Reconstitution of Actin-Based Motility with Commercially Available Proteins
Summary
This protocol describes how to produce actin comets on the surfaces of beads using commercially available protein ingredients. Such systems mimic the protrusive structures found in cells, and can be used to examine physiological mechanisms of force production in a simplified way.
Abstract
Many cell movements and shape changes and certain types of intracellular bacterial and organelle motility are driven by the biopolymer actin that forms a dynamic network at the surface of the cell, organelle, or bacterium. The biochemical and mechanical basis of force production during this process can be studied by reproducing actin-based movement in an acellular manner on inert surfaces such as beads that are functionalized and incubated with a controlled set of components. Under the appropriate conditions, an elastic actin network assembles at the bead surface and breaks open due to the stress generated by network growth, forming an “actin comet” that propels the bead forward. However, such experiments require the purification of a host of different actin-binding proteins, often putting them beyond the reach of non-specialists. This article details a protocol for reproducibly obtaining actin comets and motility of beads using commercially available reagents. Bead coating, bead size, and motility mixture can be altered to observe the effect on bead speed, trajectories, and other parameters. This assay can be used for testing the biochemical activities of different actin-binding proteins, and for performing quantitative physical measurements that shed light on active matter properties of actin networks. This will be a useful tool for the community, enabling the study of in vitro actin-based motility without expert knowledge in actin-binding protein purification.
Introduction
Actin polymerization in cells is spatially and temporally controlled by tight regulation of actin filament nucleation downstream of cell signaling1. Nucleation occurs via the formation of an actin trimer, and then both ends of the nascent filament polymerize spontaneously, although one end is more dynamic (the barbed end) than the other (the pointed end)2. When nucleation and barbed end polymerization are directed toward a surface, they produce enough force (in the pico-to-nano Newton range) to push out the cell membrane for movement and to move micron-sized objects inside the cell with ATP hydrolysis as the energy source3. Some examples include Listeria monocytogenes bacteria that use actin comets to spread from cell to cell, and mitochondria, where actin comet-based movement is important for randomized inheritance during mitosis4,5. Actin comets on endosomes and other intracellular vesicles are implicated in detachment from donor membranes6,7,8.
With the method presented here, the signaling aspects of cellular actin polymerization are bypassed, and actin polymerization is produced on micrometric polystyrene beads by coating them with activators of branched actin nucleation, specifically the active domain of the human WASP protein, VCA (also called WA or WCA)1. The coated beads are then incubated in a mix containing the ingredients necessary for actin polymerization, including the main actin polymerization nucleator in cells, the Arp2/3 complex, which is activated by VCA at the bead surface to form new filaments as branches off the sides of daughter filaments1. Actin initially polymerizes uniformly around the bead, but then spontaneously breaks symmetry to create an actin comet that pushes the bead forward, thereby recreating cell-like protrusive networks and comets in a controlled manner. Similar approaches with beads and other coated surfaces have been used in the past by us and others to study the biochemistry and biophysics of actin polymerization9,10,11,12, but extensive expertise in actin-binding proteins was required for these experiments. The protocol presented here describes how to robustly create actin comets and motility entirely with commercially available (or soon to be available) reagents, making this approach accessible to anyone, including in an educational setting for teaching biophysical concepts. Key features include the importance of gentle and reliable pipetting, the use of profilin-complexed monomer as the actin source, and the essentiality of using a highly active Apr2/3 complex activator as a bead coating reagent.
Protocol
1. Preparation of buffers
NOTE: Use ultrapure H2O for all buffers. It does not have to be sterile. Filter all the solutions described in steps 1.1-1.4 with a 0.2 µm syringe filter, aliquot in portions of 500 µL-2 mL per tube depending on usage, and store at -20 °C.
- Prepare 10% BSA by weighing 2 g of BSA into a 50 mL conical tube and filling to the 20 mL mark with H2O. Mix until the BSA is dissolved (about 30 min), and then make up the volume to 20 mL.
NOTE: Use high quality BSA (see Table of Materials). The 10% BSA solution is used in both bead preparation and the motility mix. - For bead preparation, prepare Xb buffer (10 mM HEPES, 0.1 M KCl, 1 mM MgCl2, and 0.1 mM CaCl2, pH 7.5) as a 10x solution, and dilute before use (100 µL of 10x Xb stock solution + 900 µL of H2O). Prepare Xb/1% BSA by mixing 100 µL of 10x Xb stock solution + 100 µL of 10% BSA + 800 µL of H2O.
- Prepare G-buffer (2 mM Tris, 0.2 mM CaCl2, 0.2 mM DTT, 2 mM ATP), which is the buffer used to dilute monomeric actin (G-actin). Adjust to pH 7, not pH 8 as traditionally used (see discussion).
- Prepare the motility buffer MB13 (10 mM HEPES, 1.5 mM ATP, 3 mM DTT, 1.5 mM MgCl2, 1 mM EGTA, 50 mM KCl, 1% BSA, pH 7.5). For some applications, 10x MB13 is useful. However, prepare 10x MB13 without BSA, as it leads to problems during pH adjustment. Add BSA from 10% stock solution (prepared in step 1.1) when reconstituting 1x MB13 from 10x MB13.
2. Preparation of protein solutions
NOTE: Use ultrapure H2O for all resuspensions. It does not have to be sterile. Handle all proteins on ice and aliquot into pre-chilled tubes. Manipulate gently so as not to produce bubbles, and never vortex protein solutions. For stocks to be stored at -80°C, flash freezing in liquid nitrogen is not necessary. Adapt aliquot size to avoid more than about five freeze-thaw cycles, as this does not appear to affect the activity of any of the proteins. Working aliquots can be stored at -20 °C for a few weeks.
- Prepare G-actin (rabbit skeletal muscle) solution as described below.
- Pulse centrifuge actin powder (see Table of Materials) at 4 °C to gather the solid at the bottom of the tube.
- Add H2O as per manufacturers' instructions (1 mg protein in 100 µL of cold H2O for unlabeled actin, 100 µg protein in 100 µL of cold H2O for ATTO-labeled actin).
- Let it sit on ice for at least 15 min. Mix gently by pipetting up and down, let it sit on ice at least another 15 min, and mix again. Pulse centrifuge at 4 °C to collect the solution at the bottom of the tube, and remix.
- Prepare 10-50 µL aliquots of unlabeled actin depending on use, and 20 µL aliquots of ATTO-labeled actin. Store the aliquots at -80 °C.
- To depolymerize actin oligomers that form during lyophilization and freezing, dilute an aliquot of resuspended actin ~8-fold in G-buffer spiked with additional ATP and DTT (for example, to 20µL of resuspended actin solution from step 2.1.4, add 134 µL of G-buffer, 0.32 µL of 0.2 mM ATP and 0.16 µL of 1 M DTT). For fluorescent labeling, add approximately 10% labeled actin; for example, add 5 µL of ATTO-labeled actin to 40 µL of diluted unlabeled actin. Let it depolymerize on ice with occasional mixing (pipetting) at least a few days to a week before measuring protein concentration by Bradford assay as described in step 3.
NOTE: Keep diluted unlabeled and fluorescent actin on ice in a cold room or refrigerator; never freeze or allow to warm. The preparation will continue depolymerizing over time, and when handled properly can be used for at least 6 months.
- Resuspend Arp2/3 complex (porcine brain) (see Table of Materials) following the manufacturer's instructions (20 µg protein in 20 µL cold H2O), with the sequence of pulse centrifuging, mixing, etc. on ice as described for actin in step 2.1. Combine the protein solutions from resuspending two tubes of powder to have a larger stock for reproducible experiments. Prepare 2 µL aliquots and store at -80 °C.
- Resuspend profilin (human recombinant) (see Table of Materials) at a 4x higher concentration than stipulated in the manufacturer's instructions (100 µg protein in 25 µL cold H2O), with the sequence of pulse centrifuging, mixing, etc. on ice as for actin in step 2.1. Combine the protein solutions from resuspending two tubes of powder before determining protein concentration to have a larger stock for reproducible experiments.
NOTE: Keep on ice in a cold room or refrigerator; never freeze or allow to warm. When handled properly, resuspended profilin is good for at least 6 months to 1 year. - Resuspend capping protein (α1β2, human recombinant) (see Table of Materials) following the manufacturer's instructions (50 µg protein in 50 µL cold H2O), with the sequence of pulse centrifuging, mixing etc. on ice as described for actin in step 2.1. Chill 50 µL of glycerol on ice and add the 50 µL of resuspended capping protein to it; mix gently. Store at -20 °C.
NOTE: The solution does not freeze and the activity is robust, so the solution can be kept as a single aliquot for months, or even years, if handled carefully. Mouse recombinant capping protein, the one most commonly used in the past in in vitro experiments13, will soon be commercially available. - Resuspend gelsolin (human recombinant, His-tagged) (see Table of Materials) following the manufacturer's instructions (20 µg protein in 20 µL cold H2O), with the sequence of pulse centrifuging, mixing, etc. on ice as described for actin in step 2.1. About 2 µL of gelsolin is used per day of experiments; therefore, prepare large aliquots (5-10 µL) and store at -80 °C.
NOTE: The protocol with gelsolin is provided as an alternative. The use of capping protein in the place of gelsolin is recommended, either purchased or purified as in13. - Resuspend VCA (human WASP-VCA, GST-tagged) (see Table of Materials) at 2x higher concentration than stipulated in the manufacturer's instructions (500 µg protein in 250 µL cold H2O), with the sequence of pulse centrifuging, mixing, etc. on ice as for actin in step 2.1. Make 10 µL aliquots and store at -80 °C.
NOTE: Once SpVCA (human pVCA, streptavidin and His-tagged) is commercialized or if protein purification14 is possible, the use of SpVCA instead of VCA is recommended. VCA does not reproducibly give comets under the conditions described here.
3. Measurement of protein concentrations
- Construct a Bradford standard curve made of two overlapping serial dilutions of BSA.
NOTE: The standard curve only needs to be constructed every couple of months (or even less frequently) as long as the spectrophotometer does not change.- In row 1 of a microtube rack, place tubes for BSA dilution series #1: four 2 mL tubes followed by four 1.5 mL tubes. In row 3 of the rack, place tubes for BSA dilution series #2 as for series #1. In row 5 of the rack, place one 2 mL tube for each sample to be measured along with two 1.5 mL tubes. Add a single 1.5 mL tube in row 5 for the blank.
- Measure out Bradford Reagent (see Table of Materials) into the 1.5 mL tubes.
- Fill a 15 mL conical tube to the top with Bradford Reagent for easier pipetting (excess will be returned to the stock bottle in the refrigerator) on ice. Take up 200 µL Bradford Reagent and eject it back into the conical tube to wet the pipette tip. As the solution is viscous, pipette slowly to allow the solution to enter and leave the tip completely without making bubbles.
- With the "pre-wet" tip, slowly pipette 200 µL of Bradford Reagent into each of the 1.5 mL tubes in the rack (four for each BSA dilution, one for the blank and two for each sample to be measured). Do this first to allow the Bradford Reagent to thoroughly warm to room temperature before mixing with protein solutions. Return the remaining contents of the 15 mL conical tube to the bottle.
- Measure out H2O into the 2 mL tubes. In row 1 of the rack, add 1,990 µL H2O to the first tube, and 900 µL to the three other tubes. For row 3, add 1,992.5 µL to the first tube and 900 µL to the three other tubes. Add 2,000 µL H2O to each of the sample tubes in row 5.
NOTE: For all volumes larger than 1,000 µL, use a 1,000 µL pipette, but administer the full amount by pipetting twice. It is important to get everything ready before starting the subsequent steps, in order to avoid leaving proteins highly diluted in H2O for long periods of time. - To prepare BSA dilution series #1, mix 10 µL of calibrated 2 mg/mL BSA (see Table of Materials) into the tube with 1,990 µL H2O to make a 10 µg/mL solution. From this, make three serial dilutions (5 µg/mL, 2.5 µg/mL, and 1.25 µg/mL BSA) by transferring 900 µL of each solution into the next tube (containing 900 µL H2O).
- To prepare BSA dilution series #2, mix 7.5 µL of calibrated 2 mg/mL BSA into the tube with 1,992.5 µL H2O to make a 7.5 µg/mL solution. From this, make three serial dilutions (3.75 µg/mL, 1.875 µg/mL and 0.9375 µg/mL BSA) by transferring 900 µL of each solution into the next tube (containing 900 µL H2O).
- Mix and read absorbance to generate the standard curve. Add 800 µL H2O to the Bradford reagent tube for the blank, and start the timer. As efficiently as possible without making bubbles, mix 800 µL of each BSA standard with 200 µL Bradford reagent in the prepared tubes. Once all the standards have been mixed with Bradford reagent (< 5 min), pour each standard into a disposable cuvette and read the absorbance at 600 nm in the spectrophotometer after blanking the machine.
NOTE: The same cuvette can be used to read the whole series if the least concentrated standard is read first and the cuvette is well-emptied between reads. Redo the standard curve until the linear fit has an R value of at least 0.99. Only after pipetting and gentle mixing is mastered, proceed to reading samples.
- Measure concentrations of actin and actin-binding proteins
- In the 2 mL tubes containing 2,000 µL of H2O (prepared in step 3.1.3), gently mix in the following: 2 µL each of Arp2/3 complex and profilin, 4 µL of labeled G-actin, 5 µL each of gelsolin and VCA and 8 µL of capping protein (human recombinant). Immediately take 800 µL of the solution and mix with the already prepared Bradford reagent, and then repeat to have two readings within 5%-10 % of each other for each sample. A larger difference indicates a problem with resuspension or handling. Read within a few minutes of mixing with Bradford reagent.
NOTE: If using a standard curve from another day, only the blank from step 3.1.6 has to be prepared, in addition to the samples.
- In the 2 mL tubes containing 2,000 µL of H2O (prepared in step 3.1.3), gently mix in the following: 2 µL each of Arp2/3 complex and profilin, 4 µL of labeled G-actin, 5 µL each of gelsolin and VCA and 8 µL of capping protein (human recombinant). Immediately take 800 µL of the solution and mix with the already prepared Bradford reagent, and then repeat to have two readings within 5%-10 % of each other for each sample. A larger difference indicates a problem with resuspension or handling. Read within a few minutes of mixing with Bradford reagent.
- Calculate the concentrations of actin and actin-binding proteins using the standard curve and the dilution factor. Redo the measurement for each new resuspension. Molecular weights to convert mg/mL readings obtained via Bradford assay to µM are: actin 43 kD, Arp2/3 complex 224 kD, profilin 15 kD, gelsolin 95 kD, capping protein 68 kD (human recombinant) or 63.5 kD (mouse recombinant), VCA 43 kD, and SpVCA 54 kD (monomer molecular weight).
4. Coating of beads
- Pre-chill the centrifuge to 4 °C and set the agitating dry block (see Table of Materials) to 18 °C.
- Wash the beads: pipette 50 µL of Xb buffer into a 1.5 mL microcentrifuge tube, add 9 µL of 4.5 µm diameter bead suspension or 2 µL of 1 µm diameter bead suspension (2.5 % w/v suspension) (see Table of Materials). Mix thoroughly and centrifuge the samples at 20,000 x g for 10 min at 4 °C.
NOTE: Total bead surface area of both bead sizes is 3 cm2:
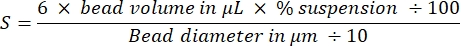
derived by calculating the number of beads and then their total surface using classic equations for the volume and surface area of spheres. Other sizes of beads can be used if amounts are adjusted to keep the total surface area constant at 3 cm2. - Coat the beads: carefully remove the supernatant without disturbing the beads, and resuspend the bead pellet in 40 µL of 2 µM SpVCA (or 7 µM VCA) in Xb buffer by gentle pipetting. Agitate at 18 °C, 1,000 rpm for 20 min.
- Wash coated beads: centrifuge the mix (20,000 x g for 10 min at 4 °C) and carefully remove the supernatant. Resuspend the beads in 50 µL of cold Xb/1% BSA, and centrifuge at 20,000 x g for 10 min at 4 °C. Remove the supernatant and repeat the washing step 1x.
- Resuspend the coated bead pellet from step 4.4 in 120 µL cold Xb/1% BSA for both sizes of beads so that the amount of bead surface area/µL of bead solution is the same. Store on ice in a refrigerator or cold room. Coated beads will continue to function normally for at least several weeks.
5. Preparing the motility mix and slides for observation
NOTE: The total volume of the motility mix is 8.4 µL to allow for about 25 µm clearance between the slide and the 18 mm x 18 mm coverslip so beads of all sizes (up to 10 µm diameter) are not squeezed. The basic motility mix is approximately 5 µM G-actin (10% labeled fluorescent actin) with 5 µM profilin, 50 nM Arp2/3 complex, and 25 nM capping protein (or 240 nM gelsolin).
- Prepare the motility reaction mixture. Exact amounts of actin (and therefore profilin) depend on the concentration calculated in step 3.3, but a representative reaction is as follows. Mix on ice, in this order: 3.2 µL of MB13, 1.5 µL of profilin at 30 µM diluted in MB13, 1 µL of capping protein at 0.21 µM diluted in MB13 or gelsolin diluted to 2 µM in MB13, 1 µL of Arp2/3 complex at 0.47 µM diluted in MB13, 0.2 µL of bead suspension (vortex just before use), and 1.5 µL of actin at 30 µM in G-buffer. Mix well but quickly and start the timer.
- Spot the entire motility reaction mixture on a slide. Cover with an 18 mm x 18 mm coverslip and seal the coverslip with melted VALAP using a small paintbrush. VALAP is a mix of lanolin, paraffin, and petroleum jelly (see Table of Materials) 1:1:1 by weight, melted and stirred together.
6. Microscopy observation
- Observe motility reactions immediately using a 100x objective on either an upright or an inverted microscope (see Table of Materials), equipped with phase contrast and/or epifluorescence microscopy (GFP cube, see Table of Materials). Observations are done at room temperature (23-25 °C).
- To obtain average displacement speeds for a whole population of beads, record phase contrast or fluorescent still images over time by scanning the entire slide. Measure comet length by hand and plot versus time. The slope of the linear fit is the average growth speed.
- To evaluate the speed of individual beads, collect time-lapse movies in phase contrast microscopy. Depending on the bead speed and the resolution required, take frames every 1-10 s. Use the tracking tool of any image processing program to obtain bead speeds and trajectories.
Representative Results
One of the key aspects of reproducibly creating actin comets on beads is gentle and precise pipetting of delicate actin-binding proteins. Generating a Bradford standard curve is a good way of evaluating pipetting skills. Figure 1A,B shows the tubes for the standard curve and an example of what the two serial dilutions of BSA look like once mixed with Bradford reagent. Note the graded blue hue (higher protein concentration gives a bluer solution). When read in the spectrophotometer and plotted, these solutions give a standard curve as shown in Figure 1C. To practice careful pipetting, the assay should be repeated until the linear correlation factor is 0.999, as shown.
Once the concentrations of commercial resuspended proteins have been carefully evaluated via the Bradford assay, coated beads and motility mix are prepared and mixed together. Figure 2A shows representative images of the different stages of comet formation: actin clouds form within minutes of mixing SpVCA-coated beads and motility medium; cloud polarization occurs at ~5 min and comet production at 15-20 min. The actin comets, visible with both epifluorescence and phase contrast microscopy (Figure 2A), continue to elongate for many hours, but a consistent speed is not maintained so bead motility is normally evaluated within 1 h. On the other hand, it takes 30 min with VCA-coated beads to obtain bright actin clouds (Figure 2B), and no comets form, although symmetry starts to break at 1-2 h (arrow in Figure 2B) and clouds show polarization after overnight incubation.
Figure 3 shows an example of bead velocity evaluation in the presence of capping protein. Since all beads break symmetry at approximately the same time, "pseudo-time-lapse" recordings are performed where the slide is scanned and pictures of the whole population of comets are taken over time (Figure 3A). Comets do not depolymerize; therefore, the increase in comet lengths measured over time can be used to calculate displacement speed (Figure 3B). Gelsolin can be used in the place of capping protein for comet formation if 10x more gelsolin is added to compensate for its reduced capping activity. Comets formed in the presence of gelsolin are qualitatively the same and move at approximately the same speeds as beads with capping protein (Figure 3C). Capping activity is key for concentrating polymerization at the surface of the bead, and when neither capping protein nor gelsolin are included in the motility mix, actin clouds never polarize to form comets, although bright actin clouds form around the beads (Figure 3D). Comets on beads can be used to measure actin-based force production in different biochemical contexts by altering the motility mix and observing the result on motility using different micromanipulation techniques, for example15.
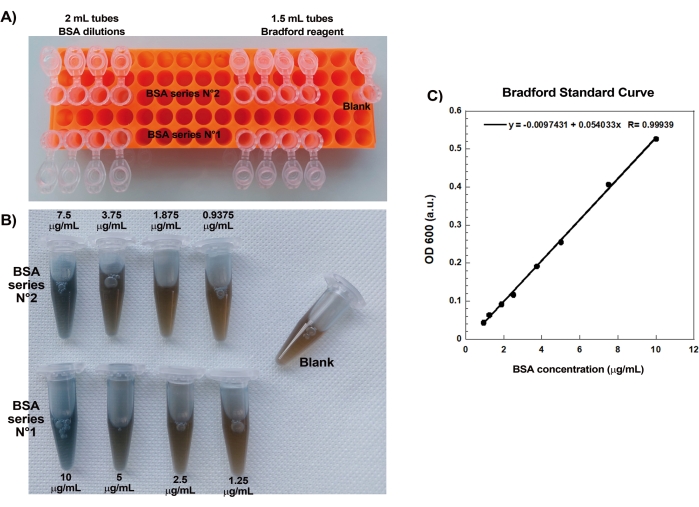
Figure 1: Bradford standard curve. (A) Picture of how to set up the tubes for making the Bradford standard curve. Sample tubes are not shown. (B) Picture of the two overlapping BSA serial dilutions once mixed with Bradford reagent. (C) The absorbances at 600 nm of the solutions shown in (B) are measured in the spectrophotometer and are plotted as a function of the protein concentrations of the BSA solutions. The linear fit is used to calculate the sample concentration. The correlation factor, R, of the linear fit is 0.999. Please click here to view a larger version of this figure.
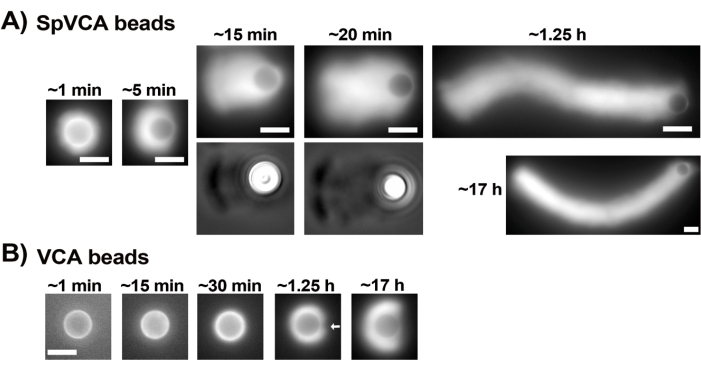
Figure 2: Comet formation on SpVCA-coated beads as opposed to VCA-coated beads. (A) Representative SpVCA-coated beads (different bead in each image) shown over time. Time from the moment of mixing is indicated. Actin clouds form immediately, and cloud polarization gives comets, which continue to elongate for hours. (B) Representative VCA-coated beads (different bead in each image) shown over time. More than 1 h is necessary to see the beginnings of actin cloud polarization (arrow) and even long incubations do not produce comets. All images are of 4.5 µm diameter beads, epifluorescence imaging of fluorescent actin paired with phase contrast visualization for the 15 and 20 min time-points in (A), scale bars = 5 µm. Please click here to view a larger version of this figure.
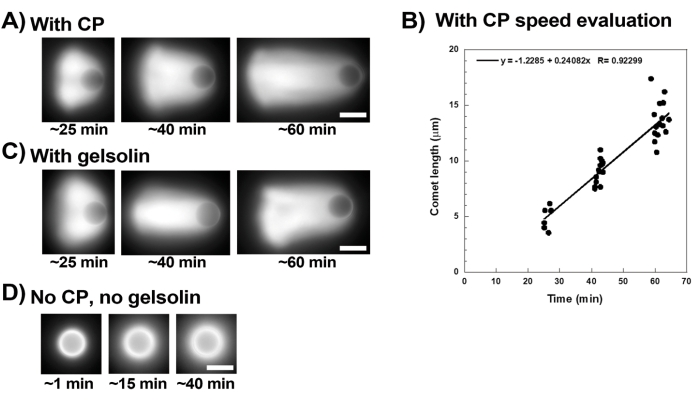
Figure 3: Comets and bead velocity analysis. (A) and (C) In the presence of either capping protein (CP) or gelsolin, actin clouds polarize to form comets in the first 20 min of reaction, and comets elongate over time. Time from mixing indicated for each image; each image is a different bead. There is some variability among beads and preparation, but on average beads move at micron/sub-micron per min speeds (0.2-1 µm/min) under the standard conditions described here. (B) Representative graph showing evaluation of comet length (whole population of beads) over time. The slope of the linear correlation corresponds to the average displacement speed, in this case 0.24 µm/min. (D) In the absence of capping activity (no capping protein or gelsolin), actin clouds form around beads, but comets do not form. All images are of 4.5 µm diameter beads, epifluorescence imaging of fluorescent actin, scale bars = 5 µm. Please click here to view a larger version of this figure.
Discussion
The protocol detailed here describes how to obtain actin network growth on bead surfaces, comet formation, and bead motility using commercially available proteins. However, sometimes comets are not reproducibly observed or are inhomogeneous between the slide and coverslip. The following discussion emphasizes some key points in the protocol and suggests some parameters that can be adjusted. One factor to keep in mind is that comet formation and bead speed are affected by temperature, with temperatures much above 25 °C or much below 23 °C negatively impacting comet formation and giving irreproducible data. Use of a temperature-controlled microscope or a microscope in a climate-controlled room is strongly recommended. Although fluorescently labeled actin is often included in the motility mix to observe comets by fluorescence microscopy, once comets are more than a bead diameter in length, they are also visible by phase contrast microscopy as a dark smear next to the bead. Phase contrast visualization is more appropriate for time-lapse imaging as some phototoxicity is associated with fluorescence imaging even via spinning disc. Because beads settle over time, an inverted microscope produces less horizontal bead drift than an upright one and is more appropriate for movies. The use of molten VALAP to seal slides is important as substances such as nail polish interfere with comet formation. Large quantities of VALAP can be made in a beaker, and then scooped out to refill smaller beakers more amenable to rapid melting. VALAP is good for years at room temperature.
Another key technical aspect is meticulous buffer and motility mix preparation. Care should be taken when preparing MB13, in particular at the pH adjustment step. The pH of MB13 should be adjusted rapidly to neutral with NaOH to avoid ATP hydrolysis, but not too quickly as the EGTA solubilizes as the pH approaches neutral. EGTA is a key ingredient because it complexes the calcium bound to actin, giving in the motility mix the more active magnesium form16. MB13 prepared too quickly or too slowly gives suboptimal comet formation or even none at all. An additional key point is to keep careful track of KCl concentration in the motility mix when playing with conditions. For example, when using 1x MB13 in the reaction mix and diluting profilin, capping protein, and the Arp2/3 complex in MB13, the final KCl concentration in the motility reaction is about 40-50 mM due to dilution by G-buffer. This concentration gives the best results in the comet assay, and any more than 60 mM KCl decreases Arp2/3 complex nucleating activity.
On the protein side of things, a critical technical aspect of obtaining actin comets is proper handling of commercial actin-binding proteins, in particular precise pipetting of microliter quantities. The linearity of the Bradford standard curve is a good test of pipetting and the curve can then be used for routine measurements of protein concentrations. Indeed, when using resuspended commercial proteins for the comet procedure, it is important to always verify protein concentrations, as batch variability and user error during resuspension can lead to differences between real and expected concentrations. Sometimes small differences in protein concentrations can lead to the complete absence of comets.
Another important aspect of the method presented here is the use of profilin-complexed G-actin as the fuel for polymerization. Historically, in vitro systems used pre-polymerized filamentous actin (F-actin) as the actin source: depolymerization in the bulk fed polymerization on the surface10,17. This had the advantage of controlling G-actin levels, but added a layer of complexity requiring additional components to catalyze depolymerization. Since turnover of the actin network is not necessary for force production and motility, which are fueled by nucleation and polymerization at the surface of the bead, while actin depolymerization factors such as ADF/cofilin act on the aged networks far from the surface18, most in vitro reconstitution of actin-based motility is now done without turnover for simplicity. However, there are some drawbacks to using G-actin. First, when using commercial actin, which has been lyophilized, oligomers are present. The depolymerization steps described here are very important in obtaining reproducible results. In particular, although G-buffer is traditionally adjusted to pH 8, lower pH (pH 7, for example) appears to work better in the assays described in this article, possibly because low pH enhances depolymerization19. Another disadvantage of using G-actin is that once placed in salt conditions permissive to polymerization, spontaneous nucleation occurs and F-actin forms in the bulk as well as on the bead surface. Complexing G-actin with profilin suppresses spontaneous nucleation in the bulk and pointed end polymerization, thereby focusing both nucleation and barbed end polymerization at the surface20. Profilin-G-actin is physiologically relevant, as much of the actin in the cell is present in this form21. Here, a 1:1 ratio of profilin:actin is used; however, higher ratios (for example 3:1) more thoroughly inhibit polymerization in the bulk, although higher ratios also inhibit the Arp2/3 complex and barbed end elongation to some extent22,23.
Capping activity is also key for comet formation since it ensures insertion of new actin at the surface via cycles of nucleation by surface-activated Arp2/3 complex24,25. Without capping, actin clouds do not break symmetry to form comets because polymerization at the surface does not build up enough tension to break open the cloud26. In the past, we have used home-purified recombinant mouse capping protein13, but tests performed for this article indicate that commercially available recombinant human capping protein is equally effective, as is commercially available gelsolin, although 10x more gelsolin has to be used, and for certain applications, it may not be appropriate as it has actin severing activity as well as capping27.
Finally the robustness of this method resides in the use of a very active Arp2/3 complex activator, streptavidin-pVCA (SpVCA)28. SpVCA includes the profilin-G-actin binding domain of WASP (the p domain) in addition to the Arp2/3 complex binding domain as this is found to be most efficient in profilin-G-actin conditions29. More importantly, the use of the streptavidin tag, originally introduced to allow surface functionalization via the biotin-streptavidin link, has the additional effect of increasing Arp2/3 complex activation, presumably due to the fact that streptavidin is a tetramer and thus clusters the activator, known to increase Arp2/3 complex activity30. Commercially-produced SpVCA is currently in development and will soon be available for purchase. It should further be noted that, although 40 µL of 2 µM SpVCA is routinely used to coat 3 cm2 of bead surface, other coating concentrations (higher and lower) also work, and playing with these conditions gives different comet growth speeds and morphologies. Indeed, when comets do not form or comet size is not homogenous on the slide, different coating conditions should be tested, as well as different KCl and profilin concentrations in the motility mix. The concentrations of actin, Arp2/3 complex, and capping protein in the motility mix can also be altered to optimize comet formation, but in our hands, changing these proportions often gives confusing results.
To conclude, the methods described here produce actin assembly on bead surfaces and motility, but any surface that can be functionalized with SpVCA can be used. In cases where adsorption as described here does not work, the streptavidin moiety can be used to attach SpVCA to the surface of interest after biotinylation. The actin structures thus formed, comets or otherwise, can be used for testing different biochemical and biophysical aspects of actin networks, and are especially appropriate for physical manipulations with micropipettes, optical tweezers, and laser ablations15,26,31,32. In addition to its uses to the research community, the approach described here is appropriate as a teaching tool for undergraduate biophysics students to study active matter concepts such as symmetry breaking and self-organization.
Offenlegungen
The authors have nothing to disclose.
Acknowledgements
We sincerely acknowledge the members of our new home at LPENS for their warm welcome, and in particular, the ABCDJ team for all their help and support. J.P. acknowledges financial support from the Foundation ARC (Grant PJA 20191209604), and C.S. acknowledges financial support from the Human Frontiers Science Program Organization (Grant RGP0026/2020).
Materials
| Actin, rabbit muscle, Alexa Fluor 488 conjugate | Invitrogen (ThermoFisher Scientific) | A12373 (recently discontinued) | This product can be replaced with ATTO-488 actin from Hypermol. |
| Actin, rabbit muscle, ATTO-488 | Hypermol | 8153 | |
| Actin, rabbit skeletal muscle | Cytoskeleton | AKL99 | |
| Arp2/3 complex | Cytoskeleton | RP01P | |
| ATP | Sigma | A7699 | |
| BioSpectrometer, basic | Eppendorf | 035739 | |
| Bradford Reagent | Bio-Rad | 500-0006 | |
| BSA, high quality | Sigma | A3059 | |
| BSA standard 2 mg/mL (Pierce) | Thermo Scientific | 23209 | |
| Capping protein (a1b2, mouse recombinant) | Home-purified (Reference 13) | This product will soon be commercially available from Cytoskeleton. | |
| Capping protein (a1b2, human recombinant) | Hypermol | 8322 | |
| Cube, GFP: U-MNIBA3 or U-MWB2 | Olympus | discontinued | Any GFP cube, adapted to the microscope being used, can be used. |
| Dry block, agitating: ThermoMixer C (refrigerated) | Eppendorf | 035963 | |
| ** with SmartBlock, 24 microtubes 2 mL | Eppendorf | 035969 | |
| Gelsolin (human recombinant, His-tagged) | Cytoskeleton | HPG6 | |
| Lanolin | Sigma | 49909 | |
| Microcentrifuge 5427R + rotor | Eppendorf | 934126 | |
| Microscope, upright: BX51 | Olympus | discontinued | Any epifluorescence upright microscope equipped with phase contrast optics can be used. |
| Microscope, inverted: IX70 | Olympus | discontinued | Any epifluorescence inverted microscope equipped with phase contrast optics can be used. |
| Paraffin | Sigma | 76244 | |
| Petroleum jelly: Vaseline | Sigma | 16415 | |
| Pipettes Research Plus | Eppendorf | Gilson pipettes don't work as well for delivery of very small volumes (0.5 µL for example). | |
| **10 µL | 933954 | ||
| **2.5 µL | 933953 | These two sizes are essential, but the use of high-quality pipettes (a full Research Plus set for example) is recommended. | |
| Polystyrene carboxylate beads | Polysciences | ||
| **approx. 1 µm diameter | 08226 | ||
| **approx. 4.5 µm diameter | 17140-5 | ||
| Profilin 1 (human recombinant, untagged) | Cytoskeleton | PR02 | |
| SpVCA (human WASP pVCA domain, N-ter His-tag, C-ter Streptavidin tag) | Home-purified (Reference 14) | This product will soon be commercially available from Cytoskeleton. | |
| VCA (human WASP VCA domain, GST-tagged) | Cytoskeleton | VCG03 |
Referenzen
- Campellone, K. G., Welch, M. D. A nucleator arms race: cellular control of actin assembly. Nature Reviews Molecular Cell Biology. 11 (4), 237-251 (2010).
- Pollard, T. D. Rate constants for the reactions of ATP- and ADP-actin with the ends of actin filaments. Journal of Cell Biology. 103, 2747-2754 (1986).
- Blanchoin, L., Boujemaa-Paterski, R., Sykes, C., Plastino, J. Actin dynamics, architecture and mechanics in cell motility. Physiological Reviews. 94 (1), 235-263 (2014).
- Tilney, L. G., Tilney, M. S. The wily ways of a parasite: induction of actin assembly by Listeria. Trends in Microbiology. 1 (1), 25-31 (1993).
- Moore, A. S., et al. Actin cables and comet tails organize mitochondrial networks in mitosis. Nature. 591 (7851), 659-664 (2021).
- Taunton, J., et al. Actin-dependent propulsion of endosomes and lysosomes by recruitment of N-WASP. Journal of Cell Biology. 148 (3), 519-530 (2000).
- Velarde, N., Gunsalus, K. C., Piano, F. Diverse roles of actin in C. elegans early embryogenesis. BMC Developmental Biology. 7, 142 (2007).
- Merrifield, C. J., et al. Endocytic vesicles move at the tips of actin tails in cultured mast cells. Nature Cell Biology. 1 (1), 72-74 (1999).
- Samarin, S., et al. How VASP enhances actin-based motility. Journal of Cell Biology. 163 (1), 131-142 (2003).
- Bernheim-Groswasser, A., Wiesner, S., Golsteyn, R. M., Carlier, M. -. F., Sykes, C. The dynamics of actin-based motility depend on surface parameters. Nature. 417 (6886), 308-311 (2002).
- Boujemaa-Paterski, R., et al. Network heterogeneity regulates steering in actin-based motility. Nature Communications. 8 (1), 655 (2017).
- Akin, O., Mullins, R. D. Capping protein increases the rate of actin-based motility by promoting filament nucleation by the Arp2/3 complex. Cell. 133 (5), 841-851 (2008).
- Palmgren, S., Ojala, P. J., Wear, M. A., Cooper, J. A., Lappalainen, P. Interactions with PIP2, ADP-actin monomers, and capping protein regulate the activity and localization of yeast twinfilin. Journal of Cell Biology. 155 (2), 251-260 (2001).
- Carvalho, K., et al. Actin polymerization or myosin contraction: two ways to build up cortical tension for symmetry breaking. Philosophical Transactions of the Royal Society B. 368 (1629), 20130005 (2013).
- Marcy, Y., Prost, J., Carlier, M. -. F., Sykes, C. Forces generated during actin-based propulsion: a direct measurement by micromanipulation. Proceedings of the National Academy of Sciences of the United States of America. 101 (16), 5992-5997 (2004).
- Carlier, M. -. F. Actin: protein structure and filament dynamics. Journal of Biological Chemistry. 266 (1), 1-4 (1991).
- Loisel, T. P., Boujemaa, R., Pantaloni, D., Carlier, M. F. Reconstitution of actin-based motility of Listeria and Shigella using pure proteins. Nature. 401 (6753), 613-616 (1999).
- Reymann, A. -. C., et al. Turnover of branched actin filament networks by stochastic fragmentation with ADF/cofilin. Molecular Biology of the Cell. 22 (14), 2541-2550 (2011).
- Wioland, H., Jegou, A., Romet-Lemonne, G. Quantitative variations with pH of Actin Depolymerizing Factor/Cofilin’s multiple actions on actin filaments. Biochemie. 58 (1), 40-47 (2019).
- Plastino, J., Blanchoin, L. Dynamic stability of the actin ecosystem. Journal of Cell Science. 132 (4), 219832 (2019).
- Pollard, T. D., Blanchoin, L., Mullins, R. D. Molecular mechanisms controlling actin filament dynamics in nonmuscle cells. Annual Review of Biophysics and Biomolecular Structure. 29, 545-576 (2000).
- Suarez, C., et al. Profilin regulates F-actin network homeostasis by favoring formin over Arp2/3 complex. Developmental Cell. 32 (1), 43-53 (2015).
- Courtemanche, N., Pollard, T. D. Interaction of profilin with the barbed end of actin filaments. Biochemie. 52 (37), 6456-6466 (2013).
- Achard, V., et al. A "primer"-based mechanism underlies branched actin filament network formation and motility. Current Biology. 20 (5), 423-428 (2010).
- Sykes, C., Plastino, J. Actin filaments up against a wall. Nature. 464 (7287), 365-366 (2010).
- vander Gucht, J., Paluch, E., Plastino, J., Sykes, C. Stress release drives symmetry breaking for actin-based movement. Proceedings of the National Academy of Sciences of the United States of America. 102 (22), 7847-7852 (2005).
- McGough, A. M., Staiger, C. J., Min, J. -. K., Simonetti, K. D. The gelsolin family of actin regulatory proteins: modular structures, versatile functions. FEBS Letters. 552 (2-3), 75-81 (2003).
- Abou-Ghali, M., et al. Capping protein is not necessary for polarized actin network growth and actin based motility. Journal of Biological Chemistry. 295, 15366-15375 (2020).
- Yarar, D., D’Alessio, J. A., Jeng, R. L., Welch, M. D. Motility determinants in WASP family proteins. Molecular Biology of the Cell. 13 (11), 4045-4059 (2002).
- Padrick, S. B., et al. Hierarchical regulation of WASP/WAVE proteins. Molecular Cell. 32 (3), 426-438 (2008).
- Bussonier, M., et al. Mechanical detection of a long-range actin network emanating from a biomimetic cortex. Biophysical Journal. 107 (4), 854-862 (2014).
- Paluch, E., vander Gucht, J., Joanny, J. -. F., Sykes, C. Deformations in actin comets from rocketing beads. Biophysical Journal. 91 (8), 3113-3122 (2006).

