well as advances in camera detection technology, fluorescence microscopy using wide-field and confocal imaging systems has significantly expanded our understanding of the spatial dynamics of cells. Using these techniques, our knowledge of the bacterial cell over the last 20 years has progressed greatly to a far more detailed view that reveals a high level of structural and protein organization within the bacterial cell1.
However, conventional methods of optical microscopy are diffraction limited, meaning that the inherent resolution is about half of the wavelength of the excitation light used. Consequently, even with the most optimal conventional optical microscopy system, the best lateral resolution is about 220-250 nm and the axial resolution is about 500 nm (for review see Schermelleh et al.2). For bacterial cell biologists in particular, this still severely limits our understanding of the spatial organization of these tiny cells; being only 1-2 µm in diameter and 1-10 µm in length. There is also the need for higher resolution techniques that can be performed on live cells where dynamic spatial behavior of a protein can be analyzed to complement in vitro data.
Over the past decade, several super-resolution fluorescence imaging techniques have been developed. Super-resolution microscopy describes imaging techniques that at least double the lateral resolution obtainable with conventional light microscopy, thereby overcoming the diffraction barrier. These techniques manipulate the illumination and/or analysis of the emitted light to create real increases in the apparent resolution. There are currently three main types of super-resolution techniques – i) stimulated emission depletion microscopy3 (STED); ii) single-molecule localization microscopy, including techniques such as photoactivation localization microscopy4,5 (PALM), stochastic optical reconstruction microscopy6 (STORM), direct STORM7 (dSTORM), and ground state depletion with individual molecule return8 (GDSIM); and iii) structured illumination microscopy9-12 (SIM). Single-molecule localization techniques offer the largest increases in lateral resolution, down to between 10-40 nm in biological samples. In many implementations of single molecule localization microscopy, depth of sampling is limited to the evanescent wave and the initial implementations of STED were also limited in the sampling depth. However, recent advancements such as the use of adaptive optics, exploitation of the astigmatism in the point spread function at different focal points, multi-plane detection and using two objectives to infer the axial position of the emitted light have seen improvements in both the axial resolution and sampling depths in both STED and single molecule localization13,14. Data acquisition rates for STED and single-molecule localization techniques are also relatively slow due to the large number of images that need to be acquired for maximal resolution (up to 50,000 for localization techniques). This slow data acquisition does not lend itself to the imaging of live samples without custom modifications that are often quite expensive. These two techniques are currently optimally suited for fixed samples (for review see 2,15), however, much work is being done to address this limitation.
Three dimensional structured illumination microscopy (3D-SIM) is a wide-field technique that improves both lateral and axial resolution resulting in resolutions of approximately 110 and 280 nm, respectively. Depth of sampling is up to 30 µm from the coverslip, allowing for imaging of whole cells. The variation of 3D-SIM developed by Sedat, Agard, and Gustaffsson on the Optical Microscope eXperimental (OMX) platform involves illuminating the sample with a known grid pattern which is moved into 15 different positions in each z plane9-11. The fringes in the resulting Moiré patterns that are created in frequency space outside the observable region are measured. The information from the frequency space is computationally reconstructed to generate a visible super-resolution image10,11. The relative speed at which the raw images can be acquired using this technique enables imaging of live cells. However, it is important to note that for biological processes occurring on a time-scale of seconds, the acquisition rate of the raw images is still too slow to capture dynamic changes. For time-series with a capture rate of seconds, the repeated acquisition without sufficient recovery time can lead to phototoxicity and photobleaching, especially during live imaging of small bacterial cells.
To overcome these problems, recent advancements for fast 3D-SIM (f3D-SIM) have been developed. Here we describe one known as OMX Blaze16 that was introduced in late 2011. This technology creates the patterned illumination without the use of a physical grid as was used in earlier systems. The use of interfering light beams and new shutter technology allows for the creation of the patterned light on the sample in a very rapid fashion. The speed of image acquisition was further enhanced with the introduction of scientific complementary metal oxide semiconductor (sCMOS) cameras, especially when compared to the existing EMCCD cameras. These changes resulted in a possible image acquisition rate of one frame per second for a sampling depth of 1 µm (512 x 512 pixels, 9 z-slices), allowing monitoring of dynamic changes within cells that occur within seconds16. The decrease in the apparent exposure (excitation) times to live cells using this method allows either extended time series to be captured or an increase in the capture rate.
Here we describe the application of f3D-SIM microscopy to examine the structure and dynamic movement of the cytokinetic Z ring in the cells of two bacterial species: the rod-shaped model organism Bacillus subtilis and the coccoid human pathogen Staphylococcus aureus. FtsZ is a tubulin-like cytoskeletal protein that plays a key role in cell division in bacteria17,18. It localizes to the division site to recruit at least 20 other division proteins, forming the division apparatus17,18, and is believed to provide the force for cell constriction19-23. This method reveals that FtsZ is heterogeneously distributed around the Z ring in both organisms in a bead-like arrangement; solving the long-held issue of whether the Z ring is a continuous uniform belt around the cell, as visualized by conventional wide-field fluorescence microscopy, or a discontinuous structure as suggested by electron cryo-tomography (ECT)24. We show how the ability of 3D-SIM can vastly improve spatial examination of tiny (1 µm diameter) round cells like S. aureus that divide in three consecutive perpendicular planes (x,y,z). Time-lapse studies using these techniques show that the structure of the Z ring is dynamic, with gross organization changes within the ring, rather than FtsZ molecules moving in and out of a fixed scaffold24.
Protocol
1. Sample Preparation
- Preparation of B. subtilis Cells for Imaging
- Inoculate 10 ml of Penassay Broth (PAB, also known as Antibiotic Medium 3) with a single colony of B. subtilis strain SU570 (168 trpC2 ftsZ::ftsZ-gfp-spec)16. Grow overnight in a low speed (100 rpm) shaker at 30 °C.
- Dilute the overnight culture to 0.05 A600 (absorbance measured at 600 nm) in fresh PAB at 30 °C with high speed (215 rpm) shaking and grow until mid-exponential phase (A600 ~ 0.4).
- Add 2 g of electrophoresis-grade agarose in 100 ml 1x growth medium (PAB) in a screw-capped glass bottle. Dissolve the agarose by autoclaving. This can be stored at 4 °C for a few months, and microwaved to melt the agarose when required. Allow to set at room temperature.
- Attach an adhesive frame (65 µl capacity; see Table of Materials) on to a standard glass microscope slide. To do this, remove the thin polyester sheet (each adhesive frame is positioned between two polyester sheets, one thin and one thick, and has the center square removed) and apply the exposed adhesive surface of the frame to the surface of the microscope slide. This effectively creates a square well on top of the slide. Leave the thick polyester sheet attached to the frame as this will prevent the coverslip sticking to it in subsequent steps.
- Melt the agarose solution in a microwave until boiling and pipette 67 µl into the adhesive frame. Immediately place a coverslip on top to create a flat surface and leave at room temperature for 5-10 min. Remove the coverslip for 1-2 min while the sample is harvested.
- Harvest a 1 ml aliquot of the sample and centrifuge at 5,900 x g for 30 sec. Decant the supernatant and resuspend the pellet in 200 µl fresh PAB.
- Take 2.5 µl of the concentrated sample and pipette onto a pre-prepared agarose pad. Spread the sample evenly along the flat surface of the agarose pad. Remove the adhesive frame from the microscope slide using tweezers while not touching the agarose pad.
- Transfer the agarose pad from the microscope slide to a 35 mm diameter Petri dish with a glass coverslip bottom. Invert the agarose pad so that the cell suspension and coverslip of the Petri dish are in direct contact with each other. The bottom of the Petri dish has a refractive index of 1.525 for high resolution imaging.
- Preparation of Live S. aureus Cells for Imaging
- Inoculate 10 ml of L-broth with a single colony of S. aureus strain SA94 (SH1000 containing pLOW-FtsZ-GFP and pGL485; Ermr, Cmr)25. Grow overnight in a low speed (100 rpm) rotary shaker at 37 °C.
- Dilute the overnight culture to 0.05 A600 (absorbance measured at 600 nm) in fresh L-broth with 0.05 mM IPTG (isopropyl β-D-1-thiogalactopyranoside) to induce production of the fusion protein.
- Incubate at 37 °C with high-speed (215 rpm) shaking and grow until mid-exponential phase (A600 ~ 0.4).
- Follow steps 1.1.3 – 1.1.8 as described for B. subtilis live-cell preparation.
Note: Add the appropriate concentration of inducer to the agarose solution in step 1.1.3 for strains that express a fluorescent fusion protein under inducible control.
2. Preparation of the Microscope
This method uses a DeltaVision OMX (Optical Microscope, eXperimental) 3D-SIM imaging system fitted with a Blaze module. For the experiments described, only one laser and one camera are used. The illumination and light path are described in Figure 1. This method assumes that the microscope has appropriate calibration, optical transfer function (OTF) files and light collimation maintenance performed.
- For live cell imaging experiments, make sure the heating stage is attached to the ceramic stage and the objective-heating collar is attached to the objective (60X 1.42 NA oil objective).
- Turn on the objective and stage heaters at least 4 hr prior to imaging and slowly ramp the temperature to the required setting at a maximum rate of 5 °C/hr. It is recommended to start this process the evening prior to imaging and ramp at 3 °C/4 hr. For live cell experiments with B. subtilis, most experiments are performed at 30 °C. For S. aureus, all experiments are performed at 37 °C.
- On the day of the experiment, turn the system on at least 1 hr prior to imaging to allow the system and lasers to fully stabilize. Load the small dish stage insert onto the heated stage to preheat.
- Turn on the master controller workstation.
- Turn on the image processing workstation.
- Turn on the camera cooler located on the floor outside the microscope enclosure.
- Turn on all of the sCMOS cameras (even though they will not all be used).
- Inside the laser and electronics enclosure, turn on:
- Instrument controller chassis.
- Nanomotion chassis.
- Jena controller for Z piezo.
- All 4 galvo motor controllers.
- Primary laser control module.
- Secondary laser control module.
- All camera workstations.
- OMXIC workstation.
- On the master controller workstation, open the OMX software by clicking the icon. Set the file path for data saving to an appropriate data folder.
- To initialize the hardware, go to the Hardware menu, choose Hardware and click the Restart Hardware button. Close window.
- Start the softWoRx software.Turn on the lasers required for the experiments (488 nm).
- Turn on the key switch for the laser safety interlock.
- Ensure the correct dichroic filter drawer is inserted below the objective. For GFP, it is the standard (or fixed) drawer.
- In OMX SF, go to the File menu, choose Settings and select the Fixed drawer from the Drawer dropdown menu. Close window.
- Do the following from the main screen of the software, in the Light Parameters section:
- Activate the appropriate camera for the excitation laser and emission filter (FITC) camera from the Channel selection.
- Check the Image Mode is set to Sequential.
- Check the Light Path is set to SI.
- Check the Mode is set to 95 MHz.
- Check the 488 laser is selected in Excitation.
- Set the Size a 512 x 512 for the window size and Binning a 1 x 1.
- In the Experiment tab, ensure Tipo is set to SI from the dropdown menu.
- Ensure Sectioning is activated and that the Optical section spacing is set to 0.125.
- Ensure Focus point when scan starts is middle.
3. Image Acquisition
- Apply a drop of oil to the top of the objective. For 37 °C, the recommended starting oil has a refractive index of 1.518 to best match the refractive index of the gel pad used to mount the bacterial cells at 37 °C (refer to step 1.1.3).
- Place the sample Petri dish containing the prepared cells into the stage inset and cover with the circular heating lid. Lower the stage until the oil touches the bottom of the sample Petri dish.
- Allow the dish to heat-equilibrate for 15 min in the stage. Using a low Exposure setting of 5 msec or less (located in the Light Parameters section), focus on the sample using the nano positioning stage controls (dZ, located in the Nano Positioning section). Focus up and down through the sample to determine if the light is spread evenly above and below the sample focal plane. If the point spread function is not even (spherical aberration), clean the bottom of the sample dish and the objective with chloroform. Change the oil and repeat this step until a suitable match is found between the oil and sample refractive index to minimize spherical aberration.
- Using 0.5 µm dZ step sizes, mark the top and bottom of the image stack. Return to the middle of the sample. Click the Get Thickness button in the Experiment tab to set the sample acquisition thickness. Keep the stack height to a minimum whilst being careful to not cut off the top and bottom of the Z-rings. Typical thicknesses for B. subtilis samples are 2-3 µm.
- Determine the maximum intensity value (Max) of the sample image with the current Exposure and %T settings. This value is found below the image window. For time-lapse imaging, adjust the Exposure and %T to obtain a maximum intensity of 1,100-1,400 above background for the image. A minimum intensity of 1,000 above background is needed to obtain image reconstruction. For a one-off image, increase the maximum intensity to 4,000-5,000.
- Activate the Time-lapse section in the Experiment tab. Set the required frame rate and total time. In Data File, enter an appropriate file name. Click the Run button to begin the image acquisition.
Note: Image acquisition is complete when the green Progress bar is finished and a log file appears for the data set in the appropriate data folder.
4. Checking Data Quality
- At the start of the image acquisition, note the maximum intensity value of the image at the beginning of the image acquisition for the first frame of the time series. At the end of the acquisition of the first frame, check that there has not been more than a 10% decrease in signal intensity through the z-stack for this first frame. If there has been more than this level of photobleaching in the first time point, the data through the time series will be of insufficient quality for analysis and the times series acquisition can be stopped.
- At the end of the acquisition of the time series, open the resulting raw image file with .dv suffix and ascertain the decrease in maximum signal intensity from the beginning of the time series to the final image. Discard any time points where there has been more than 30% photobleaching.
5. Reconstructing 3D-SIM images
- From the Process menu, activate the SI Reconstruction window. Use the pre-existing settings for all of the parameters.
- Activate the Use Channel Specific OTF button, and set to the Standard filter set. Activate the Use Channel Specific KO angles button and set to the Standard filter set.
- Drag and drop the raw (.dv) file into the Input file space. Click Do it. The output file will have the same name as the input file with _SI added at the end.
Note: This reconstruction process can be run as a task using the Task Builder function from the Process menu if multiple files are to be processed.
6. Data Export and Analysis
- Use Imaris to visualize 3D images in surpass mode and export image data as tiff (300 dpi) or avi files (no compression). Size measurements in Imaris can be performed using the measurements tool in surpass mode.
- Generating 3D intensity plots of the Z ring from 3D-SIM image data:
- Examine the distribution of FtsZ around the Z ring and to create 3D intensity plots
- Open a 3D image file to view the individual z-slices. Use the volume viewer tool located under the view menu tab to crop a region of interest.
- Enter the window number for this 3D image into the input window and enter a new and unused window number into the output window. The select region button will become available to crop an individual Z ring of interest from the original 40 × 40 μm field of view.
- Adjust the desired axis and degree of rotation in the rotation tab. Click the Do It button. Scroll through the volume to obtain the desired perspective of the Z ring.
- Use the data inspector tool and adjust the column/row dimensions to create a new region of interest around an individual Z ring. Click the center of the image to position this new region of interest. The text image data is an automatically generated table that displays the fluorescence intensity of each pixel within the region of interest.
- Click the 3D graph button to initially view the intensity plot.
- Alternatively the text image data can be exported by clicking the save table data button in the file menu.
- Copy the image data from the saved text file into a new spread sheet. Select all the cells within the spread sheet and create a new 3D-Surface graph. Format the 3D intensity plot graph for publication.
- For time-lapse data repeat steps 6.2.1.1 – 6.2.1.7 on each of the different time-points from the 3D image file.
- Examine the distribution of FtsZ around the Z ring and to create 3D intensity plots
- Tracking Average Fluorescence Intensity Over Time
- Use the text image data to track the fluorescence intensity over time in a region of interest.
- Select the cells which correspond to the region of interest.
- Calculate the average fluorescence intensity at each time point for this region of interest.
- Use the average fluorescence intensity values to generate a separate line graph.
Representative Results
In this study we visualized the bacterial cell division protein FtsZ, expressed in live cells as an FtsZ-GFP fusion, to illustrate the powerful advantages of f3D-SIM over conventional fluorescence microscopy for studying bacterial protein localization and dynamics. FtsZ is a tubulin-like cytoskeletal protein that polymerizes into a dynamic ring structure around the circumference of the cell. The Z ring has an important function in cell division: its assembly marks the future site of division, it recruits all cell division proteins to this site, and Z ring constriction appears to provide the force to allow inward movement of the cell envelope during cytokinesis17,18.
When visualized by conventional wide-field fluorescence microscopy, the Z ring appears as a single transverse band of fluorescence in rod-shaped bacteria, including the most well studied organisms such as B. subtilis (Figure 2A), E. coli, and C. crescentus. Importantly, the fluorescence intensity throughout this band appears to be more or less uniform, and the localization pattern offers very little insight into the structural organization and distribution of FtsZ polymers within the Z ring. When the same cells are examined by using the f3D-SIM method described here (Figure 2B), however, the advantages of this technique are immediately apparent. First, rotation of the 3D-SIM image around the z-axis enables visualization of the Z ring as a genuine ring structure in three dimensional space (Figures 2C and 3A). Together with the increase in resolution, this reveals that the distribution of FtsZ in the Z ring is in fact highly heterogeneous (Figure 3). The improvement in resolution also allows us to see Z rings that have begun the process of constriction to form a division septum (indicated by the invagination of cell membrane in Figure 2C and 2D).
Additional examples of B. subtilis Z rings visualized by this technique are shown in Figure 3Bi – 3Biv and illustrate how the fluorescence intensity around the Z ring is never the same. Moreover, regions of very low fluorescence intensity are often observed. We refer to these regions as gaps (white arrowheads). We observe these gaps in 15% of all Z rings examined (n = 84) and predominantly in Z rings having a diameter of 800 – 900 nm (93%). To quantify these observations, 3D intensity plots were created of the Z ring and are shown in Figure 3Ci and 3Cii using the Z rings shown in Figures 3Biii and 3Biv. The intensity plots show that typically the amount of fluorescence in the Z ring can vary up to 3 – 4 fold in different regions of the B. subtilis Z ring. Furthermore the 3D intensity plots show that the gaps of fluorescence inside the Z ring almost read baseline levels of background fluorescence (black arrow in Figure 3Ci). These above examples depict good reconstructions of the Z ring and are dependent upon sufficient light being emitted from the sample without significant photobleaching. This is accomplished through the brightness of the GFP fluorophore fused to FtsZ and its abundance in the cell under these conditions (high signal to noise ratio). In other cells where the signal to noise ratio is low the reconstruction of the image is poor. To simulate this occurrence with FtsZ-GFP, the amount of emission signal collected from the B. subtilis cells was reduced by lowering the excitation energy. As shown in Figure 3D a 100 fold reduction of excitation energy results in an image of the Z-ring that has significant artifacts. This occurs as a consequence of insufficient local contrast between the FtsZ-GFP signal and the background leading to poor reconstruction of the image.
Interestingly, B. subtilis Z rings showed more pronounced gaps and heterogeneity in the top and bottom regions of the ring but not in the sides of the ring (Figure 3B). This occurs because of the difference in resolution achieved by this variation of 3D-SIM in the lateral plane compared to the axial plane. This results in the top and bottom regions of the Z ring (lateral plane) being imaged with optimal resolution. The gaps in the Z-ring are too small to be detected in the sides of the Z ring as visualized in the axial plane, which has about half the resolution of the lateral plane. Bacteria that have a coccoid morphology, such as S. aureus, have Z rings which are positioned in all planes. So imaging the Z ring when it lies in the lateral plane (or close to it) provides the ability to image all regions of the ring with optimal resolution. Taking advantage of this, we examined the structure of the Z ring in live S. aureus cells, utilizing a strain that contains a plasmid-bearing an ftsZ-gfp fusion. Production of this fusion is under the control of a Pspac inducible promoter (see step 1.2.1). When imaged with in the axial plane, S. aureus Z rings appeared identical to B. subtilis Z rings which are present in the same plane (Figure 4Ai). However, imaging Z rings in the lateral plane confirmed that the entire Z ring appeared both bead-like and heterogeneous. About 26% of these rings also showed at least one visible “gap” of fluorescence (Figure 4Aii, 4B). Similar to B. subtilis FtsZ, fluorescence intensity plots show that the intensity of fluorescence in the Z ring varies as much as 4 – 6 fold in different areas of the S. aureus Z ring (Figure 4C).
The length and width of the beads can be measured (refer to schematic in Figure 4D). This showed that 80% of the Z rings (n = 43) that were examined had a bead length of 200 nm, reaching a maximum length of 400 nm. The width of beads, on the other hand, measured up to 200 nm. There was on average 12 ± 2 (SEM) FtsZ beads per Z ring. Similar localization data was observed when native FtsZ was visualized with these methods using immunofluorescence microscopy (IFM) showing that this heterogeneous and bead-like localization pattern was genuine and not an artifact caused by expression of ftsZ-gfp in addition to native FtsZ (data not shown). This advancement of 3D-SIM is able to show that the Z ring appears as a heterogeneous structure and is possibly discontinuous in nature.
To address the question of how this heterogeneous Z-ring structure undergoes constriction and facilitates cell division, a time-lapse analysis of Z ring dynamics can be performed. One of the main challenges with fluorescence microscopy time-lapse studies is in the minimization of sample photobleaching and phototoxicity during imaging. 3D-SIM requires a large number of raw images to be obtained which means that photobleaching of a sample over time will result in a poor signal-to-noise ratio leading to insufficient signal to allow appropriate image reconstructions. The application of the new technology minimizes the amount of excitation energy used for each exposure and hence a decrease in the energy entering the live cells. It does this by a combination of light beam interferometry to create the structured pattern upon the sample and the use of scientific CMOS cameras which increase sensitivity and quantum efficiency. This combination results in z-stack acquisition of 1 µm per sec (512 × 512 pixels x 8 z-slices) compared to 1 µm per 5-8 sec (512 x 512 pixels x 8 z-slices) obtained with our previous iteration of 3D-SIM (described in Riglar et al.26). Decreasing the image acquisition time and exposure settings can also help minimize phototoxicity of live cells which is particularly relevant in time-lapse studies. The new technology presented here also addresses another problem in live-cell imaging that we refer to as ‘motion blur’ which in this context refers to the localization of proteins changing during image acquisition. By decreasing the image acquisition time the amount of ‘motion blur’ is minimized thus creating a more accurate image reconstruction.
Previous studies that have addressed how FtsZ is organized within the Z ring have not been able to show how Z ring structure changes over time. To investigate this aspect we performed time-lapse using f3D-SIM. This enabled acquisition of a 3D-SIM image of the Z ring in B. subtilis every 5-10 sec over a period of 50 sec, which was previously not possible on this time scale. Shown in Figure 5A is an example of the rapid changes in FtsZ distribution within the Z ring over time (diameter of 890 nm). Other Z rings that were visibly in the process of constriction were also shown to have similar dynamics that affected the distribution of FtsZ around the Z ring (not shown). 3D intensity plots can also be generated for each of the time points to quantify and appreciate the continual fluorescence intensity changes and thus relative abundance of FtsZ in different regions of the Z ring on a time scale of seconds (Figure 5B). Regions of interest can be identified from these intensity plots to track the fluctuations of fluorescence intensity (Figure 5C). Tracking these changes in the Z ring structure under these conditions would be virtually impossible without the advanced f3D-SIM technology. In addition, this work revealed that in S. aureus the dynamic movement of FtsZ is similar to that observed in B. subtilis. S. aureus RN4220 cells producing FtsZ-GFP were imaged for 50 sec at 10 sec time intervals. Changes to the heterogeneity of the Z ring in S. aureus with f3D-SIM were also similar to B. subtilis (see arrowheads and arrows in Figure 6). These time lapse studies support a model of Z ring constriction (the iterative pinching model) that was predicted through examination of fixed C. crescentus cells, but could not be demonstrated due to the need to examine Z-ring dynamics in live cells27.
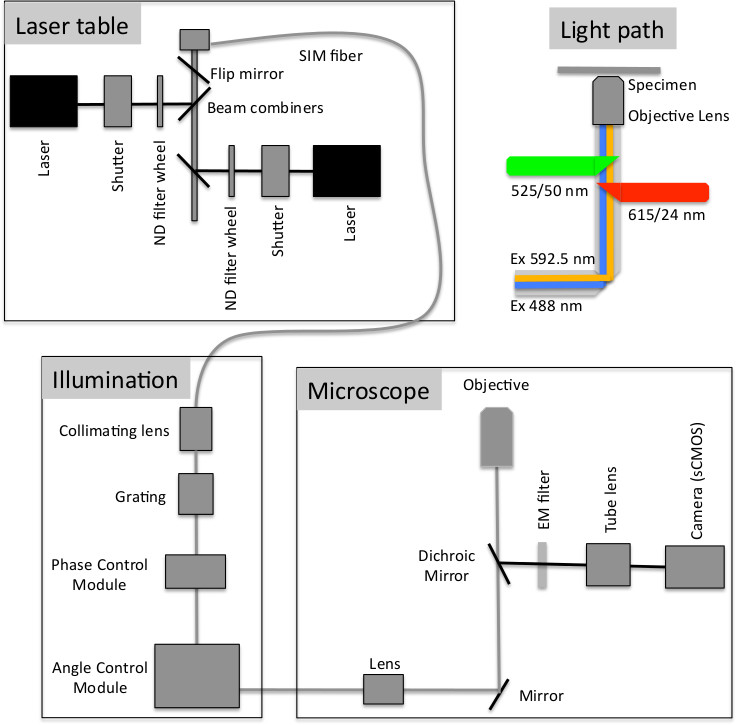
Figure 1. Schematic of the optical configuration of the f3D-SIM used in this study. Laser light from the laser table is directed through a SIM fiber to a collimating lens and fixed grating, then into the phase control module that creates both the wavelength specific optimal spacing of the ± 1st order beams from the zero order central beam and the phase steps for the SIM collection. The beams then enter the angle control module where the beams are reflected to the 3 different clusters to create the 3 angles of illumination. The beams then enter the microscope and light path as shown (Modified with permission from Paul Goodwin, API).
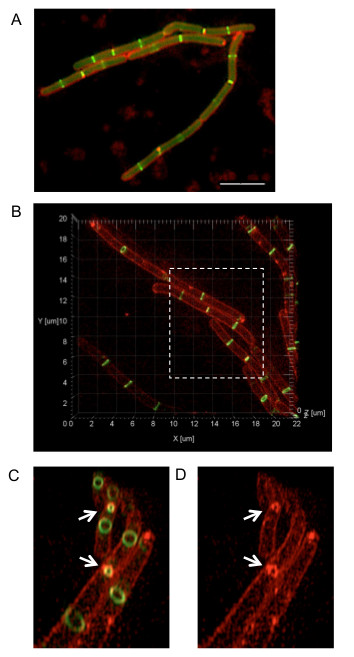
Figure 2. Comparison of conventional wide-field fluorescence and 3D-SIM imaging of B. subtilis cells expressing ftsZ-gfp. (A) Conventional wide-field fluorescence imaging of B. subtilis cells expressing ftsZ-gfp (SU570; green) as a single copy were also stained with the membrane dye FM4-64 (red). Scale bar = 5 µm. The image was acquired using an upright fluorescence microscope. (B) 3D-SIM imaging of the same B. subtilis strain expressing ftsZ-gfp, stained with FM4-64. Regions of interest from the image can be selected (dashed box) to zoom in. The image can also be rotated around the z-axis to view 3D FtsZ rings in the axial plane. (C, D) The removal of out-of-focus light and improved image resolution provided by 3D-SIM allows clear visualization of Z rings (including those that are constricting) and the inner cell membrane during division (indicated by white arrows). Note that the protocol text describes the visualization of a single fluorophore. However, the procedure can be adapted for acquiring up to four different wavelengths simultaneously. This figure has been reprinted from Strauss et al16.
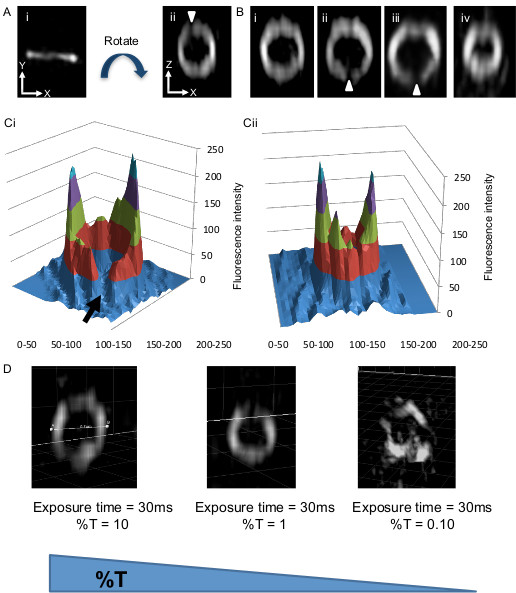
Figure 3. Representative images of the Z ring in live rod-shaped bacterial cells (B. subtilis) expressing FtsZ-GFP using 3D-SIM. (Ai) The Z ring is observed as a band of fluorescence perpendicular to the long axis of the cell which is in the x-y plane. (Aii) The image has been rotated around the z-axis using Imaris 3D visualization software (refer to protocol step 6.1) so that one is looking through the ring structure to clearly see how FtsZ is distributed within the Z ring. This image, now in the z-x plane shows that the ring has a heterogeneous distribution of FtsZ with regions of no or very little fluorescence (white arrowheads). The image must be rotated to observe these ‘gap’ regions of fluorescence. Bi-Biv show further examples of rotated Z rings that now lie in the z-x plane. Z rings in (Bi–iii) have a diameter of ~0.9 µm. (Biv) shows a Z ring with a diameter of only 0.65 µm undergoing constriction. (Ci) This typical 3D intensity profile illustrates the fluorescence intensity (i.e. concentration) differences in FtsZ-GFP around the Z ring having a diameter of 0.85 µm (refer to protocol steps 6.1-6.3). The level of fluorescence in these gaps is low and approaches background fluorescence levels (black arrow). (Cii) A similar 3D intensity profile of a Z ring in the process of constriction. FtsZ-GFP concentration is also non-uniform ; diameter, 0.65 µm. Panels A, B, CI and Cii have been reprinted from Strauss et al16.
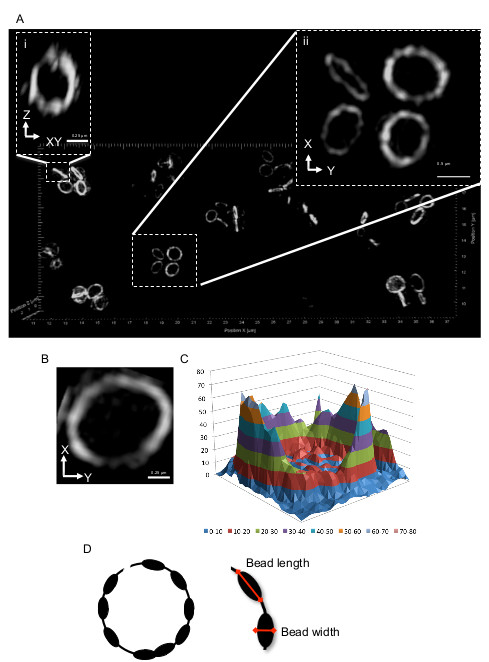
Figure 4. Representative 3D-SIM images of S. aureus cells producing FtsZ-GFP. Since these cells are round (cocci), the Z ring will have various orientations. Cells that show a band of fluorescence in the normal viewing orientation (that is, in the x-y plane) as shown in (Ai), look very similar to those in rod-shaped cells when rotated around the Z axis as for Figure 1Ai. In (Aii) the Z ring is already in the x-y plane; lateral orientation and resolution is maximal over the entire ring which now gives a bead-like structure (B). (C) A 3D intensity plot of the relative abundance of FtsZ-GFP in the Z ring. (D) Schematic representation of the S. aureus Z ring. Red arrows indicate bead length and width dimensions (refer to protocol step 6.1). This figure has been modified from Strauss et al16.
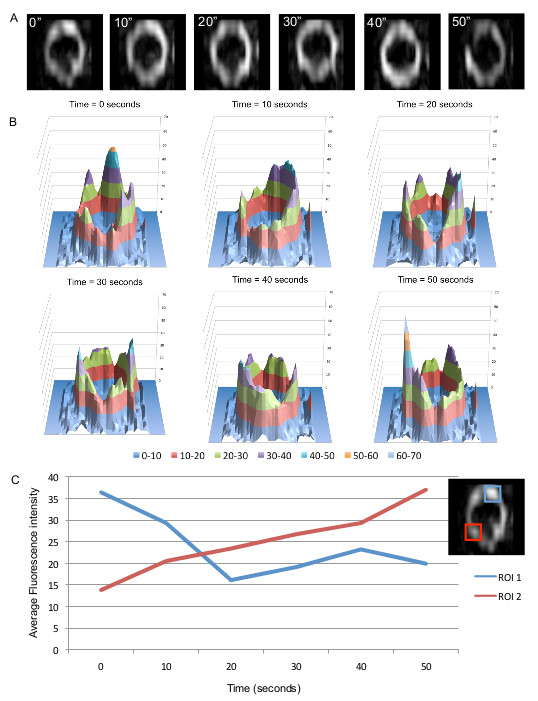
Figure 5. f3D-SIM allows fast analysis of FtsZ localization over time in 3D-SIM. (A) Changes in FtsZ-GFP distribution within the Z ring in rod-shaped B. subtilis cells can be visualized to indicate the dynamics of the ring. Time (sec) is indicated on the upper left corner of each image. (B) 3D fluorescence intensity plots show the non-uniform and dynamic distribution of FtsZ in the Z ring. (C) FtsZ dynamics (FtsZ-GFP fluorescence changes in the ring) can also be examined in more detail by quantifying fluorescence intensity over time in two regions of interest. This figure has been reprinted from Strauss et al16.
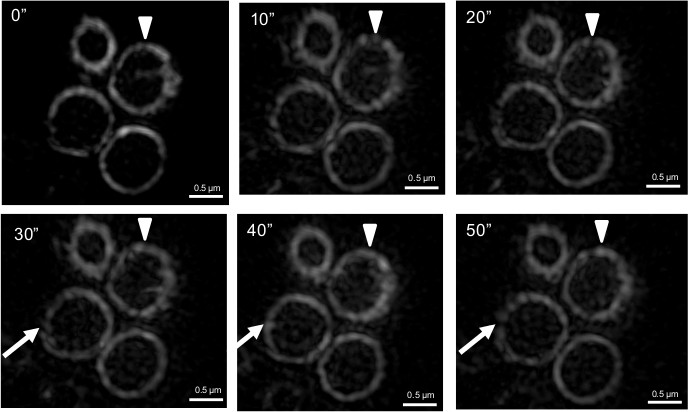
Figure 6. f3D-SIM time-lapse images show changes in FtsZ localization within the ring over time in S. aureus. A white arrowhead indicates a gap when it is initially detected in the Z ring. The appearance and disappearance of gaps at the same position (as marked by the white arrowhead) over time illustrates how FtsZ is redistributed around the Z-ring. White arrows indicate the formation of gaps in the Z ring at later time points. Time (sec) is shown on the upper left-hand side of the images. This figure has been modified from Strauss et al16.
Discussion
Fluorescent live-cell imaging is a powerful tool that enables observation of dynamic changes within cells over time. Live imaging of bacterial cell biology has been challenging because of the small cell size and reduced ability of the bacterial cells to withstand repeated exposure to excitation light. f3D-SIM has expanded the tools available to interrogate all cell biology but it is necessary to carefully perform this technique as artifacts can be introduced if poorly implemented. There are a number of important considerations for the successful use of this technique. As it is a wide-field fluorescence imaging method that relies on the use of the point spread function of the emitted light, refractive index mismatch and spherical aberration of the emitted light must be avoided to enable accurate image reconstruction. The use of coverslips of 0.17 mm thickness of a high quality to avoid variation of signal intensity due to glass thickness variability in the coverslip is highly recommended. It is critically important to carefully evaluate the spherical aberration of the emitted light during the initial stages of all experiments and adjust the immersion oil refractive index as needed. This may vary from day-to-day as it is dependent on both the temperature and the mounting media/substrate of the sample. The use of the incorrect oil will result in inferior reconstruction of the images with artifacts such as halos and echo signals.
At the end of the image reconstruction process, for each phase a measure of the quality of the reconstruction can be obtained by the “best fit for kO angle” for each phase/angle of the illumination. If this value is below 1 for each angle, then the quality of the reconstructed will most probably be high. If this value is above 6, it is most likely that the reconstructed image will contain artifacts. Common artifacts include i) the appearance of stripes in the image that correspond to one of the angles of the grid. This may result from lower signal from that grid angle; ii) haloing around structures. This may result from very uneven levels of signal from bright structures compared to surrounding structures, a mismatch of refractive index of the oil and the sample or may be due to the structures in the image being too amorphous and not containing enough “structure” to be recognized by the algorithm; iii) ‘honeycombing’ which results from a low signal to noise ratio in the image, usually due to high background signal; iv) structures that are stretched/elongated in xy which may be due to refractive index mismatch of oil and sample. This artifact is difficult to discern for an inexperienced user. It is recommended that new users consult an experienced SIM researcher for advice on image interpretation and analysis when starting to use this technique.
As with all fluorescent live-cell imaging experiments, it is important to carefully balance the excitation energy input in order to minimize the degree of photobleaching and phototoxicity of the cells with sufficient emission signal needed to reconstruct images with minimal artifact. Excessive photobleaching (greater than 30% across a single z-stack acquisition) will lead to a striping pattern in the reconstructed image. With the use of sCMOS cameras, super-resolution images can be successfully reconstructed from raw images with emission signal as low as 1,000 counts above background. This can allow for very short exposure times thereby reducing photobleaching and enabling rapid capture for each frame. It also increases the amount of time between frames for cells to recover from the excitation energy input. Additionally, as the capture time for each individual frame can be very short, the degree of artifact introduced by ‘motion blur’ during an individual capture is reduced with the use of this method.
This variation of 3D-SIM offers a number of other advantages over other available super- and high-resolution imaging technologies for live imaging by bacterial cell biologists. The 2 fold increase in resolution in all 3 dimensions and the ability to image up to 30 µm into a sample enables imaging of whole bacterial (and mammalian) cells during an experiment. Techniques such as single-molecule localization that are commonly performed in the evanescent wave cannot achieve the depth of penetration into a sample and do not offer information on whole cells. Recent advances that permit higher sampling depths for this technique may result in better axial resolution of whole cells. Additionally, single-molecule localization techniques that require thousands of raw images to be acquired are also limited in the frame rate that can be captured and may require compromise between capture frame rate and the ultimate achieved spatial resolution, as the spatial resolution is dependent on the number of events recorded within a defined space. Decreasing the number of captured frames will result in a lower spatial resolution but may be sufficient to achieve the temporal resolution required for the biological process of interest. The amount of recovery time between frames may also need to be increased to minimize phototoxicity in live cells thereby limiting the duration and frequency at which time-series can be obtained. High resolution techniques such as electron microscopy (EM) or electron cryotomography (ECT) offer increased resolution over 3D-SIM (and other super-resolution optical microscopy techniques) but must be performed on fixed samples. The fixing and processing may introduce artifacts and the lack of labeling may make interpretation difficult. Label-free ECT has poor contrast and can achieve sample penetration depths of around 500-200 nm24, so even if the protein of interest can be identified, the structure of the protein in a whole bacterial cell cannot be observed. Even when labeling can be used in EM, such as with immunogold, the imaging is only performed in 2D. 3D is only achievable with thin sectioning and computational reconstruction of the sections. The thin sectioning requires an experienced technician but can be problematic and lead to artifact. With live or fixed cells, 3D-SIM does not need to be embedded in a specific manner and cells can be rapidly prepared for imaging in a near native state.
This method is relatively easy to implement by researchers with minimal experience in fluorescent microscopy. Experiments can be performed in media that support healthy cell function as long as it does not fluoresce or generate undue background signal. For bacterial cells, this can allow the use of many types of complex and minimal media or the use of solid or semi-solid media to control bacterial cell motility. Many biologists who have already engineered fluorescent protein (FP) fusions and tested the functionality of these fusions can take up this technology rapidly without further engineering and validation with new photoactivatable FP fusions. We found, as a general observation, that S. aureus was more susceptible to photobleaching during imaging with FP fusions than B. subtilis. Using our original 3D-SIM methods, where the grid pattern was generated by a physical grating, we were unable to perform live cell imaging with a number of S. aureus FP fusions. However, with this advanced f3D-SIM method, we were able to image these FP fusions and perform short time series to capture the dynamics of these tagged proteins. This is most probably due to the reduced exposure times needed for imaging and increased time between frames for cell recovery.
OMX Blaze 3D-SIM is a commercially available technology that can be relatively easily implemented in a single laboratory or a core imaging facility. Care must be taken to perform the technique to avoid artifacts due to poor sample preparation or poor image capture. Once the technique is mastered, studies of protein structure and dynamics in small organisms such as bacteria can be performed in single or multi-color fluorescence.
Divulgaciones
The authors have nothing to disclose.
Acknowledgements
The authors thank P. Levin, S. Moriya, S. Foster, and M. Pinho for the gift of strains and antibodies. They also thank L. Ferrand, P. Goodwin, J. Cooper, and B. Dougherty for expert technical assistance. CBW is supported by an Australian National Health and Medical Research Council Senior Research Fellowship (571905). LT was supported by a UTS Chancellors Postdoctoral Fellowship. MPS and ATFL were supported by Australian Postgraduate Awards. This work was supported by an Australian Research Council Discovery Project Grant (DP120102010) to EJH.
Materials
| Name of Material/ Equipment | Company | Catalog Number | Comments/Description |
| Agarose I (Biotecnology grade) | AMRESCO | 0710-500G | Used for constructing 2% agarose pads. Use microwave to dissolve agarose in growth media |
| N-(3-Triethylammoniumpropyl)-4-(6-(4-(Diethylamino) Phenyl) Hexatrienyl) Pyridinium Dibromide (FM 4-64) | Life Technologies | T-13320 | Lipophylic dye used for staining cell membranes. |
| 1.5 ml microtube, graduated Natural | Scientific specialties Inc. | 1210-00 | Made from Homopolymer polypropylene, Used to harvest bacterial cells prior to being placed on agarose pads for visualization |
| Luria broth base, Miller | BD | 241420 | Prepared according to manufacturer's instructions. Add 2%?? after autoclaving |
| BBL Antibiotic assay broth, Antibiotic medium 3 | BD | 210932 | Prepared according to manufacturer's instructions. |
| Adhesive frame | Thermo-Fisher Scientific | ABGAB-0577 | Known as Gene Frame with this company. Used to prepare 2% agarose pads (Dimensions: 15x16mm) |
| Microscope cover glass | Thermo-Fisher Scientific | 111512-9 | 22×22 No 1 Thickness |
| 35 mm diameter petri dish with a glass coverslip bottom | World precision instruments | FD35-100 | Known as FluoroDish with this company. Dish: Ø35mm, Glass: Ø 23mm, Glass thickness: 0.17 mm, non pyrogenic |
| Chloramphenicol | Sigma-Aldrich | C0378-25G | Make a 10 mg/ml stock in 95% Ethanol |
| Lincomycin hydrochloride | Sigma | L6004-5MU | Make a 25 mg/ml stock in 50% Ethanol |
| Erythromycin | Boehringer Mannheim GmbH | 12390120-14 | Make a 10 mg/ml stock in 95% Ethanol |
| Combined Orbital/Linear Shaking waterbath | Grant scientific Instruments | OLS-200 | Shaking speed used for cultures: 215 rpm |
| UV visible Spectrophotometer | Shimadzu | UV-1601 | 600 nm wavelength was used for measuring Optical density of bacterial cultures |
| Centrifuge | Eppendorf | 5415R | Pellets of bacterial culture were obtained by spidding down mid exponentially growing cultures at 8000 for 30 seconds |
| Isopropyl β-D-1-thiogalactopyranoside (IPTG) | Sigma | 15502-10G | Prepare 1M stock and filter sterilze prior to use |
| softWoRx software | Applied Precision Inc/A GE Healthcare Company | version 5.0 | |
| Imaris data analysis software | Bitplane Scientific Software, Bitplane AG | version 7.0.0 | Matlab XTensions included |
| DeltaVision OMX 3D-SIM imaging system fitted with a Blaze module. | Applied Precision Inc/A GE Healthcare Company | Blaze module fitted on OMX V3 | |
| Upright fluorescence microscope | Zeiss Axioplan 2 |
Referencias
- Harry, E., Monahan, L., Thompson, L., & Kwang, W. J. Bacterial Cell Division: The Mechanism and Its Precison. Int Rev Cytol. 253, 27-94 (2006).
- Schermelleh, L., Heintzmann, R., & Leonhardt, H. A guide to super-resolution fluorescence microscopy. J Cell Biol. 190, 165-175, doi:10.1083/jcb.201002018 (2010).
- Hell, S. W., & Wichmann, J. Breaking the diffraction resolution limit by stimulated emission: stimulated-emission-depletion fluorescence microscopy. Opt. Lett. 19, 780-782 (1994).
- Betzig, E. et al. Imaging intracellular fluorescent proteins at nanometer resolution. Science. 313, 1642-1645 (2006).
- Hess, S. T., Girirajan, T. P. K., & Mason, M. D. Ultra-High Resolution Imaging by Fluorescence Photoactivation Localization Microscopy. Biophys j. 91, 4258-4272, doi:10.1529/biophysj.106.091116 (2006).
- Rust, M. J., Bates, M., & Zhuang, X. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM). Nat Meth. 3, 793-796, doi:10.1038/nmeth929 (2006).
- Heilemann, M. et al. Subdiffraction-resolution fluorescence imaging with conventional fluorescent probes. Angew. Chem. Int. Ed. (English). 47, 6172-6176, doi:10.1002/anie.200802376 (2008).
- Bretschneider, S., Eggeling, C., & Hell, S. W. Breaking the Diffraction Barrier in Fluorescence Microscopy by Optical Shelving. Phys. Rev. Lett. 98, 218103 (2007).
- Gustafsson, M. G. Surpassing the lateral resolution limit by a factor of two using structured illumination microscopy. J. Microsc. 198, 82-87 (2000).
- Gustafsson, M. G. L. et al. Three-Dimensional Resolution Doubling in Wide-Field Fluorescence Microscopy by Structured Illumination. Biophys J. 94, 4957-4970, doi:10.1529/biophysj.107.120345 (2008).
- Schermelleh, L. et al. Subdiffraction multicolor imaging of the nuclear periphery with 3D structured illumination microscopy. Science. 320, 1332-1336, doi:10.1126/science.1156947 (2008).
- Heintzmann, R., & Cremer, C. G. Lateral modulated excitation microscopy: Improvement of resolution by using a diffraction grating. Proc. SPIE. 3568, 185-195, doi:10.1117/12.336833 (1999).
- Gould, T. J., Burke, D., Bewersdorf, J., & Booth, M. J. Adaptive optics enables 3D STED microscopy in aberrating specimens. Opt. Express. 20, 20998-21009, doi:10.1364/OE.20.020998 (2012).
- Urban, Nicolai T., Willig, Katrin I., Hell, Stefan W., & Nägerl, U. V. STED Nanoscopy of Actin Dynamics in Synapses Deep Inside Living Brain Slices. Biophys. J. 101, 1277-1284, doi:10.1016/j.bpj.2011.07.027 (2011).
- Galbraith, C. G., & Galbraith, J. A. Super-resolution microscopy at a glance. J Cell Sci. 124, 1607-1611, doi:10.1242/jcs.080085 (2011).
- Strauss, M. P. et al. 3D-SIM Super Resolution Microscopy Reveals a Bead-Like Arrangement for FtsZ and the Division Machinery: Implications for Triggering Cytokinesis. PLoS biol. 10, e1001389, doi:10.1371/journal.pbio.1001389 (2012).
- Adams, D. W., & Errington, J. Bacterial cell division: assembly, maintenance and disassembly of the Z ring. Nature Rev. Microbiol. 7, 642-653, doi:10.1038/nrmicro2198 (2009).
- Boer, P. A. J. Advances in understanding E. coli cell fission. Curr. Opin. Microbiol. 13, 730-737, doi:10.1016/j.mib.2010.09.015 (2010).
- Erickson, H. P. Modeling the physics of FtsZ assembly and force generation. Proc. Natl. Acad. Sci. U S A. 106, 9238-9243, doi:10.1073/pnas.0902258106 (2009).
- Erickson, H. P., Anderson, D. E., & Osawa, M. FtsZ in Bacterial Cytokinesis: Cytoskeleton and Force Generator All in One. Microbiol. Mol. Biol. Rev. 74, 504-528, doi:10.1128/MMBR.00021-10 (2010).
- Lan, G., Wolgemuth, C. W., & Sun, S. X. Z-ring force and cell shape during division in rod-like bacteria. Proc. Natl. Acad. Sci. U S A. 104, 16110-16115, doi:10.1073/pnas.0702925104 (2007).
- Osawa, M., Anderson, D. E., & Erickson, H. P. Reconstitution of Contractile FtsZ Rings in Liposomes. Science. 320, 792-794, doi:10.1126/science.1154520 (2008).
- Osawa, M., & Erickson, H. P. Liposome division by a simple bacterial division machinery. Proc. Natl. Acad. Sci. U S A. 110, 11000-11004, doi:10.1073/pnas.1222254110 (2013).
- Murphy, G. E., & Jensen, G. J. Electron cryotomography. BioTechniques. 43, 413-415 (2007).
- Liew, A. T. F. et al. A simple plasmid-based system that allows rapid generation of tightly controlled gene expression in Staphylococcus aureus. Microbiology. 157, 666-676, doi:10.1099/mic.0.045146-0 (2011).
- Riglar, D. T. et al. Super-Resolution Dissection of Coordinated Events during Malaria Parasite Invasion of the Human Erythrocyte. Cell Hos., & Microbe. 9, 9-20, doi:10.1016/j.chom.2010.12.003 (2011).
- Li, Z., Trimble, M. J., Brun, Y. V., & Jensen, G. J. The structure of FtsZ filaments in vivo suggests a force-generating role in cell division. EMBO J. 26, 4694-4708 (2007).

