Generation of Fluorescent Protein Fusions in Candida Species
Summary
PCR-mediated gene modification can be used to generate fluorescent protein fusions in Candida species, which facilitates visualization and quantitation of yeast cells and proteins. Herein, we present a strategy for constructing a fluorescent protein fusion (Eno1-FP) in Candida parapsilosis.
Abstract
Candida species, prevalent colonizers of the intestinal and genitourinary tracts, are the cause of the majority of invasive fungal infections in humans. Thus, molecular and genetic tools are needed to facilitate the study of their pathogenesis mechanisms. PCR-mediated gene modification is a straightforward and quick approach to generate epitope-tagged proteins to facilitate their detection. In particular, fluorescent protein (FP) fusions are powerful tools that allow visualization and quantitation of both yeast cells and proteins by fluorescence microscopy and immunoblotting, respectively. Plasmids containing FP encoding sequences, along with nutritional marker genes that facilitate the transformation of Candida species, have been generated for the purpose of FP construction and expression in Candida. Herein, we present a strategy for constructing a FP fusion in a Candida species. Plasmids containing the nourseothricin resistance transformation marker gene (NAT1) along with sequences for either green, yellow, or cherry FPs (GFP, YFP, mCherry) are used along with primers that include gene-specific sequences in a polymerase chain reaction (PCR) to generate a FP cassette. This gene-specific cassette has the ability to integrate into the 3′-end of the corresponding gene locus via homologous recombination. Successful in-frame fusion of the FP sequence into the gene locus of interest is verified genetically, followed by analysis of fusion protein expression by microscopy and/or immuno-detection methods. In addition, for the case of highly expressed proteins, successful fusions can be screened for primarily by fluorescence imaging techniques.
Introduction
Candida species are commensal fungi that colonize the intestinal and genitourinary tracts of all humans. Under conditions of immunodeficiency, such as that occur with premature birth or immunosuppressive effects from treatments for cancer, Candida species can become opportunistic pathogens. Of the Candida species, Candida albicans is the most prevalent fungal colonizer and causes the majority of invasive fungal infections. Other Candida species such as C. glabrata, C. parapsilosis, C. tropicalis, and C. kruseii also cause serious infections in immunocompromised patients, with some exhibiting intrinsic resistance to commonly used anti-fungal antibiotics such as fluconazole and amphotericin B. Hence, infections with some of these species are being observed more frequently, especially in patients being treated prophylactically with anti-fungal agents. Even with appropriate and timely anti-fungal treatment, invasive Candida infections continue to be associated with significant morbidity and mortality1. Because of the significance of Candida species in human health, there is a need for readily available molecular tools that allow the study and elucidation of their pathogenesis mechanisms.
One important tool that allows researchers to visualize and quantify microbial cells and the proteins that they express is FP fusion technology. Polymerase chain reaction (PCR)-mediated gene modification, as described in this paper, allows the construction of fusions, between FP sequences and a Candida protein coding sequence of interest at its genomic locus. Stable integration of the construct facilitates analysis of protein expression as well as protein localization dynamics. Plasmids containing FP sequences, optimized for expression in Candida albicans and that can be used in the PCR-mediated gene modification strategy, have been previously constructed2,3,4,5. Plasmids contain FP transformation "cassettes": a FP sequence linked to a nutritional marker gene that facilitates the transformation of C. albicans and C. parapsilosis2,3,4,5,6,7. Currently available plasmids contain a variety of selectable nutritional marker genes (URA3, HIS1, ARG4) for transformation of auxotrophic strains as well as a dominant drug resistance marker (NAT1), which facilitates transformation of clinical strains lacking auxotrophies. In addition, plasmids contain options for up to four different FP sequences (green [GFP], yellow [YFP], cyan [CFP], and cherry [mCherry]) and either an ADH1 termination sequence for construction of carboxy-terminus protein fusions, or a promoter sequence for construction of amino-terminus protein fusions. Primers are designed with homology to the plasmid DNA surrounding the FP cassette. In addition, the primers also contain 5'-extension sequences bearing homology to the yeast gene of interest to be tagged, which facilitates integration of the cassette into the genomic locus via homologous recombination (Figure 1). Gene-specific FP cassettes are generated by PCR and then transformed into Candida cells made competent for uptake of DNA by treatment with lithium acetate.
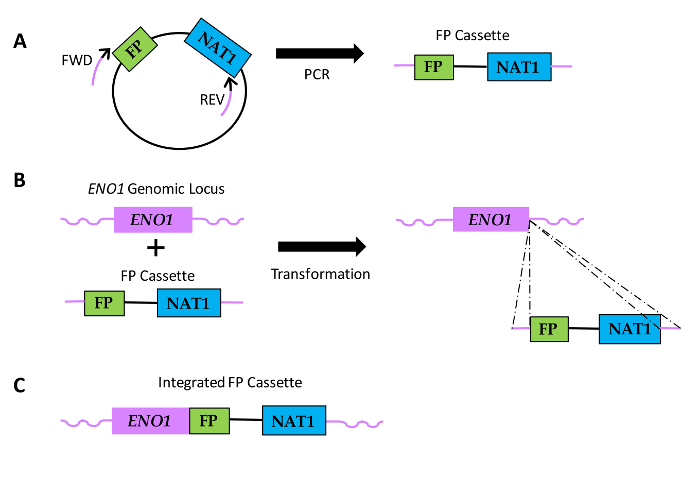
Figure 1: Diagram of how FP sequence fusions are generated in Candida species. (A) Plasmid DNA includes a FP sequence and a sequence encoding nourseothricin resistance (NAT1). Relative locations of Forward (FWD) and reverse (REV) primers are shown, with black portions of the primers indicating the region of homology to the plasmid sequence and the purple portions denoting the gene-specific homology region or primer extension. (B) FP cassettes are transformed into Candida and integrate within the ENO1 genomic locus via homologous recombination (dotted lines). (C) Resulting FP fusion sequence at the 3'end of ENO1. Please click here to view a larger version of this figure.
Herein, we present an example of protein fusion (Eno1-FP) constructions in Candida species. We use tagging plasmids containing the NAT1 transformation marker gene along with sequences encoding GFP, YFP, or mCherry (Figure 2). These plasmids are used along with primers in PCR to generate gene-specific cassettes that facilitate fusion of FPs to the 3'-end of ENO1, resulting in expression of Eno1 fused to FPs at its carboxy-terminus.

Figure 2: Maps of FP cassette-containing plasmids. Forward (F) and reverse (R) primers used to generate the cassettes from the plasmids are indicated along with the relative location of their homology to the plasmids. Primer sequences are as listed in Table 1. F1 and R1 were also used to generate the pYFP-NAT1 cassette. The plasmid containing the YFP-NAT1 cassette (pMG2263) is identical to pMG2120 with the exception of YFP in place of the GFP sequence. Cassette sizes: GFP-NAT1, 3.7 kbp; mCherry-NAT1, 3.2 kbp; YFP-NAT1, 3.7 kbp. This figure has been modified from Gerami-Nejad, et al.4 Please click here to view a larger version of this figure.
Protocol
1. Isolate Template Plasmids from E. coli
- Grow E. coli containing the template plasmid overnight in 10 ml lysogeny broth (LB) + 200 mg/L ampicillin (AMP) at 37 °C with shaking.
- Harvest cells by centrifuging at 6,000 x g for 2 min.
- Decant liquid, isolate and purify DNA from E. coli cells by a standard method as described previously in Ausubel et al.8.
- Resuspend DNA in Tris-EDTA (TE; 10 mM Tris, pH 8.0, 1 mM EDTA, pH 8.0) at a working concentration of 50-100 ng/ml.
2. Design Primers
| Primer | Primer Sequence |
| F1 | 5' GAGAATTGAAGAAGAGTTGGGAGACAATGCTATCTATGCTGGTAAGGACTTCCACAATGCTCAAACTTTG GGTGGTTCTAAAGGTGAAGAATTATT 3' |
| R1 | 5' GAGCGTTTGCACCAACAGGCCATCATTTGTGACGAGAGAAGACCTGACGTCATTAGATTGGCACCTTTGC GTAAAACGACGGCCAGTGAATTC 3' |
| F2 | 5' GAGAATTGAAGAAGAGTTGGGAGACAATGCTATCTATGCTGGTAAGGACTTCCACAATGCTCAAACTTTG GGTGGTGTTTCAAAAGGTGAAGAAGATAAT 3' |
| R2 | 5' GAGCGTTTGCACCAACAGGCCATCATTTGTGACGAGAGAAGACCTGACGTCATTAGATTGGCACCTTTGC ACTGGATGGCGGCGTTAGTATC 3' |
Table 1: Primer sequences used in this study. Bold italicized text indicates homology to the ENO1 genomic locus, normal font regions are homologous to plasmid DNA.
- Design primers to be homologous to plasmid sequences bordering the cassette to be amplified as well as to the 3'-end of the target gene of interest (e.g. ENO1) to facilitate recombination into the gene's genomic locus (Figure 2 and Table 1).
- Ensure that the forward primer sequences match the last 70 base pairs (bp) of the gene of interest, 5'- 3', minus the stop codon, to maintain the coding frame, plus the first approximately 30 bp of the plasmid sequence to be amplified. As a note, the GGTGGTGGTT in each primer is a poly-glycine linker with no FP homology. Of note, in the absence of a linker, there can be direct fusion of functional domains, which can theoretically lead to protein misfolding, low yield in protein production, or impaired bioactivity.
- Ensure that the reverse primer sequences are 70 bp just downstream of gene, 3'- 5', not including any gene sequence, plus the last approximately 30 bp of the nutritional or drug resistance marker used in the plasmid.
- Use F1 and R1 to generate GFP-NAT1 and YFP-NAT1 cassettes with pMG2120 and pMG2263, respectively and F2 and R2 to generate mCherry-NAT1 with pMG2343.
3. Generate FP Cassettes by PCR (Day 1)
- Prepare reagents for PCR. Make a master mix (500 µl final volume) by adding the following volumes and concentrations to a 1.5 ml tube: 50 µl PCR buffer (500 mM potassium chloride, 100 mM Tris pH 8.0 in water), 20 µl deoxynucleotides (dNTPs; stock mixture of nucleotides at 10 mM each), 40 µl 25 mM magnesium chloride, 20 µl purified plasmid (from ~50-100 ng/µl stock solution), 10 µl each forward and reverse primer (from 10 mM stock solutions), 30 µl Taq polymerase (generic, 5,000 units/ml), and 320 µl water.
- Aliquot 50 µl of master mix into each of 10 PCR-compatible 0.5 ml tubes.
- Place PCR tubes in thermocycler and run the following steps: 1 cycle of 5 min at 94 °C to denature dsDNA; 40 cycles sequentially of 45 sec at 94 °C, 30 sec at 55 °C to allow primers to anneal to plasmid template DNA, and 4 min at 68 °C for extension of DNA products; and 1 final extension cycle of 15 min at 72 °C.
NOTE: The PCR master mix and cycling parameters may need to be modified based on the particular Taq polymerase used. - Pool all products from the 10 PCR reactions in a 1.5 ml tube.
- Subject 5 µl of pooled PCR product to agarose gel electrophoresis to verify amplicon size and obtain an estimate of product concentration, based on comparison to a DNA ladder. Generally, use ~250 µg of cassette DNA in each subsequent transformation mix.
- Precipitate DNA by adding 50 µl 3 M sodium acetate followed by 750 µl 95% ethanol to the products and incubate at least 30 min at -20° C.
- Harvest the PCR products by centrifuging the tube at 16,000 x g for 10 min. Carefully remove and discard the supernatant and dry the pellet overnight. Resuspend the dried DNA cassette pellet in 40 µl TE pH 8.0 and store at room temperature until use.
4. Transform Candida Cells with FP DNA Cassettes
- On Day 1, recover yeast strain to be transformed from a 15% glycerol frozen (-80 °C) stock by streaking a few scraped crystals onto yeast peptone dextrose with adenine (YPAD) agar and incubate at 30 °C. After recovery of colony growth, inoculate a single colony into 2 ml liquid YPAD medium in a glass culture tube with a breathable cap and incubate overnight at 30 °C with agitation.
- On Day 2, dilute 300 µl of overnight yeast culture into 50 ml fresh YPAD (to a final OD600 of ~0.2) in a 125 ml Erlenmeyer flask with a breathable cap. Shake at 30 °C for ~3 hr (to a final OD600 ~0.6-0.8).
- Pour the overnight culture into a 50 ml conical tube and pellet the cells by spinning for 5 min at 1 500 x g in a table top centrifuge.
- Pour off and properly discard the supernatant. Resuspend the cell pellet in 5 ml water. Re-pellet the cells by centrifuging again for 5 min at 1,500 x g in a table top centrifuge.
- Pour off and properly discard the supernatant. Resuspend the cells in 500 µl TELiAc (TE lithium acetate: 10 mM Tris, pH 8.0, 1 mM EDTA, pH 8.0, 0.1 M lithium acetate) while transferring to a 1.5 ml tube. Centrifuge the tube for 2 min at 3,000 x g in a microcentrifuge.
- Resuspend cells in 250 µl TELiAc. The total volume including the pellet should be ~300 µl.
- To a clean (different microfuge tube than in 4.2.4), add 5 µl carrier DNA (10 mg/ml) and 150 µl of prepared Candida cells (from 4.2.4). This is the negative control for transformation.
- To a second clean microfuge tube, add 5 µl of denatured carrier DNA (that has been boiled at 90 °C for 10 min and cooled to 4 °C), all 40 µl of the prepared PCR product (from 3.3.1) and 150 µl of prepared Candida cells (from 4.2.4).
- Incubate the two transformation mixes for 30 min at room temperature.
- To each transformation mix tube, add 700 µl PLATE mix (10 mM Tris, pH 8.0, 1 mM EDTA, pH 8.0, 0.1 M lithium acetate in 50% polyethylene glycol 3350). Invert the tubes to mix and incubate them overnight at RT.
- On Day 3, incubate the transformation mixes at 42 °C for 1 hr (heat shock).
- Centrifuge the transformation mixes for 30 sec at 16,000 x g in a microcentrifuge. Remove and properly discard the supernatant. Resuspend each of the cell pellets in 150 µl water by gently pipetting up and down so as to not to damage the cells.
- For transformations utilizing auxotrophic marker genes (e.g. URA3), plate each entire mixture by pipetting the solutions onto the appropriate selective media agar (e.g. lacking uridine) and spread the mixture evenly using sterile glass beads.
- For transformations utilizing the nourseothricin resistance marker gene (NAT1), as described here, plate transformation mixes first onto non-selective YPAD agar and incubate at 30 °C for 6-12 hr. This step aids in cell recovery, post heat shock, before nourseothricin stress is applied.
- After partial growth recovery, replica plate the Candida cells onto YPAD containing 400 µg/ml nourseothricin. For transformations utilizing nutritional marker genes (e.g. URA3), this intermediate plating step is not needed and cells can be directly plated onto selective yeast media (e.g. YPAD lacking uridine) as described in 4.4.2.
NOTE: If the transformation is successful, colonies should appear within one to three days (potentially up to five days of outgrowth for selection on nourseothricin-containing agar). No colonies should appear on plates spread with transformation mixes containing carrier DNA alone (negative control).
- For auxotrophic and nourseothricin marker selection, streak putative transformants as single colonies to fresh selective media agar plates and incubate at 30 °C to propagate yeast cells that can be screened for successful construction of FP fusions.
- Screen transformants for correct integration of the tagging cassette (see Representative Results for a detailed example). If the gene of interest is expressed at sufficient amounts, whole colony fluorescence may occur such that it is possible to detect potential candidate integrants using a plate imaging system with fluorescence detection ability.
- Check putative integrants by PCR using primers homologous to sequences outside of the region of integration to confirm fusion to the target gene.
- In addition, consider Western blot analysis to determine the expression and size of the fusion protein, as well as by fluorescence microscopy of single cells for visual confirmation of protein localization, if known.
Representative Results
As an example, we used the protocol described above to construct GFP and mCherry fusions to Eno1 in a C. parapsilosis laboratory strain. Each putative transformant was initially restreaked for growth. In this example, since the resultant fusion protein is highly expressed (enolase) and the FPs are bright, we were able to screen transformants by fluorescence microscopy prior to performing diagnostic PCR (Figure 3)6.
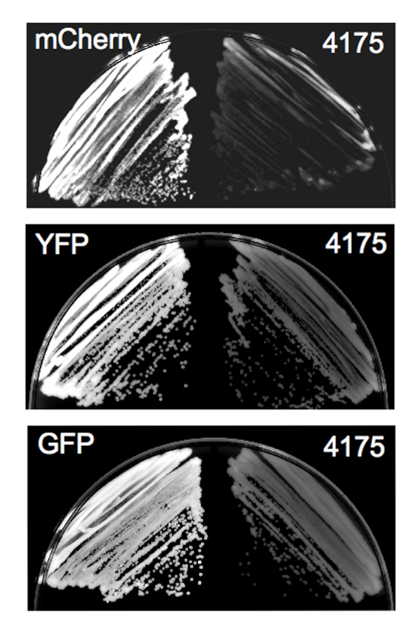
Figure 3: Colony fluorescence exhibited by Candida transformants. Representative images of yeast plate growth obtained using a laser scanning system equipped with a Cy3 filter to image the mCherry-expressing strain and a Cy2 filter to image the YFP- and GFP-expressing strains. Left panel in each pair, FP-expressing strains; right panel in each pair, non-transformed parent strain (4175) imaged with the same filter as the corresponding FP-expressing strain. Following visualization, the images were inverted and brightness and contrast were adjusted identically for FP-expressing and control strains. This figure is reprinted with permission from Gonia, et al.6 Please click here to view a larger version of this figure.
Genomic DNA was isolated9 from each of these putative transformants and used as the DNA template, along with primers complementary to sequences flanking the sites of cassette integration (i.e. sequences outside of where F and R primers anneal within the intended genomic locus to be tagged), in diagnostic PCRs to verify correct integration of the FP cassette just prior to the stop codon of the gene of interest. Of note, for lithium acetate transformations with C. parapsilosis, we observed lower efficiency (~1-2% of colonies screened contained a correct integration) as compared to our experience with C. albicans transformations.
To analyze enolase localization, we observed yeast cells expressing Eno1-FP constructs by fluorescence microscopy using a photomicroscope equipped with a 100 W mercury lamp and epifluorescence illumination with GFP, YFP and Texas Red filter sets. A charge-coupled device (CCD) camera was used to collect images that were processed with image analysis software (Figure 4A)6. In addition, we found that different FP colors were useful to visually distinguish different yeast strains within a mixture (Figure 4B)6. Because enolase is highly expressed and located in a large portion of the yeast cell, FP fusions to it can be used to generate fluorescent yeast strains that can be easily visualized, for example, in analyses of Candida-host interactions.
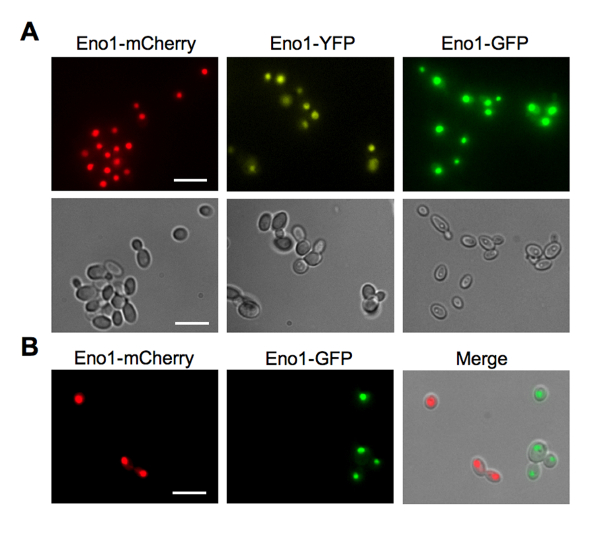
Figure 4: Expression and localization of Eno1-FP fusions by fluorescence microscopy. (A) Fluorescence (top) and differential interference contrast (DIC, bottom) images for Candida strains expressing Eno1-FPs. Eno1-FPs in C. albicans and C. parapsilosis tend to concentrate in the nucleus as well as in the cytoplasm, but to a much lower degree. (B) Microscopic images of a mixed culture of Candida strains expressing Eno1-mCherry and Eno1-GFP. Right panel, Texas red filter; center panel, GFP filter; right panel, merge of both fluorescent panels with the DIC image. Scale bars = 10 µm. This figure is reprinted with permission from Gonia, et al.6 Please click here to view a larger version of this figure.
FP fusions also facilitate analysis of protein expression levels by Western blotting. Using yeast cells generated by the protocol described above, proteins were isolated from cell lysates, separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and transferred to a polyvinylidene difluoride (PVDF) membrane10. Eno1-FP fusions were detected on the blots using commercially available antibodies as previously described6. Briefly, mouse anti-GFP (1:2,000 dilution), followed by horseradish peroxidase-conjugated goat anti-mouse IgG (1:10,000 dilution) antibodies were used to probe for GFP- and YFP-tagged enolase. Rabbit anti-mCherry (1:2,000 dilution) followed by horseradish peroxidase-conjugated goat anti-rabbit IgG (1:10,000 dilution) antibodies were used to probe for mCherry-tagged enolase. All fusion proteins were readily detected from lysates of successful transformants (Figure 5, lanes 2, 5, and 7)6 and were of expected size (~ 75 kDa) as compared to a protein ladder standard and C. albicans enolase-FP standards (Figure 5, lanes 1 and 6)6. No signal was observed for the untagged parent yeast strain (Figure 5, lanes 3 and 4)6.
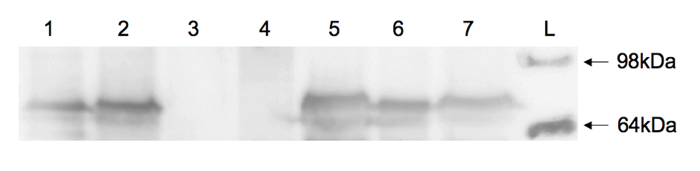
Figure 5: Immunoblot of cell lysates from Candida strains. Lane 1, C. albicans containing ENO1-mCherry fusion (J. Berman, University of Minnesota) (positive control); lane 2, C. parapsilosis containing ENO1-mCherry fusion; lanes 3 and 4, untransformed C. parapsilosis parent strain 417511 (negative control); lane 5, C. parapsilosis containing ENO1-YFP fusion; lane 6, C. albicans containing ENO1-GFP fusion (J. Berman, University of Minnesota) (positive control); lane 7, C. parapsilosis containing ENO1-GFP fusion; lane L, protein ladder with 98 and 64 kDa protein standards indicated. Lanes 1-3 were incubated with anti-mCherry antibody and lanes 4-7 were incubated with anti-GFP antibody. This figure is reprinted with permission from Gonia, et al.6 Please click here to view a larger version of this figure.
| Protocol Step/Item | Modification | Potential Improvement (cautions) |
| Primer design | Lengthen primers by including longer region of homology to genomic locus of interest | Improve binding efficiency and homologous recombination (modifying primer length may change optimal annealing temperatures) |
| PCR (amplicon quality) | Use higher fidelity Taq Gel purify PCR product |
Reduce introduction of errors into cassette sequences to improve targeting of primer to genome sequence and transformation efficiency (gel purification may decrease amplicon quantity and hence, transformation efficiency) |
| PCR (amplicon quantity) | Increase number of PCR reactions and pool products | Increase number of transformed cells (too much amplicon can also result in reduced transformation efficiency) |
| Cassette precipitation | Perform centrifugation at 4 °C, keep all reagents and tubes on ice | Increase DNA yield and, hence, transformation efficiency |
| Heat Shock | Shorten the time | Improve viability of stressed yeast cells (shorter heat shock times could also result in decreased uptake of DNA cassette by cells) |
| Recovery Growth1 | Allow for longer recovery on YPAD agar before plating to agar containing nourseothricin | Stressed yeast cells may need longer recovery times when exposed to nourseothricin (longer incubation times may also increase contaminant and false-positive transformant growth) |
| Colony Outgrowth1,2 | Incubate plates for longer times (~ 5 d) | Stressed yeast cells may need longer incubation times for colony growth (longer incubation times may also increase contaminant and false-positive transformant growth) |
| 1 Specific to transformations using NAT1 selection marker or 2 transformations of yeast strains containing genetic mutations that affect growth | ||
Table 2: Guide for troubleshooting and optimization.
Discussion
Construction of epitope tagged sequences in Candida species using the PCR-mediated gene modification strategy described above can be summarized as a three-step process. First, a cassette is made by PCR that encodes both the sequence desired for integration and regions homologous to the locus of insertion into the yeast genome. Second, the yeast cells to be transformed are made chemically competent with lithium acetate and co-incubated with the cassette. Third, the cells are plated on selective media to recover transformants and resulting colonies are tested for correct integration of the cassette into the desired genomic locus.
There are several critical steps within this protocol to improve the success rate of obtaining a fluorescent fusion protein in C. albicans and C. parapsilosis. First, use of primers that have been purified by polyacrylamide gel electrophoresis (PAGE; by the manufacturer) is highly recommended. Without purification, there is an increased likelihood of obtaining primers of incorrect lengths in the final product, which could decrease the efficiency with which the cassette integrates into the intended genomic locus by homologous recombination. Second, we recommend verifying that the PCR-generated tagging cassette is of high quality, by analyzing ~5 µl of cassette DNA by agarose gel electrophoresis to ensure correct size of the PCR product. Third, to obtain yeast cells that are optimized for transformation, cells should be in the log phase of growth, with yeast buds evident by microscopy prior to their use. Fourth, it is imperative to use gentle pipetting technique during resuspension of transformed yeast cells after heat shock and before plating onto selective media. Fifth, for transformant selection on nourseothricin, which is toxic to yeast cells, allowing for more time for growth on YPAD agar before replicating onto nourseothricin-containing medium may be beneficial to promote recovery of transformants. Last, intact plasmid and PCR cassette integration outside of the intended genomic location does occur at low frequencies in Candida and will also result in colony growth. Thus, it is necessary to analyze all transformed colonies by PCR using primers outside of the sequence used for integration. For transformants that are verified by PCR but do not exhibit fluorescence by microscopy, Western blot analysis of cell lysates and sequencing should be considered to further investigate the reason for construction or visualization failure. In addition to these critical steps, there are several steps in the protocol that can be modified, if needed, to improve transformation efficiency and increase the frequency of correct cassette integration. A troubleshooting guide is presented in Table 2.
There are potential limitations of this technique that could be optimized for future applications. The lithium acetate method used for DNA transformation results in a lower efficiency (~1-2% of colonies screened contained a correct integration) for C. parapsilosis12, as compared to C. albicans. Electroporation is an alternative approach to lithium acetate transformation that may improve transformation efficiency13. Use of the ADH1 terminator, while necessary for a universal tagging method, may change the stability and/or regulation of the generated mRNA from that which would have occurred with the native terminator sequence. This potential issue needs to be considered when the native function of a protein is important to the specific research question. For fluorescent fusion proteins that are expressed at low levels, whole colony microscopy to identify or screen for successful transformants may not be an option. In this case, PCR and Western blot analyses will provide evidence of successful fusion protein sequence/protein construction. A limitation, common to all fusion protein constructions, is that epitope tags could interfere with the native function of the protein, resulting in unintended mutant phenotypes, including abnormal localization of the fusion protein. Consideration of these issues is important and appropriate controls should be included as indicated.
Plasmids useful for generating epitope tags in Candida by this PCR-mediated approach, in addition to those mentioned here, have previously been generated by our research groups and are available to researchers through the Fungal Genetics Stock Center (http://www.fgsc.net). More information about several of these plasmids can also be found at (http://www6.tau.ac.il/berman/). The available plasmids contain a myriad of combinations of nutritional and drug resistance markers, FP sequences to generate different colored yeast, other epitope tag options (HA, myc, V5, HIS9) to generate proteins that are tagged at either the carboxy- or amino-termini, and promoter sequences for constitutive and regulatable expression of proteins. Plasmids containing drug resistance markers are particularly beneficial to the field of fungal pathogenesis as this tool allows for the transformation and study of clinical yeast strains that are prototrophic, rendering conventional nutritional markers unusable. The construction of clinical Candida strains that fluoresce at different wavelengths will be useful for a variety of cell biological assays that require the ability to visualize and differentiate multiple yeast strains in co-culture.
Invasive fungal disease due to Candida species continues to be a significant human health challenge that is difficult to treat and is associated with high morbidity and mortality, especially in immunocompromised patients14,15. Thus, molecular tools that allow researchers to more quickly and easily study Candida species-host interactions are vitally important for furthering our understanding of Candida biology and pathogenesis mechanisms.
Offenlegungen
The authors have nothing to disclose.
Acknowledgements
We thank N. Dean for providing the original mCherry FP sequence, M. Gerami-Nejad for construction of plasmids, B. Larson for technical assistance, and T. Heisel for helpful advice during the development of this project. J.B. was supported by the European Research Council Advanced Award 340087 (RAPLODAPT). Microscopy and imaging systems were provided by the University of Minnesota Pediatrics Foundation and the University of Minnesota Imaging Center.
Materials
| 100W mercury lamp | CHIU Technical Corporation | M-100T | |
| 95% Ethanol | Any | NA | |
| Adenine | Any | NA | |
| Ampicillin | Any | NA | |
| Carrier DNA | Ambion | AM9680 | Sheared Salmon Sperm DNA 10 mg/ml |
| CCD Camera | Photometrics | CoolSNAP HQ | |
| Conical Tube | Corning | 430828 | 50ml |
| Culture Tube Rotator | New Brunswick | 2013923 | TC-8, or Any Culture Tube Rotator |
| Deoxynucleotides (dNTP) PCR Grade | Any | NA | |
| Eppendorf Tubes | Eppendorf | 022363719, 022363212 | 0.5ml, 1.5ml |
| Erlenmeyer Flask | Fisher Scientific | 7250089 | 125ml |
| Ethylenediaminetetraacetic Acid (EDTA) | Any | NA | |
| Freezer (-80 °C ) | Thermo Electron Corporation | ULT-1386-9-V | Revco Ultima II |
| GFP, YFP and Texas Red Filter Sets | Chroma Technology Corporation | 49002, 86004v2, 49008 | |
| Glass culture tubes | Fisher Scientific | 1496126 | 75mm |
| HRP goat anti-mouse antibody | Santa Cruz Biotechnology | SC-2005 | |
| HRP goat anti-rabbit antibody | Santa Cruz Biotechnology | SC-2301 | |
| Incubator (30 °C ) | Any | NA | |
| Lithium Acetate | Any | NA | |
| Lysogeny Broth (LB) Media | Any | NA | |
| Magnesium Chloride | Any | NA | |
| Microcentrifuge | Eppendorf | 5415 D | |
| Microscope | Nikon | E600 | Nikon Eclipse E600 |
| Microscope Image Analysis Software | Universal Imaging Corporation | 6.3r7 | MetaMorph Software Series 6.3r7 |
| Mouse anti-GFP antibody | Roche | 11814460001 | |
| Nourseothricin | Fisher Scientific | 50997939 | |
| PCR Thermocycler | Applied Biosystems | 9700 | GeneAmp PCR System |
| PCR tubes | BioExpress, GeneMate | T-3035-1 | 0.2ml |
| Polyethylene Glycol 3350 | Any | NA | |
| Potassium Chloride | Any | NA | |
| Rabbit anti-mCherry antibody | BioVision | 5993-100 | |
| Refrigerator (4°C) | Any | NA | |
| Sodium Acetate | Any | NA | |
| Stereomicroscope | Nikon | SMZ1500 | |
| Table Top Centrifuge | Labnet | Z 400 | Hermle Z 400 |
| Taq DNA Polymerase | Any | NA | |
| Tris(hydroxymethyl)aminomethane (Tris) | Any | NA | |
| Vortex Mixer | Scientific Industries | SI-0236 | Vortex Genie 2 |
| Yeast Extract Peptone Dextrose (YPD) Media | Any | NA |
Referenzen
- Bendel, C. M. Colonization and epithelial adhesion in the pathogenesis of neonatal candidiasis. Semin. Perinatol. 27 (5), 357-364 (2003).
- Gerami-Nejad, M., Berman, J., Gale, C. A. Cassettes for PCR-mediated construction of green, yellow, and cyan fluorescent protein fusions in Candida albicans. Yeast. 18 (9), 859-864 (2001).
- Gerami-Nejad, M., Dulmage, K., Berman, J. Additional cassettes for epitope and fluorescent fusion proteins in Candida albicans. Yeast. 26 (7), 399-406 (2009).
- Gerami-Nejad, M., Forche, A., McClellan, M., Berman, J. Analysis of protein function in clinical C. albicans isolates. Yeast. 29 (8), 303-309 (2012).
- Gerami-Nejad, M., Hausauer, D., McClellan, M., Berman, J., Gale, C. Cassettes for the PCR-mediated construction of regulatable alleles in Candida albicans. Yeast. 21 (5), 429-436 (2004).
- Gonia, S., Larson, B., Gale, C. A. PCR-mediated gene modification strategy for construction of fluorescent protein fusions in Candida parapsilosis. Yeast. 33 (2), 63-69 (2016).
- Milne, S. W., Cheetham, J., Lloyd, D., Aves, S., Bates, S. Cassettes for PCR- mediated gene tagging in Candida albicans utilizing nourseothricin resistance. Yeast. 28 (12), 833-841 (2011).
- Ausubel, F. M., et al. . Current Protocols in Molecular Biology. , (1995).
- Wilson, R. B., Davis, D., Mitchell, A. P. Rapid hypothesis testing with Candida albicans through gene disruption with short homology regions. J. Bacteriol. 181 (6), 1868-1874 (1999).
- Pulver, R., et al. Rsr1 focuses Cdc42 activity at hyphal tips and promotes maintenance of hyphal development in Candida albicans. Eukaryotic Cell. 12 (4), 482-495 (2013).
- Falgier, C., et al. Candida species differ in their interactions with immature human gastrointestinal epithelial cells. Pediatr. Res. 69 (5), 384-389 (2011).
- Nosek, J., et al. Genetic manipulation of the pathogenic yeast Candida parapsilosis. Curr. Genet. 42 (1), 27-35 (2002).
- Zemanova, J., Nosek, J., Tomaska, L. High-efficiency transformation of the pathogenic yeast Candida parapsilosis. Curr. Genet. 45 (3), 183-186 (2004).
- Benjamin, D. K., et al. Neonatal candidiasis among extremely low birth weight infants: risk factors, mortality rates, and neurodevelopmental outcomes at 18 to 22 months. Pediatrics. 117 (1), 84-92 (2006).
- Kullberg, B. J., Arendrup, M. C. Invasive Candidiasis. N Engl J Med. 373 (15), 1445-1456 (2015).

