Nonspecific Proteolysis-Based Glycomics Strategy for Bacterial Glycoproteins
Summary
This study presents a method for glycomics analysis of glycoproteins by a combination of pronase E digestion, permethylation, and mass spectrometry analysis. This method is capable of analyzing all types of N-linked glycans, including bacterial N-glycans.
Abstract
Protein glycosylation is one of the most common and complex post-translational modifications. Many techniques have been developed to characterize the specific roles of glycans, the relationship between their structures and their impact on the functions of proteins. A common method for glycan analysis is to employ exoglycosidase cleavage to release N-linked glycans from glycoproteins or glycopeptides using Peptide-N-Glycosidase F (PNGase F). However, the glycan-protein linkages in bacteria are different and there is no enzyme available to release glycans from bacterial glycoproteins. In addition, free glycans have also been described in mammalian cells, bacteria, yeast, plants, and fish. In this article, we present a method that can characterize the N-linked glycosylation system in Campylobacter jejuni by detecting asparagine (Asn)-linked and free glycans that are not attached to their target proteins. In this method, total proteins from C. jejuni were digested by Pronase E with a higher enzyme to protein ratio (2:1−3:1) and a longer incubation time (48−72 h). The resulted Asn-glycans and free glycans were then purified using porous graphitic carbon cartridges, permethylated, and analyzed by mass spectrometry.
Introduction
Protein N-glycosylation is one of the most common and complex post-translational modifications in eukaryotes1. N-glycans play an essential role in protein folding and also have an impact on protein sorting in biosynthetic traffic2. Mass spectrometry has been widely used in the analysis of glycans released by exoglycosidase cleavage (glycomics). The remaining deglycosylated peptides (glycoproteomics) have been commonly used to identify the sequences of glycosylated peptides in eukaryotes3. The identification of N-linked glycans in Campylobacter jejuni suggested that N-glycosylation is not restricted to eukaryotes4,5,6,7,8. However, for bacteria, there is a lack of effective exoglycosidases or endoglycosidases to release oligosaccharides for glycan analysis.
An alternative approach has been developed to characterize glycosylation sites and glycan structures in glycoproteins, which is based on the use of nonspecific proteolysis to digest most peptides' backbone and generate pseudo-oligosaccharides that only contain a few amino acids. Various non-specific enzymes have been used to generate pseudo-oligosaccharides and it has been found that Pronase E offered the most efficient and reproducible digestion9. We have developed a glycomics strategy based on Pronase E to analyze C. jejuni N-glycans10,11. The pseudo-oligosaccharides were analyzed directly by capillary electrophoresis mass spectrometry (CE-MS) and/or by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) after permethylation.
Here, a universal method is described for glycomics analysis that uses nonspecific Pronase E digestion and permethylation to study glycosylation from mucosal pathogen C. jejuni. This method is capable of characterizing N-linked glycans expressed by both eukaryotic and bacterial systems and is also useful in identifying novel intermediates in N-linked glycosylation pathways.
Protocol
1. Culture of C. jejuni 11168H and extraction of total proteins
NOTE: C. jejuni 11168H is a hypermotile clonal derivative of NCTC 1116812.
- Start the cell culture by rehydrating the cell pellet (approximately 0.15 g, NCTC) with approximately 5 mL of Mueller-Hinton culture medium. Use several drops of the primary broth tube to inoculate a Mueller-Hinton agar plate. Incubate at 37 °C for 18 h under a microaerophilic atmosphere, i.e., 5% O2, 10% CO2, and 85% N2.
- Harvest bacterial lawns with 1 mL of Brain Heart Infusion (BHI) broth. Dilute the harvest to reach approximately 1 x 105 c.f.u./mL with BHI broth. Grow cells for 18 h with shaking (100 rpm) in a microaerophilic atmosphere at 37 °C.
- Cool the cells by putting the plates on ice for 10 min and harvest cells by centrifugation (4,500 x g at 4 °C). Discard the supernatant and wash the cells 2x in 3 L of ice-cold 0.1 M Tris-HCl (pH 7.8).
- Resuspend cells (108-1011 C. jejuni cells) in 8 mL of ice-cold 0.1 M Tris-HCl (pH 7.8) and lyse by sonication (Table of Materials). Perform sonication as follows: 4 s pulse, 1 s off, 1 min each time and three times total.
- Centrifuge at 9,500 x g for 45 min at 4 °C and transfer the supernatant to another tube. Dialyze proteins in the supernatant 2x against 0.1 M Tris-HCl (pH 7.5) at 4 °C for 3 h.
- Determine the protein concentrations with a commercially available Protein Assay13 (should be in a range from 4 to 6 µg/µL).
- Dissolve 1 mg of proteins in 1 mL of 0.1 M Tris-HCl buffer (pH 7.5) and add 2.5 mg of Pronase E to the solution. Incubate at 37 °C for 48 h, followed by boiling the solution at 100 °C for 5 min to deactivate the enzyme.
- Store the protein digest solution at -80 °C for further use.
2. Purification using porous graphitic carbon (PGC)
- Activate PGC cartridges (100 mg, 5 mL) by adding 1 mL of 80% (v/v) acetonitrile (ACN) containing 0.1 % trifluoacetic acid (TFA) and repeat 2x.
- Wash the PGC cartridge with 1 mL of water 3x.
- Load the Pronase E digestion from 1 mg of protein on PGC (volume is dependent on protein concentration).
- Wash the cartridge 3x with 1 mL of water each time to remove salts.
- Elute Asn-glycans with 1 mL of 25% ACN in 0.1 % TFA, followed by 1 mL of 50% ACN in 0.1 % TFA.
- Pool both fractions and dry the samples with the refrigerated vacuum concentrator (Table of Materials) for further processing.
NOTE: Use MS grade solvents for Asn-glycan purification.
3. Permethylation of Asn-glycans
- Solubilize dried Asn-glycans with 100 µL of dimethyl sulfoxide (DMSO) in a 1.5 mL tube.
- Prepare DMSO-NaOH slurry by mixing two pellets of NaOH with 2 mL of DMSO. Add 200 µL of DMSO-NaOH slurry and 100 µL of methyl iodide to the sample.
- Cap the tube tightly and stir at 3,000 rpm for 10 min at room temperature with a vortex mixer.
- Quench the permethylation by adding 1 mL of water.
- Add 1 mL of chloroform and vortex for 1 min to extract permethylated Asn-glycans.
- Centrifuge at 9,000 x g for 3 min. Remove the supernatant and keep the lower chloroform phase with permethylated Asn-glycans.
- Wash the organic layer by adding 1 mL of water with vortexing for 1 min. Centrifuge at 9,000 x g for 3 min and remove the supernatant. Repeat 2x.
- Place the organic phase in a fume hood and let it dry with a nitrogen stream at room temperature.
- Store permethylated Asn-glycans at -80 °C for mass spectrometric analysis.
4. Electrospray Ionization Mass Spectrometry (ESI-MS) and ESI-MS/MS analysis
- Dissolve permethylated Asn-glycans in 20 µL of 50% 2-propanol in water (v/v).
- Perform CE-ESI-MS analysis using a 90 cm bare fused-silica capillary with a sheath flow of 50% 2-propanol in water at 1 µL/min.
NOTE: CE-MS is a system that combines capillary electrophoresis (CE) and a mass spectrometry. CE separate ions based on their ion mobility. In this protocol, the CE system is employed to introduce minute volume of samples and perform on-line desalting. - Set the ESI voltage at 5 kV in positive mode.
- Load permethylated Asn-glycan samples at 500 mbar for 0.2 min, corresponding to approximately 100 nL.
- Acquire MS data using a mass spectrometer (Table of Materials) using the following acquisition parameters.
- For full MS, set dwell time at 2.0 ms per step of 0.1 m/z unit. In the MS2 and MS3 experiments with enhanced product ion scan (EPI), set the scan speed to 4,000 Da/s, with Q0 trapping. Set the trap fill time as Dynamic and the resolution of Q1 as Unit. For MS3 experiments, set the excitation coefficient to make sure that only the monoisotopic peak for a single charged precursor is selected. Set the excitation times at 100 ms.
5. Analysis of permethylated glycans by MALDI-TOF MS
- Dissolve permethylated Asn-glycans in 10 µL of 50% methanol/water (v/v).
- Mix 1 µL of dissolved Asn-glycans with 2 µL of 2,5-dihydroxybenzoic acid solution (10 mg/mL in 50% acetonitrile/water (v/v)).
- Add 1 µL of mixed sample to a 384-well MALDI plate and wait until the sample is completely dried.
- Insert the plate into MALDI-TOF mass spectrometer equipped with a pulsed nitrogen laser (337 nm). Set the accelerating voltage at 20 kV in the positive mode.
- Set laser power to an arbitrary scale of 6,000 and acquire data in positive reflectron mode from m/z 1,000 to 5,000.
Representative Results
The method based on the combination of pronase E digestion and permethylation was applied to N-glycan analysis from total protein extracts of C. jejuni 11168H. Figure 1 shows a flowchart of the experimental procedure. In typical digestion, the ratio of enzyme to glycoprotein was set between 1:100 to 1:20. Here, the ratio of Pronase E to protein was increased to 2:1-3:1 and longer digestion was employed to obtain exhaust digestion by incubation at 37 °C for 48 h. The Asn-glycans from exhaust digestion were then purified by PGC and permethylated with a routine DMSO-NaOH approach. The MALDI-MS analysis of the permethylated Pronase E digests is presented in Figure 2. Three permethylation reaction times, i.e., 5 min, 10 min, and 20 min, were investigated. The obtained MS spectra indicated that the combination of Protonase E digestion and permethylation is reproducible. While the ion at m/z 1755 corresponds to Glc1GalNAc5Bac, the ion at m/z 1866 was only 111 Da higher. To verify if this ion corresponds to the permethylated product of Glc1-GalNAc5Bac-Asn, the NMR analysis of the permethylated Glc-Asn standard was performed10. The results indicated that a double bond was formed to replace the NH2 group in Asn for Glc-Asn and Glc1GalNAc5Bac-Asn. Interestingly, the data reveals the presence of free N-glycan, i.e., m/z 1755, and its relative quantity is much higher than that of Asn-linked glycan, i.e., m/z 1866. Therefore, the combination of Pronase E digestion and permethylation is not only a universal glycomics technique, but it can also distinguish free glycans from amino acid-linked glycans.
The MS and MS/MS spectra of permethylated Glc1GalNAc5Bac and Glc1-GalNAc5Bac-Asn from ESI-MS and ESI-MS/MS analysis were shown in Figure 3. The ions at m/z 1755 and m/z 1866 correspond to Glc1GalNAc5Bac and Glc1GalNAc5Bac-Asn, respectively (Figure 3A). The Glc1GalNAc5Bac-Asn structure was confirmed by annotating the MS/MS experiment (Figure 3B). Free heptasaccharide structure was further confirmed from the fragment spectrum of m/z 1755 (Figure 3C). Moreover, one can easily distinguish free N-glycans from Asn-linked glycans by observing the mass interval of 111 Da.
Figure 1: Flowchart of the experimental procedure. Please click here to view a larger version of this figure.
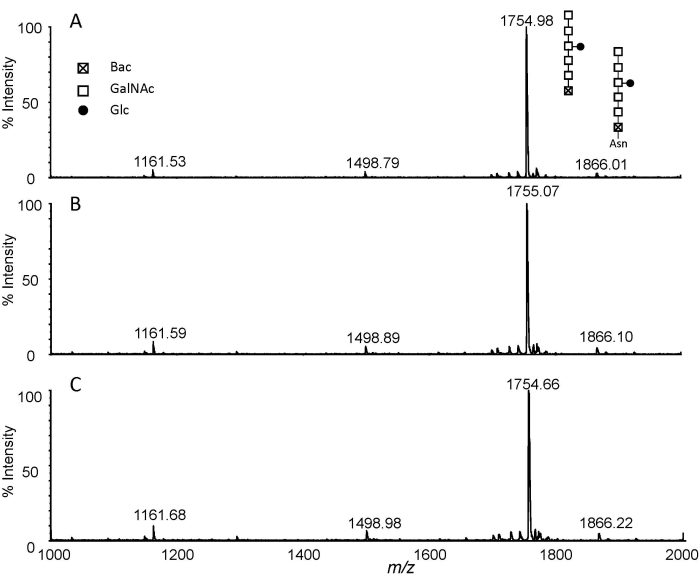
Figure 2: MALDI-TOF analysis. Extracted spectra of permethylated glycans isolated from pronase E digested C. jejuni 11168H (1 mg) with different permethylation reaction times. (A) 5 min; (B) 10 min; and (C) 20 min. Abbreviations: Glc, glucose; GalNAc, N-acetylgalactosamine; Bac, bacillosamine; Asn, asparagine. Please click here to view a larger version of this figure.
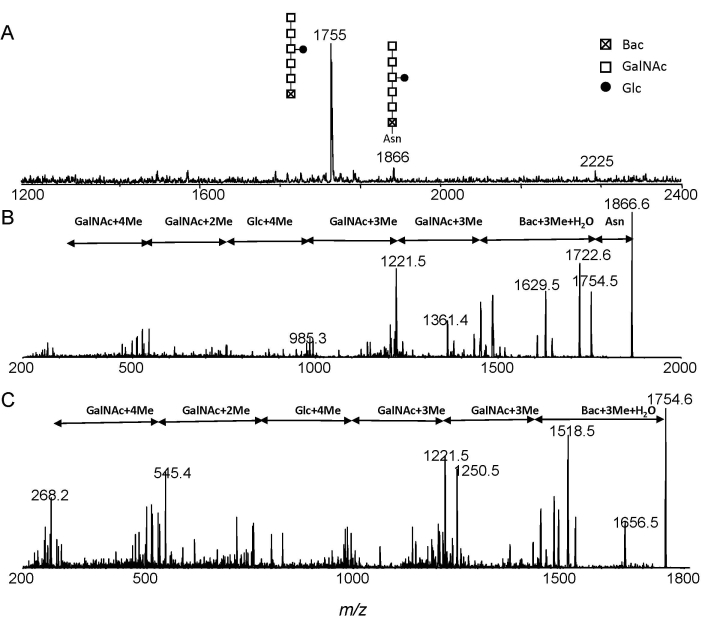
Figure 3: ESI-MS analysis. Extracted spectra of N-linked glycans from C. jejuni after Pronase E digestion. (A) Extracted MS; (B) MS/MS of precursor ion at m/z 1866; (C) MS/MS of precursor ion at m/z 1755. Abbreviations: Glc, glucose; GalNAc, N-acetylgalactosamine; Bac, bacillosamine; Asn, asparagine. Please click here to view a larger version of this figure.
Discussion
There are two critical steps in the protocol for the implementation of this universal glycomics strategy. The first critical step is the completion of exhaust digestion. This method strongly depends on the completion of Pronase E digestion. It is thus essential to use a long digestion time and a high enzyme-to-protein ratio. Typically, it is suggested to use a ratio of 2:1-3:1 for Pronase E/protein, and an incubation time of 48 h. It has been demonstrated that this proposed exhaust digestion produced the glycans with a single amino acid as the major component10. The products from exhaust digest need to be purified by a PGC cartridge. It did not only retain glycans but also the undigested enzymes. However, the enzyme proteins will not be eluted from the cartridge due to their high hydrophobicity.
The second critical step is the completion of methylation. Although MALDI-MS can be used to analyze glycans in their native forms, it is advantageous to convert glycans into their methylated derivatives. The analysis of permethylation glycans makes it feasible to determine branching and interglycosidic linkages. Permethylation can also help produce more predictable ion products during MS/MS fragmentation by stabilizing the sialic acid in oligosaccharides with terminal sialylation. It is worth mentioning that the molecular masses for permethylated Pronase E released Asn-glycans were only 111 Da higher compared to free oligosaccharides. This is due to molecular rearrangements. NMR spectroscopy of permethylated Asn-glycans confirmed that the NH2 group in Asn had been replaced with a double bond10.
A few experimental conditions can be modified when applied to different samples. Firstly, digestion time and enzyme-to-protein ratio can be optimized. Nonspecific proteolytic digestion of glycoproteins can be used in glycomics and glycoproteomics with different enzyme/protein ratios and digestion times, although it may result in producing Asn-glycans with different amino acid residues. For pronase E digestion, glycoproteins are digested to small Asn-glycans having one to six amino acid residues, which can also be analyzed using mass spectrometry. In addition, microwave irradiation can be employed to accelerate the proteolytic cleavage of glycoproteins mediated by Pronase E14. Secondly, different approaches can be used for the derivatization of glycans. In addition to the permethylation of glycans, a simple and rapid “one-pot” methylation method can be used to esterify sialic acids and construct a permanent charge for Asn-linked glycan analysis15. Briefly, the carboxylic acids of Asn and sialic acids were converted to Na+ form by passing through the cation exchange column. Converted Asn-glycans were then methylated with methyl iodide and the derivatized glycans can be purified with a hydrophilic interaction chromatography cartridge. Finally, other ESI mass spectrometers from different vendors can be used to analyze underivatized or permethylated glycans. MALDI-TOF/TOF can be used for analyzing permethylated glycans.
Unlike conventional glycomics strategy, this method is capable of differentiating free glycans and amino acid-linked glycans. For example, we characterized free glycans and N-linked glycans in C. jejuni. When glycans were directly isolated from C. jejuni 11168 whole-cell lysate, free glycans could be quantified in different culture conditions11. The results indicated that the quantity of free oligosaccharide (fOS) was inversely proportional to the solute concentration in the growth media. The higher salt and sucrose concentration resulted in lower quantity of fOS11.
There are also a few limitations of the method. The analytical throughput is limited by the long times for digestion (1-3 days) compared to routine digestion procedures (usually overnight). In addition, time-consuming permethylation can negatively impact the throughput.
The importance and potential of this method are the usefulness for screening N-linked glycans expressed by eukaryotes and bacteria and for detecting novel intermediates of N-linked glycosylation pathways.
Offenlegungen
The authors have nothing to disclose.
Acknowledgements
The authors thank Kenneth Chan, David J. McNally, Harald Nothaft, Christine M. Szymanski, Jean-Robert Brisson, and Eleonora Altman for assistance and helpful discussions.
Materials
| 2,5-dihydroxybenzoic acid (DHB), | Sigma-Aldrich (St. Louis, MO) | 149357 | |
| 2-Propanol | Fisher Scientific( Ottawa, Ontario,Canada) | AA22906K7 | |
| 4000 Q-Trap | AB Sciex (Concord, Canada) | Mass spectrometer | |
| 4800 MALDI-TOF/TOF | Applied Biosystems (Foster City, CA) | Mass spectrometer | |
| acetonitrile (ACN) | Fisher Scientific( Ottawa, Ontario,Canada) | A996-4 | |
| Brain Heart Infusion (BHI) broth | Sigma-Aldrich (St. Louis, MO) | 5121 | |
| C18 Sep-Pak cartridges | Waters (Milford, MA). | WAT036945 | |
| chloroform | Sigma-Aldrich (St. Louis, MO) | CX1054 | |
| Difco | Fisher Science (Ottawa, Ontario,Canada) | DF0037-17-8 | Fisher Science |
| dimethyl sulfoxide (DMSO) | Sigma-Aldrich (St. Louis, MO) | 276855 | |
| Eppendorf tube | Diamed (Missisauga, Ontario,Canada) | SPE155-N | |
| glacial acetic acid | Sigma-Aldrich (St. Louis, MO) | A6283 | |
| methanol | Fisher Scientific, Ottawa, Ontario,Canada) | A544-4 | |
| methyl iodide | Sigma-Aldrich (St. Louis, MO) | 289566 | |
| PGC cartridge | Thermo Scientific(Waltham,MA) | 60106-303 | Porous graphitic carbon |
| Pronase E | Sigma-Aldrich (St. Louis, MO) | 7433-2 | |
| Protein Assay Kit | Bio-rad (Mississauga, Ontario, Canada) | 5000001 | |
| Refrigerated vacuum concentrator | Thermo Scientific(Waltham,MA) | SRF110P2-230 | |
| sodium hydroxide | Sigma-Aldrich (St. Louis, MO) | 367176 | |
| Sonicator Ultrasonic Processor | Mandel Scientific (Guelph, Ontario,Canada) | XL 2020 | |
| Trifluoacetic acid (TFA) | Fisher Scientific( Ottawa, Ontario,Canada) | A11650 | |
| Tris-HCl | Sigma-Aldrich (St. Louis, MO) | T3253 |
Referenzen
- Dwek, R. A. Glycobiology: more functions for oligosaccharides. Science. 269 (5228), 1234-1235 (1995).
- Scheiffele, P., Peränen, J., Simons, K. N-glycans as apical sorting signals in epithelial cells. Nature. 378 (6552), 96-98 (1995).
- Hofmann, J., Hahm, H. S., Seeberger, P. H., Pagel, K. Identification of carbohydrate anomers using ion mobility-mass spectrometry. Nature. 526 (7572), 241-244 (2015).
- Kowarik, M., et al. N-linked glycosylation of folded proteins by the bacterial oligosaccharyltransferase. Science. 314 (5802), 1148-1150 (2006).
- Young, N., et al. Structure of the N-linked glycan present on multiple glycoproteins in the Gram-negative bacterium, Campylobacter jejuni. Journal of Biological Chemistry. 277 (45), 42530-42539 (2002).
- Wacker, M., et al. N-linked glycosylation in Campylobacter jejuni and its functional transfer into E. coli. Science. 298 (5599), 1790-1793 (2002).
- Szymanski, C. M., Wren, B. W. Protein glycosylation in bacterial mucosal pathogens. Nature Review Microbiology. 3, 225-237 (2005).
- Linton, D., et al. Functional analysis of the Campylobacter jejuni N-linked protein glycosylation pathway. Molecular Microbiology. 55 (6), 1695-1703 (2005).
- Nwosu, C. C., et al. Simultaneous and Extensive Site-specific N- and O-Glycosylation Analysis in Protein Mixtures. Analytical Chemistry. 85, 956-963 (2013).
- Liu, X., et al. Mass spectrometry-based glycomics strategy for exploring N-linked glycosylation in eukaryotes and bacteria. Analytical Chemistry. 78 (17), 6081-6087 (2006).
- Nothaft, H., Liu, X., McNally, D. J., Li, J., Szymanski, C. M. Study of free oligosaccharides derived from the bacterial N-glycosylation pathway. Proceedings of the National Academy of Sciences of the United States of America. 106 (35), 15019-15024 (2009).
- Macdonald, S. E., et al. Draft genome sequence of Campylobacter jejuni 11168H. Genome Announcements. 5 (5), e01556 (2017).
- Bradford, M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Analytical Biochemistry. 72, 248-254 (1976).
- Liu, X., Chan, K., Chu, I. K., Li, J. Microwave-assisted nonspecific proteolytic digestion and controlled methylation for glycomics applications. Carbohydrate Research. 343 (17), 2870-2877 (2008).
- Liu, X., et al. 34;One-pot" methylation in glycomics application: Esterification of sialic acids and permanent charge construction. Analytical Chemistry. 79 (10), 3894-3900 (2007).

.
