Automatically Generated
Profiling of the Human Natural Killer Cell Receptor-Ligand Repertoire
Summary
Here we design two complementary mass cytometry (CyTOF) panels and optimize a CyTOF staining protocol with the aim of profiling the natural killer cell receptor and ligand repertoire in the setting of viral infections.
Abstract
Natural killer (NK) cells are among the first responders to viral infections. The ability of NK cells to rapidly recognize and kill virally infected cells is regulated by their expression of germline-encoded inhibitory and activating receptors. The engagement of these receptors by their cognate ligands on target cells determines whether the intercellular interaction will result in NK cell killing. This protocol details the design and optimization of two complementary mass cytometry (CyTOF) panels. One panel was designed to phenotype NK cells based on receptor expression. The other panel was designed to interrogate expression of known ligands for NK cell receptors on several immune cell subsets. Together, these two panels allow for the profiling of the human NK cell receptor-ligand repertoire. Furthermore, this protocol also details the process by which we stain samples for CyTOF. This process has been optimized for improved reproducibility and standardization. An advantage of CyTOF is its ability to measure over 40 markers in each panel, with minimal signal overlap, allowing researchers to capture the breadth of the NK cell receptor-ligand repertoire. Palladium barcoding also reduces inter-sample variation, as well as consumption of reagents, making it easier to stain samples with each panel in parallel. Limitations of this protocol include the relatively low throughput of CyTOF and the inability to recover cells after analysis. These panels were designed for the analysis of clinical samples from patients suffering from acute and chronic viral infections, including dengue virus, human immunodeficiency virus (HIV), and influenza. However, they can be utilized in any setting to investigate the human NK cell receptor-ligand repertoire. Importantly, these methods can be applied broadly to the design and execution of future CyTOF panels.
Introduction
Natural killer (NK) cells are innate immune cells whose primary role is to target and kill malignant, infected, or otherwise stressed cells. Through their secretion of cytokines such as IFNγ and TNFα, as well as their cytotoxic activity, NK cells can also shape the adaptive immune response to pathogens and malignancies. The NK response is mediated in part by the combinatorial signaling of germline-encoded inhibitory and activating receptors, which bind a myriad of ligands expressed on potential target cells. Several NK cell receptors have more than one ligand with new receptor-ligand pairs being identified regularly.
There is a particular interest in studying NK cells in the context of viral infections, where their ability to rapidly respond to stressed cells may limit viral spread or promote the development of NK cell evasion strategies. This interest in NK cell biology extends to the field of cancer immunotherapy where researchers are investigating the role of NK cells in tumor immunosurveillance and in the tumor microenvironment1. However, the ability to profile NK cell-target cell interactions is complicated by the fact that human NK cells can express over 30 receptors which in turn can interact with over 30 known ligands2. The simultaneous detection of multiple NK cell receptors and their cognate ligands is, therefore, necessary to capture the complexity of the receptor-ligand interactions that control NK function. Consequently, we turned to mass cytometry (CyTOF), which allows for the simultaneous detection of over 40 markers at the single cell level. Our goal was to create two CyTOF panels to profile the NK cell receptor-ligand repertoire. We also wanted to design a protocol for effective processing and staining of clinical samples. Clinical human samples provide a wealth of information on how the body responds to viral infection. Therefore, we developed this protocol to investigate expression of NK cell receptors and their cognate ligands in parallel for better standardization, improved recovery, reduced reagent consumption, and limited batch effects.
Several flow cytometry panels designed to characterize the phenotype of human NK cells have been published previously3,4,5,6,7,8. Most of these panels are limited in their ability to capture the breadth of the receptor-ligand repertoire, only allowing for the detection of a limited selection of markers. Moreover, these panels are limited by signal overlap between fluorochromes. CyTOF uses antibodies conjugated to metal isotopes, which are read out by time-of-flight mass spectrometry, thus dramatically reducing spillover between channels.
Like us, other researchers have turned to CyTOF to study NK cells9,10,11,12,13,14, though generally with fewer NK cell markers, which reduces the depth of phenotyping. While the general staining protocols used by these groups are similar to ours, there are some key differences. Other protocols do not involve isolating NK cells prior to staining even though the researchers are only interested in that subset13,14. Given that NK cells only make up 5-20% of peripheral blood mononuclear cells (PBMCs), staining whole PBMCs rather than isolated NK cells means that most of the collected events will not be NK cells. This reduces the amount of data generated on the subset of interest and results in inefficient use of machine time. Additionally, while many of these panels interrogate expression of NK cell receptors such as killer Ig-like receptors (KIRs), NKG2A/C/D, and the natural cytotoxicity receptors (NKp30, NKp44, and NKp46), expression of these markers is not put into a broader context due to the absence of data on expression of their respective ligands. Consequently, while these previously published methods for investigating NK cells via CyTOF are sufficient for broad NK cell phenotyping, used in isolation, they cannot provide a comprehensive picture of NK cell activity. This brings us to the major advantage of the methods described here, which is that up to this point there are no published flow cytometry or CyTOF panels focused on exploring the expression of ligands for NK cell receptors. Importantly, our ligand panel has several open channels to allow for the addition of markers to suit the unique needs of each experiment.
Considering that one of the main limitations of CyTOF is the inability to recover the sample after analysis, this method may not be appropriate for researchers who have limited samples with which they are interested in performing additional experiments. Additionally, the low throughput nature of CyTOF means that the data generated will be of poor quality if the starting number of cells is low. Barring these two limitations, this method will perform well in any setting to investigate receptor-ligand interactions between NK cells and target cells.
Protocol
Anonymized healthy adult PBMCs were obtained from leukoreduction system chambers purchased from the Stanford Blood Center. PBMCs from de-identified healthy pediatric donors and pediatric acute dengue patients were obtained from Gorgas Memorial Institute of Health Studies in Panama City, Panama and hospitals belonging to the Ministry of Health, the Social Security System in Panama City, and suburban areas. The dengue study protocol was approved by the IRB of Hospital del Niño (CBIHN-M-0634), then approved by the committees of ICGES, CSS, Santo Tomas Hospital, and Stanford University. PBMCs from HIV-infected patients on antiretroviral treatment were obtained from ACTG study A5321.
1. Antibody labeling, panel preparation, and storage
- Antibody labeling with metal isotopes
NOTE: To increase inter-experimental standardization of staining, it is recommended to perform multiple conjugations for each antibody and then combine the products into a single master mix for long-term storage as described below.- Determine the concentration of each antibody by measuring the absorbance at 280 nm prior to conjugation. Antibodies used for this protocol are commercially available and were purchased from the vendors listed in the Table of Materials.
- Label antibodies with metal isotopes using commercially available antibody labeling kits according to the manufacturer's instructions. Use 100 µg of antibody for each reaction.
- Determine the final concentration of the recovered antibody by measuring the absorbance at 280 nm. Store antibodies for short-term at 4 °C.
- Antibody titrations
NOTE: CyTOF technology is very sensitive to potential contaminating signals from environmental metals. Therefore, all buffers/reagents used should be prepared with ultrapure water and stored in plastic or glass containers that have never been washed with soap.- Prepare centrifuge tubes for each donor containing 9 mL of warm, complete RPMI (RPMI 1640, 10% FBS, 1% L-glutamine, 1% penicillin/streptomycin) and 20 µL of benzonase per vial of PBMCs to be thawed. Thaw the PBMCs in a water bath and add to tubes.
NOTE: Benzonase decreases viscosity and background from free DNA from lysed cells. - Centrifuge at 300 x g at room temperature for 5 min. Resuspend the PBMCs in 5 mL of complete RPMI media and count.
- For each panel titration, plate 2-4 million PBMCs/well in 6 wells of a round bottom 96-well plate (one well for each titer and one for unstained). Centrifuge the plate at 600 x g at room temperature for 3 min. Flick the plate to remove the supernatant. Resuspend each well in 200 µL of CyPBS.
- Perform cisplatin viability staining as described below.
NOTE: Cisplatin is used to discriminate live from dead cells in mass cytometry.- Resuspend cells in 100 µL of 25 µM cisplatin stock. Incubate at room temperature for 1 min.
- Quench the cisplatin reaction by adding 100 µL of FBS to each well and mixing. Centrifuge and flick the plate.
NOTE: Perform all subsequent centrifuge steps at 4 °C. - Wash cells twice with 200 µL of CyFACS (1x PBS without heavy metal contaminants in ultrapure water with 0.1% BSA, 0.05% sodium azide). Centrifuge and flick the plate each time.
- Titrate the surface antibody panel as described below.
NOTE: Separate master mixes should be made for the NK surface panel and the ligand panel.- Make a master mix of all the surface antibodies at a concentration of 10 µg/mL using CyFACS. Aim for a final volume of 150 µL. Make serial 1:2 dilutions using CyFACS, to obtain the following concentrations: 10, 5, 2.5, 1.25, and 0.625 µg/mL.
- Filter antibody cocktails through a centrifugal filter unit (0.1 µm pore size) at 10,600 x g for 3 min prior to staining.
- Resuspend the plated cells in 50 µL of the surface antibody cocktail at the respective titer. Resuspend the unstained well in CyFACS. Incubate at 4 °C for 30 min.
- Wash cells with 150 µL of CyFACS. Centrifuge and flick the plate.
- Wash cells with 200 µL of CyFACS. Centrifuge and flick the plate.
- Perform fixation of cells by resuspending each well in 100 µL of 2% paraformaldehyde (PFA) in CyPBS. Incubate the plate at room temperature in the dark for 20 min. Wash cells with 100 µL of CyFACS. Centrifuge at 700 x g for 5 min.
CAUTION: PFA is suspected of causing genetic defects as well as cancer. Additionally, it is harmful if it gets in contact with the eyes, skin, or is inhaled. Handle appropriately by ensuring good ventilation, opening the receptacle carefully, and preventing the formation of aerosols.
NOTE: All subsequent centrifuge spins are performed at 700 x g for 5 min at 4 °C. - Permeabilize cells by resuspending in 200 µL of 1x Permeabilization Buffer (Perm buffer) diluted in ultrapure water. Centrifuge and flick the plate. Wash cells with 200 µL of Perm buffer. Centrifuge and flick the plate.
NOTE: Incubation in the Perm buffer is not necessary. - Intracellular antibody panel titration
- Make a master mix of all the intracellular antibodies at a concentration of 10 µg/mL using Perm Buffer. Aim for a final volume of 150 µL. Make serial 1:2 dilutions using Perm Buffer to obtain the following concentrations: 10, 5, 2.5, 1.25, and 0.625 µg/mL.
- Filter antibody cocktails through a centrifugal filter unit (0.1 µm pore size) at 10,600 x g for 3 min prior to staining.
- Resuspend the plated cells in 50 µL of the intracellular antibody cocktail at the respective titer. Resuspend the unstained well in Perm Buffer. Incubate at 4 °C for 45 min.
NOTE: If an intracellular panel is not going to be titrated, resuspend wells in 50 µL of the Perm buffer. - Wash cells with 150 µL of Perm buffer. Centrifuge and flick the plate.
- Wash cells with 200 µL of Perm buffer. Centrifuge and flick the plate.
- Wash cells twice with 200 µL of CyFACS. Centrifuge and flick the plate.
- DNA intercalator staining
NOTE: Intercalator binds to cellular nucleic acid and is used to identify nucleated cells in mass cytometry.- Resuspend cells in 200 µL of intercalator diluted 1:10,000 in CyPBS and 2% PFA. Incubate the plate overnight at 4 °C.
- Store plates, if needed, at 4 °C covered with paraffin film for up to a week.
NOTE: Perform all subsequent centrifuge steps at 700 x g for 5 min at 4 °C. - Before running the samples on CyTOF, remove the paraffin film from the plate and centrifuge at 700 x g for 5 min at 4 °C. Flick the plate. Wash cells once with 200 µL of CyFACS. Centrifuge and flick the plate.
- Wash cells three times with 200 µL of ultrapure water. Centrifuge and flick the plate. Resuspend cells in 200 µL of ultrapure water. Immediately before running the sample, adjust the concentration to approximately 6 x 105 cells/mL in normalization beads diluted to a 1x concentration in ultrapure water.
- Run the samples on CyTOF.
- Analyze data and choose appropriate titers for each antibody by selecting the lowest antibody titer which results in the highest signal intensity and the best separation between positive and negative populations based on visual assessment.
NOTE: Titrations for the NK and ligand panels are shown in Figure 1 and Figure 2 respectively. If a clear distinction between positive and negative populations is not identified, titers can be assessed on multiple cell types or on cell lines, to allow for identification of both positive and negative cell populations. - Antibody panel storage
- Combine titrated antibodies into a master mix and filter through a sterile 0.1 µm syringe filter unit. Separate master mixes should be made for the NK surface panel, the NK intracellular panel, and the ligand panel.
- For the long-term storage of panels follow one of the two acceptable options:
- Send master mix to a third-party company for lyophilization. This method is used for the NK panel. Antibodies not conjugated in-house cannot be lyophilized, due to the presence of antibody stabilizer, which interferes with the lyophilization process. These antibodies are added to the panel on the day of staining.
- Use a repeater pipette to make 50 µL aliquots of each master mix and store them at -80 °C.
- Prepare centrifuge tubes for each donor containing 9 mL of warm, complete RPMI (RPMI 1640, 10% FBS, 1% L-glutamine, 1% penicillin/streptomycin) and 20 µL of benzonase per vial of PBMCs to be thawed. Thaw the PBMCs in a water bath and add to tubes.
2. Staining protocol
- Thaw peripheral blood mononuclear cells (PBMCs) as described in steps 1.2.1 and 1.2.2. Set aside at least 1 million PBMCs for ligand panel staining in a 15 mL centrifuge tube. Keep these PBMCs on ice during the NK cell isolation.
- NK cell isolation
NOTE: The following NK cell isolation steps are a modified version of a specific vendor's protocol. However, any kit or protocol that performs magnetic-based negative selection of NK cells would be a suitable alternative. Additionally, this step is optional as this protocol is also suitable for the phenotyping of NK cells from whole PBMCs.- Spin the remaining PBMCs at 450 x g for 5 min. Resuspend cell pellet in 40 µL of MACS buffer (PBS, 0.5% BSA, 2 mM EDTA) per 107 total cells.
- Add 10 µL of NK cell Biotin-Antibody Cocktail per 107 total cells. Mix well and incubate for 5 min on ice.
- Add 30 µL of MACS buffer per 107 total cells and 20 µL of NK cell MicroBead Cocktail per 107 total cells. Mix well and incubate for 10 min on ice.
- Prepare elution columns by rinsing with 500 µL of MACS buffer. Add 2 mL of cold complete RPMI to 15 mL collection tubes.
- Add MACS buffer to tubes containing cells to bring the volume up to 500 µL. Pipette the entire 500 µL volume onto the prepared elution column. Rinse out the tube with another 500 µL of MACS buffer and transfer to the column.
- After flow has stopped, rinse the elution column with 500 µL MACS buffer twice. After flow has stopped, count NK cells.
- Plate and wash cells.
- Centrifuge isolated NK cells and PBMCs at 300 x g at room temperature for 10 min. Resuspend cells at a concentration of 5 million cells/mL in CyPBS (1x PBS without heavy metal contaminants in ultrapure water). Plate cells in a U-bottom, 96-well plate.
NOTE: Each aliquot of the NK cell panel can stain up to 3 million cells. If six individual NK cell samples are being barcoded and pooled prior to staining, the combined total number of NK cells should not exceed 3 million. The ligand panel can stain up to 6 million PBMCs per sample. If six individual samples are being barcoded and pooled prior to staining, the combined total number of PBMCs should not exceed 6 million. - Centrifuge plate at 600 x g at room temperature for 3 min. Flick the plate to remove supernatant.
NOTE: Perform all subsequent centrifuge spins at 600 x g for 3 min until step 2.7. - Resuspend cells in 200 µL of CyPBS. Centrifuge and flick the plate.
- Centrifuge isolated NK cells and PBMCs at 300 x g at room temperature for 10 min. Resuspend cells at a concentration of 5 million cells/mL in CyPBS (1x PBS without heavy metal contaminants in ultrapure water). Plate cells in a U-bottom, 96-well plate.
- Perform cisplatin viability staining as described in step 1.2.4.
- Barcoding staining
NOTE: These panels are used in conjunction with a modified two-of-four, Palladium-based barcoding method on live unfixed cells to minimize batch effects and maximize cell recovery15. However, this step is optional as barcoding is not necessary to obtain quality data.- Resuspend each well in 50 µL of respective premixed barcode and incubate at 4 °C for 30 min. Wash cells with 150 µL of CyFACS. Centrifuge and flick the plate.
- Wash cells twice with 200 µL of CyFACS. Centrifuge and flick the plate. Resuspend all wells in 30 µL of CyFACS. Combine up to six wells of cells stained with unique barcodes into one well and perform centrifugation and flicking of the plate.
- Surface staining
- Dissolve the surface NK panel lyosphere in 50 µL of CyFACS with additional surface antibodies spiked in (anti-CD16, anti-HLA-DR, anti-LILRB1). Thaw ligand panel stored at -80 °C and spin down tube using a mini-centrifuge. Spike in additional ligand panel surface markers (anti-CD16, anti-CD19).
NOTE: Any antibody cocktail that has not been previously filtered (i.e., prior to lyophilization or freezing) should be filtered through a centrifugal filter unit (0.1 µm pore size) at 10,600 x g for 3 min prior to staining. - Resuspend each well in 50 µL of the respective panel. Incubate at 4 °C for 30 min. Wash cells with 150 µL of CyFACS. Centrifuge and flick the plate.Wash cells again with 200 µL of CyFACS. Centrifuge and flick the plate.
- Dissolve the surface NK panel lyosphere in 50 µL of CyFACS with additional surface antibodies spiked in (anti-CD16, anti-HLA-DR, anti-LILRB1). Thaw ligand panel stored at -80 °C and spin down tube using a mini-centrifuge. Spike in additional ligand panel surface markers (anti-CD16, anti-CD19).
- Fix cells as described in step 1.2.6.
NOTE: Perform all subsequent centrifuge spins at 700 x g for 5 min at 4 °C. - Permeabilize cells as described in step 1.2.7.
- Intracellular staining
- Dissolve the intracellular NK panel lyosphere in 50 µL of Perm buffer. Prepare an intracellular antibody cocktail for PBMC samples if so desired.
NOTE: Any antibody cocktail that has not been previously filtered (i.e., prior to lyophilization or freezing) should be filtered through a centrifugal filter unit (0.1 µm pore size) at 10,600 x g for 3 min prior to staining. - Resuspend wells in 50 µL of the respective intracellular panels. If an intracellular panel is not used in conjunction with the ligand surface panel, resuspend PBMC wells in 50 µL of the Perm buffer. Incubate at 4 °C for 45 min.
- Wash cells with 150 µL of Perm buffer. Centrifuge and flick plate. Wash cells with 200 µL of Perm buffer. Centrifuge and flick the plate.
- Wash cells twice with 200 µL of CyFACS. Centrifuge and flick the plate.
- Dissolve the intracellular NK panel lyosphere in 50 µL of Perm buffer. Prepare an intracellular antibody cocktail for PBMC samples if so desired.
- DNA intercalator staining. Perform DNA intercalator staining as described in step 1.2.9.1. Incubate plate overnight at 4 °C.
NOTE: Intercalator binds to cellular nucleic acid and is used to identify nucleated cells in mass cytometry. Plates can be stored covered with paraffin film for up to a week at 4 °C. - Before running the samples on CyTOF, wash cells as described in steps 1.2.9.3 and 1.2.9.4.
- Run samples on CyTOF.
Representative Results
Antibodies were conjugated to metal isotopes using commercially available labeling kits, according to the manufacturer's instructions. Antibody clones were validated by flow cytometry and mass cytometry prior to use in this panel. An initial list of clones was selected based on review of the literature and antibody availability. The expression levels of some ligands for NK cell receptors are low or undetectable on healthy PBMCs. Therefore, positive staining for some antibodies was validated by staining healthy PBMCs, chronic myeloid leukemia K562 cells, acute lymphoblastic leukemia NALM6 cells, or B cell acute lymphoblastic leukemia 697 cells (Supplemental Figure 1). Clones selected for the NK cell panel that did not produce an adequate stain or were too expensive were substituted for different ones, as detailed in Supplemental Table 1 and shown in Supplemental Figure 2.
Metal-isotope pairing with antibodies for these panels was performed using the principles outlined by Takahashi et al.16. Lineage markers were of medium to high intensity. Consequently, they were mainly conjugated to low and medium sensitivity masses leaving high sensitivity masses available for conjugation to antibodies against more dimly expressed markers. A publicly available panel designer software was used to detect abundance sensitivity (M ± 1 bleed) or oxidation (M + 16 bleed) issues and antibody-metal pairs were re-assigned accordingly. Additionally, several markers were conjugated on different metals with minimal differences noted on signal intensities (Supplemental Table 2 and Supplemental Figure 3). Antibody-metal pairings and clone information for the NK and ligand panels are listed in Table 1 and Table 2 respectively.
In-house conjugated antibodies were titrated on PBMCs at five different titers: 0.625, 1.25, 2.5, 5, and 10 µg/mL. The lowest antibody titer which resulted in the highest signal intensity and the best separation between positive and negative populations was selected based on visual assessment. Titrations for the NK and ligand panels are shown in Figure 1 and Figure 2 respectively. For certain markers, a clear distinction between positive and negative populations was not identified, due to the marker being dimly expressed or universally positive. To determine the most accurate working dilution for these antibodies, titers were assessed on multiple cell types (PBMCs, T cells, B cells, or NK cells), or on cell lines, to allow for identification of both positive and negative cell populations (Supplemental Figure 4). The staining index (SI) for each marker was not calculated as this metric is not applicable to CyTOF data17,18.
The panels described here were designed to be compatible with sample barcoding. There are several barcoding methods available for CyTOF. The most commonly used are a commercially available Palladium-based kit, which requires fixation prior to barcoding, and the CD45-based barcoding method described by Mei et al.15, which allows the barcoding of live cells. To assess which barcoding method best fit our needs, we tested the stability of NK cell marker staining after fixation in an early version of the NK cell panel (Supplemental Figure 5). We found that the expression of a majority of NK cell markers was affected by fixation. Consequently, we decided to use a modified two-of-four, CD45-based barcoding method on live cells15. This barcoding method uses 102Pd, 104Pd, 106Pd, and 108Pd, and differs from the three-of-six method originally described by Mei et al., which used 104Pd, 106Pd, 108Pd, 110Pd, 113In, and 115In. Indium channels were not included in our barcoding scheme, as they interfered with the signal from 115In-CD3. 110Pd was not included as it interfered with the signal from the HLA-DR Qdot and the CD19 Qdot in the NK cell and ligand panels, respectively.
Although we recommend NK cell purification prior to staining, the NK cell panel is designed to allow for the phenotyping of NK cells from whole PBMCs. An example of our NK cell gating strategy is shown in Figure 3A using PBMCs from a healthy donor. Staining and gates for each of the NK markers are shown on healthy, isolated NK cells in Figure 3B. The ligand panel is designed to detect the expression of NK cell ligands on whole PBMCs. Figure 4A illustrates the gating strategy used to identify CD4+ T cells, CD8+ T cells, NK cells, monocytes, and CD19+ B cells in PBMCs from a healthy donor. Representative staining examples for each ligand are shown in Figure 4B using PBMCs from acute dengue patients and HIV-infected individuals who were virologically suppressed.
To ensure panel stability over time, our protocol includes two possible options: lyophilization through a third-party company into single use beads or freezing of pre-made aliquots at -80 °C. For this protocol, the NK panel was lyophilized, and the ligand panel was frozen. Both methods were validated prior to using each panel on clinical samples.
We produced over 700 reactions of the NK panel from one master mix by performing multiple conjugations of each antibody in the panel. After validation and titration of each conjugated antibody, the antibodies were combined into a master mix, filtered through a sterile 0.1 µm syringe filter unit, and sent to a third party company for lyophilization. Two sets of single-stain lyospheres were made, one for surface staining, and one for intracellular staining. Antibodies not conjugated in-house (HLA-DR and CD16) could not be added to the lyosphere, due to the presence of antibody stabilizer, which interferes with the lyophilization process. These antibodies are added to the panel on the day of staining. A comparison between stains obtained pre- and post-lyophilization is shown in Supplemental Figure 6. The clone of LILRB1 antibody initially used in the lyospheres did not produce a sufficiently strong stain (Supplemental Table 1 and Supplemental Figure 2). A better clone was subsequently identified, conjugated and added to the panel on the day of staining (Table 1). The KIR2DS2 polyclonal antibody used in the lyopsheres was noted to produce a non-specific stain after lyophilization and we do not recommend its use for subsequent analyses (Supplemental Table 1 and Supplemental Figure 2). Most intracellular stains slightly increased in intensity following lyophilization (Supplemental Figure 6).
Prior to storage of the ligand panel master mix at -80 °C, we tested two different storage conditions. We prepared a smaller master mix of this panel and stored aliquots at -80 °C and in liquid nitrogen for approximately two months. After two months we stained whole PBMCs with the frozen aliquots. We compared the staining to that of PBMCs from the same donor stained with the freshly prepared panel (Supplemental Figure 7). We found storage at -80 °C and in liquid nitrogen does not change the signal intensity for most markers. In fact, the signal intensity of anti-pan HLA class I, anti-CD7, anti-CD4, anti-HLA-Bw4, anti-CD14, anti-CD11b, and anti-LFA-3 is higher upon freezing, particularly in the case of samples stored at -80 °C. We could not determine whether the signal intensities of anti-LLT-1, anti-Nectin-1, anti-MICA/B, anti-DR4/5, anti-ULBP-1,2,5,6, anti-Nectin-2, anti-CD155, and anti-B7-H6 were affected by freezing, due to the fact that healthy PBMCs do not express high levels of these markers. However, validation of these markers on cell lines (Supplemental Figure 1) was performed using conjugated antibodies stored at -80 °C. Consequently, we were confident that freezing did not result in a significant loss of signal. Signal intensity did decrease upon freezing for five markers: anti-CD8, anti-ICAM-1, anti-CCR2, anti-CD33, and anti-CD56. However, in all of these cases the clear separation between the positive and negative populations remained. Given that metal-conjugated antibodies are not stable at 4 °C for long periods of time, freezing was necessary to preserve panel stability long-term, and despite a decrease in staining intensity in a subset of markers, we were able to retain sufficient staining separation. Importantly, the loss of signal intensity of anti-CD8, anti-CCR2, and anti-CD56 was greater in the samples stored in liquid nitrogen compared to those stored at -80 °C. Based on this data, we decided to store the panel at -80 °C.
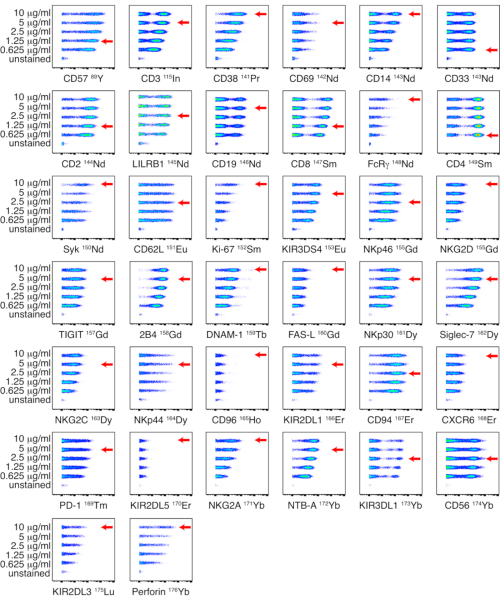
Figure 1: Titration of in-house conjugated antibody-metal conjugates for NK panel. Titrations of in-house conjugated antibodies were performed on PBMCs from a healthy donor using five different concentrations: 0.625, 1.25, 2.5, 5, and 10 µg/mL. Titers for anti-CD3, anti-CD14, anti-CD33, anti-CD19, anti-PD-1 and anti-CD56 were determined by gating on live cells. Titers for anti-CD4 and anti-CD8 were determined by gating on T cells. Titers for the remaining antibodies were determined by gating on NK cells. Since NKp44 is not expressed on resting NK cells, titers were determined on PBMCs stimulated with IL-2 and shown on NK cells. The red arrows indicate the titer selected for each antibody. Please click here to view a larger version of this figure.
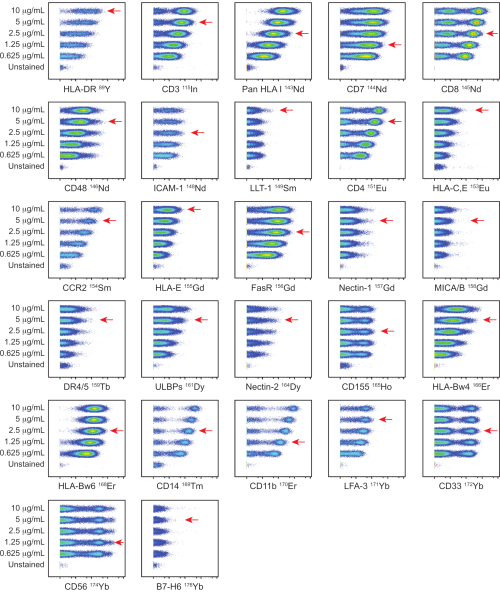
Figure 2: Titration of in-house conjugated ligand panel antibodies. Titrations of in-house conjugated antibodies were performed on PBMCs from a healthy donor using five different concentrations: 0.625, 1.25, 2.5, 5, and 10 µg/mL. Titers for anti-HLA-DR, anti-ICAM-1, anti-CCR2, anti-CD14, anti-CD11b, and anti-LFA-3 were determined by gating on CD3–CD7– cells. Titers for anti-CD3, anti-pan HLA class I, anti-CD7, anti-CD48, anti-LLT-1, anti-HLA-C,E, anti-HLA-E, anti-FasR, anti-Nectin-1, anti-MICA/MICB, anti-DR4/DR5, anti-ULBP-1,2,5,6, anti-Nectin-2, anti-CD155, anti-HLA-Bw4, anti-HLA-Bw6, anti-CD33, anti-CD56, and anti-B7-H6 were determined by gating on live cells. Titers for anti-CD4 and anti-CD8 were determined by gating on CD3+ cells. The red arrows indicate the titer selected for each antibody. Please click here to view a larger version of this figure.
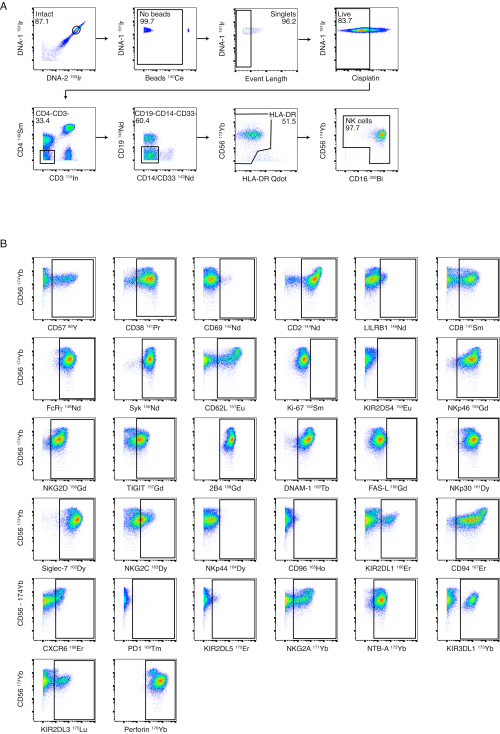
Figure 3: NK panel gating strategy and performance. (A) Serial negative gating from whole PBMCs to NK cells is shown in a healthy donor. Intact, bead and event-length gates ensure successful gating to single cells. Cisplatin staining was performed as a Live/Dead stain. T cells and B cells were excluded using CD3 and CD19. Monocytes were excluded by negative gating on CD4 and CD14/CD33 and by further negative gating of CD56-/HLA-DRbright cells. CD56 and CD16 were used to identify different subsets of NK cells (CD56bright, CD56dim and CD56–). (B) Examples of expression of NK cell receptors on NK cells from one healthy donor purified by magnetic-bead isolation. Please click here to view a larger version of this figure.

Figure 4: Ligand panel gating and performance. (A) Gating of major immune cell subsets from PBMCs derived from a healthy donor following normalization, calibration bead removal, and debarcoding. (B) Expression of ligands for NK cell receptors as well as several myeloid markers on live PBMCs. Staining for all ligands except Nectin-1 and B7-H6 is shown on PBMCs from acute dengue patients. Staining for Nectin-1 and B7-H6 is shown on PBMCs from HIV-infected individuals who were virologically suppressed. Please click here to view a larger version of this figure.
Supplemental Figure 1: Verification of antibody clones for ligand panel on cell lines. Antibodies against ligands for NK cell receptors that are expressed at low levels on healthy PBMCs were validated by staining cell lines. Chronic myeloid leukemia K562 cells were stained with anti-ICAM-1, anti-MICA/MICB, anti-DR4/DR5, anti-ULBP-1,2,5,6, anti-Nectin-2, anti-CD155, and anti-B7-H6. Acute lymphoblastic leukemia NALM6 cells were stained with anti-LLT-1 and B cell acute lymphoblastic leukemia 697 cells were stained with anti-Nectin-1. Dot plots and histograms showing staining on healthy PBMCs are in blue. Dot plots and histograms showing staining on the respective cell line are in red. The percentage of cells of the respective cell line that are positive for a given marker are provided. Please click here to download this file.
Supplemental Figure 2: Validation of antibody clones. (A) Different antibody clones were tested in healthy donors to identify the clone with the best specificity. 2B4, CXCR6, KIR2DS4, NKG2A and TIGIT are shown on NK cells. CD56 and LILRB1 are shown on live cells. (B) The KIR2DS2 antibody clone showed a non-specific stain after lyophilization. An example of staining on the same donor is provided pre- and post-lyophilization. Please click here to download this file.
Supplemental Figure 3: Optimization of antibody/metal pairs. Staining of PBMCs from healthy donors are shown. (A) Antibody/metal pairs tested for the panel. (B) Antibody/metal pairs used in the panel. LILRB1 and PD1 are shown on live cells. All the other markers are shown on NK cells. Please click here to download this file.
Supplemental Figure 4: Titration of dimly expressed and mostly positive NK cell markers. Titers for antibodies against NK cell markers that did not show a clear positive and negative population were assessed both on NK cells (red) and on either B cells (blue) or PBMCs (grey) from healthy donors. The arrows indicate the titer selected for each antibody. Please click here to download this file.
Supplemental Figure 5: Optimization of barcoding protocol. Epitope stability was tested before and after fixation with 2% paraformaldehyde on PBMCs from a healthy donor. (A) CD3, CD14 and CD56 staining was similar before (red) and after (blue) fixation, CD4 and CD16 staining was significantly affected by fixation. (B) Many NK cell markers were affected by fixation, including CD2, CD38, KIR3DL2, CD62L, KIR2DS4, NKp46, NKG2C, NKp30, NKG2D, KIR3DL1, TIGIT, KIR2DL1, KIR2DL3 and NTB-A. Please click here to download this file.
Supplemental Figure 6: Confirmation of panel stability after lyophilization. The stability of in-house antibody conjugates was confirmed by staining PBMCs from the same blood bank donor pre-lyophilization (blue) and post-lyophilization (red). Anti-CD3, anti-CD14, anti-CD33, anti-CD19, anti-PD-1, anti-CD56 stains are shown on live cells. Anti-CD4 and anti-CD8 are shown on CD3+ cells. Titers for the remaining antibodies are shown on NK cells, gated according to the gating scheme shown in Figure 1. Notably, anti-KIR2DS2 stained non-specifically after lyophilization and therefore has not been used for subsequent analyses. Please click here to download this file.
Supplemental Figure 7: Confirmation of ligand panel stability at -80 °C. The stability of in-house antibody conjugates was confirmed by staining healthy PBMCs from the same donor with a freshly prepared master mix as well as the same mix after storage at -80°C or in liquid nitrogen. Anti-HLA-DR, anti-ICAM-1, and anti-LFA-3 staining is shown on CD3–CD7– cells. Anti-CD3, anti-pan HLA class I, anti-CD7, anti-CD48, anti-LLT-1 anti-HLA-E, anti-FasR, anti-Nectin-1, anti-MICA/B, anti-DR4/5, anti-ULBP-1,2,5,6, anti-Nectin-2, anti-CD155, anti-HLA-Bw4, anti-HLA-Bw6, and anti-B7-H6 staining is shown on live cells. Anti-CD8 and anti-CD4 staining is shown on CD3+ cells. Anti-HLA-C,E, anti-CCR2, anti-CD11b, and anti-CD33 staining is shown on CD3–CD7–CD14+ cells. Anti-CD14 staining is shown on CD3–CD7–CD33+ cells and anti-CD56 staining is shown on CD3–CD7+CD14–HLA-DR– cells. Histograms showing staining with the freshly prepared panel are in red. Histograms showing staining with the panel after storage at -80°C for approximately two months are in blue. Histograms showing staining with the panel after storage in liquid nitrogen (LN2) for approximately two months are in green. Samples were stained and run on different days. Files were normalized and beads were removed using the premessa package. Please click here to download this file.
| Specificity | Clone | Isotope | Purpose | Surface/Intracellular |
| CD57 | HCD57 | 89Y | Maturity/Memory | surface |
| CD45 | HI30 | 102Pd, 104Pd, 106Pd, 108Pd | Barcoding | surface |
| HLA-DR | Tü36 | Qdot 655 (112Cd-114Cd) | Activation/Lineage | surface |
| CD3 | UCHT | 115In | T cell lineage | surface |
| CD38 | HIT2 | 141Pr | Activation Marker | surface |
| CD69 | FN50 | 142Nd | Activation Marker | surface |
| CD33 | WM53 | 143Nd | Monocyte lineage | surface |
| CD14 | M5E5 | 143Nd | Myeloid lineage | surface |
| CD2 | RPA-2.10 | 144Nd | Activation/Maturity | surface |
| LILRB1 | MAB20172 | 145Nd | Inhibitory Receptor | surface |
| CD19 | HIB19 | 146Nd | B cell lineage | surface |
| CD8 | SK1 | 147Sm | T cell lineage and NK cell Activation/Maturity | surface |
| FcRg | polyclonal | 148Nd | Maturity/Adaptive | intracellular |
| CD4 | SK3 | 149Sm | T cell lineage | surface |
| Syk | 4D10.2 | 150Nd | Signaling | intracellular |
| CD62L | DREG-56 | 151Eu | Activation | surface |
| ki-67 | Ki-67 | 152Sm | Proliferation | intracellular |
| KIR2DS4 | 179315 | 153Eu | Activating Receptor | surface |
| NKp46 | 9.00E+02 | 155Gd | Activating Receptor | surface |
| NKG2D | 1D11 | 156Gd | Activating Receptor | surface |
| TIGIT | 741182 | 157Gd | Inhibitory Receptor | surface |
| 2B4 | C1.7 | 158Gd | Activating Receptor | surface |
| DNAM-1 | DX11 | 159Tb | Activating Receptor | surface |
| FAS-L | NOK-1 | 160Gd | Apoptosis | surface |
| NKp30 | P30-15 | 161Dy | Activating Receptor | surface |
| Siglec-7 | S7.7 | 162Dy | Inhibitory Receptor | surface |
| NKG2C | 134522 | 163Dy | Maturity/Memory | surface |
| NKp44 | P44-8 | 164Dy | Activating Receptor | surface |
| CD96 | NK92.39 | 165Ho | NKG2 Co-receptor | surface |
| KIR2DL1/KIR2DS5 | 143211 | 166Er | Inhibitory Receptor | surface |
| CD94 | DX22 | 167Er | Activating Receptor | surface |
| CXCR6 | K041E5 | 168Er | Memory | surface |
| PD1 | EH12.2H7 | 169Tm | Inhibitory Receptor | surface |
| KIR2DL5 | UP-R1 | 170Er | Inhibitory Receptor | surface |
| NKG2A | 131411 | 171Yb | Inhibitory Receptor | surface |
| NTB-A | NT-7 | 172Tb | Activating Receptor | surface |
| KIR3DL1 | DX-9 | 173Yb | Inhibitory Receptor | surface |
| CD56 | NCAM16.2 | 174Yb | NK cell lineage | surface |
| KIR2DL3 | 180701 | 175Lu | Inhibitory Receptor | surface |
| Perforin | B-D48 | 176Yb | Cytolytic Protein | intracellular |
| DNA-1/DNA-2 | NA | 191Ir/193Ir | Nucleated cells | surface |
| Cisplatin | NA | 194Pt/195Pt | Viability | surface |
| CD16 | 3G8 | 209Bi | FcgRIII receptor | surface |
Table 1: NK panel. Markers are ordered according to the isotopic mass of the metal to which they were conjugated. 191Ir/193Ir is the natural abundance of the nucleic acid intercalator. 194Pt/195Pt is the natural abundance of cisplatin.
| Specificity | Clone | Isotope | Purpose | Surface/Intracellular |
| HLA-DR | L243 | 89Y | Antigen presenting cells, activation marker | surface |
| CD45 | HI30 | 102Pd, 104Pd, 106Pd, 108Pd | Barcoding | surface |
| CD19 | SJ25-C1 | Qdot 655 (112Cd-114Cd) | Lineage | surface |
| CD3 | UCHT1 | 115In | Lineage | surface |
| Pan HLA class I | W6/32 | 143Nd | KIR ligands | surface |
| CD7 | CD7-6B7 | 144Nd | Lineage | surface |
| CD8 | SK1 | 145Nd | Lineage | surface |
| CD48 | BJ40 | 146Nd | 2B4 ligand | surface |
| ICAM-1 | HA58 | 148Nd | LFA-1 ligand | surface |
| LLT-1 | 402659 | 149Sm | CD161 ligand | surface |
| CD4 | OKT4 | 151Eu | Lineage | surface |
| HLA-C,E | DT9 | 153Eu | KIR ligands | surface |
| CCR2 | K036C2 | 154Sm | Monocyte functional marker | surface |
| HLA-E | 3D12 | 155Gd | NKG2A/CD94 and NKG2C/CD94 ligand | surface |
| Fas (CD95) | DX2 | 156Gd | FasL receptor | surface |
| Nectin-1 | R1.302 | 157Gd | CD96 ligand | surface |
| MICA/B | 159227/236511 | 158Gd | NKG2D ligands | surface |
| DR4/5 | DJR1/DJR2-2 | 159Tb | TRAIL receptors | surface |
| ULBP-1/2,5,6 | 170818/165903 | 161Dy | NKG2D ligands | surface |
| Nectin-2 | TX31 | 164Dy | DNAM-1, TIGIT, and CD96 ligand | surface |
| CD155 | SKII.4 | 165Ho | DNAM-1, TIGIT, and CD96 ligand | surface |
| HLA-Bw4 | REA274 | 166Er | KIR3DL1 ligand | surface |
| HLA-Bw6 | REA143 | 168Er | KIR null allele | surface |
| CD14 | M5E2 | 169Tm | Lineage | surface |
| CD11b | ICRF44 | 170Er | Lineage | surface |
| LFA-3 | TS2/9 | 171Yb | CD2 ligand | surface |
| CD33 | WM53 | 172Yb | Lineage | surface |
| CD56 | NCAM16.2 | 174Yb | Lineage | surface |
| B7-H6 | 875001 | 176Yb | NKp30 ligand | surface |
| DNA-1/DNA-2 | NA | 191Ir/193Ir | Nucleated cells | surface |
| Cisplatin | NA | 194Pt/195Pt | Viability | surface |
| CD16 | 3G8 | 209Bi | Lineage | surface |
Table 2: Ligand panel. Markers are ordered according to the isotopic mass of the metal to which they were conjugated. 191Ir/193Ir is the natural abundance of the nucleic acid intercalator. 194Pt/195Pt is the natural abundance of cisplatin.
| Specificity | Clone | Vendor | Catalog Number | Surface/ Intracellular | Notes |
| 2B4 | 2-69 | BD Biosciences | 550814 | surface | new clone validated with improved staining |
| CD56 | N901 | Beckman Coulter | 6602705 | surface | new clone validated with improved staining |
| CXCR6 | 56811 | R&D Systems | MAB699 | surface | new clone validated with lower cost |
| KIR2DS2 | polyclonal | Abcam | ab175486 | surface | non specific staining noted after lyophilization – not used for analyses |
| KIR2DS4 | FES172 | Beckman Coulter | A60796 | surface | new clone validated with improved staining/lower cost |
| LILRB1 | GHI/75 | Biolegend | 333702 | surface | new clone validated with improved staining |
| NKG2A | Z199 | Beckman Coulter | IM2750 | surface | new clone validated with lower cost |
| TIGIT | MBSA43 | Thermo Fisher Scientific | 16-9500-82 | surface | new clone validated |
Supplemental Table 1: Antibodies for NK cell panel that were tested, but not used.
| Isotope | Specificity | Clone | Vendor | Catalog Number | Surface/ Intracellular |
| 143Nd | NKG2C | 134522 | R&D systems | MAB1381 | surface |
| 145Nd | CD38 | HIT2 | Biolegend | 303502 | surface |
| 146Nd | CD8 | SK1 | Biolegend | 344702 | surface |
| 149Sm | CD2 | RPA-2.10 | Biolegend | 300202 | surface |
| 151Eu | Siglec-7 | S7.7 | Biolegend | 347702 | surface |
| 154Sm | LILRB1 | 292319 | R&D systems | MAB20172 | surface |
| 163Dy | KIR3DL1 | DX-9 | BD biosciences | 555964 | surface |
| 168Er | CD62L | DREG-56 | Biolegend | 304802 | surface |
| 171Yb | PD1 | EH12.2H7 | Biolegend | 329902 | surface |
| 176Yb | CD69 | EH12.2H7 | Biolegend | 329902 | surface |
Supplemental Table 2: Antibodies for NK cell panel that were tested with a different antibody/metal pairing.
Discussion
Here we describe the design and application of two complimentary CyTOF panels aimed at profiling the NK cell receptor-ligand repertoire. This protocol includes several steps that are critical to obtaining quality data. CyTOF uses heavy metal ions, rather than fluorochromes, as label probes for antibodies19. This technology is therefore subject to potential contaminating signals from environmental metals20. Potential sources of metal impurities include laboratory dish soap (barium) and lab buffers (mercury, lead, tin). For this reason, it is advised that all buffers be prepared with ultrapure water, and that all reagents be stored in plastic or glass containers that have never been washed with soap. Another critical step in this protocol is the viability stain, which uses a cisplatin-based method as described by Fienberg et al.21. This method includes a one-minute incubation step, during which cisplatin preferentially labels non-viable cells. Cisplatin staining must be performed in the absence of FBS. Consequently, cells must be thoroughly washed with CyPBS prior to staining. Additionally, to avoid off-target staining of viable cells, the cisplatin stain needs to be quenched with FBS after precisely one minute. This protocol was optimized for maximum cell recovery and staining performance. Therefore, the fixation and permeabilization steps are also significant. Several fixation and permeabilization reagents are compatible with CyTOF. However, we found that fixation with 2% PFA, followed by permeabilization with a specific permeabilization buffer detailed in the Table of Materials, resulted in maximal cell recovery. As this is a transient permeabilization method, intracellular staining needs to be performed in the permeabilization buffer to ensure adequate antibody penetrance. Cells also need to be washed thoroughly with the permeabilization buffer following intracellular staining to remove unbound antibodies.
This protocol allows for several possible modifications. The CyTOF panels detailed here can be customized to include additional markers or substitute existing ones. In particular, the ligand panel was designed with several open channels to allow for flexibility in panel design. Any change or addition to the panel may require additional troubleshooting. In particular, any antibody/metal isotope pair should be thoroughly validated as described above, to avoid any issue of signal spillover to the existing channels. These panels were also designed to be compatible with sample barcoding. Barcoding decreases the possibility of batch effects and sample-to-sample carryover, while minimizing reagent consumption22. Although barcoding typically results in overall increased data quality, this step is not necessary for the acquisition of good quality CyTOF data and can be skipped entirely. Similarly, although we recommend NK cell purification prior to staining, the NK cell panel is compatible with the phenotyping of NK cells from whole PBMCs.
This method has some limitations. Due to the inherently low throughput nature of CyTOF, this method is not suitable for samples with low cell counts. Such samples are unlikely to yield data of sufficient quality for analysis. Additionally, given that these panels were specifically designed to interrogate NK cell-target cell interactions, they are limited in their ability to assess interactions between other cell types, such as CD8+ T cells and myeloid cells. Similarly, these panels were designed for direct ex vivo immunophenotyping and were not tested or validated for use under activating conditions, such as cytokine stimulation. Moreover, although these panels cover a comprehensive list of NK cell receptors and ligands, they are not fully exhaustive, and several potentially important markers were not included due to space limitations. Some of these markers include, but are not limited to KLRG1, CRACC, TIM-3, LAIR-1 in the NK panel and PD-L1 in the ligand panel. Finally, this method is not suitable for use in fixed samples, given that the majority of NK cell markers' epitopes are affected by fixation.
The protocol described here has significant benefits compared to other methods. Other groups have described flow cytometry panels aimed at the study of NK cells3,4,5,6,7,8,23. Compared to flow cytometry, the use of CyTOF eliminates issues related to fluorophore compensation, allowing for the simultaneous detection of a high number of markers. Although others have also developed CyTOF panels to study NK cells9,10,11,12,13,14, here we describe the use of two complementary CyTOF panels, which interrogate the expression of both NK cell receptors and their ligands, therefore providing a more detailed picture of NK cell function.
Our group has used this protocol and one or both of these panels to characterize the human NK cell response in healthy donors, and in a variety of disease settings including HIV infection and dengue virus infection24,25,26,27,28. Despite being designed for the purpose of studying viral infections, these panels lend themselves to the study of NK cells in other conditions given the breath of proteins they cover. In fact, our group also used these panels to characterize the NK cell receptor-ligand repertoire in patients with immunodeficiencies and multiple sclerosis25,27,29,30, as well as in humanized mice31,32. As such, the use of these panels can be extended to other contexts. For example, many NK cell receptors and cognate ligands implicated in the setting of cancer are included in these panels, making these panels excellent tools for future studies on the role of NK cells in the anti-tumor response. More broadly, our protocol for mass production and storage of CyTOF panels as well as parallel processing of samples can be applied to the execution and application of any CyTOF panel.
Disclosures
The authors have nothing to disclose.
Acknowledgements
The authors would like to thank all current and former members of the Blish Laboratory who contributed to this panel. Thank you to the AIDS Clinical Trials Group and the ACTG A5321 team as well as Dr. Sandra López-Vergès and Davis Beltrán at Gorgas Memorial Institute for Health Studies for sample curation. Finally, thank you to Michael Leipold, Holden Maecker, and the Stanford Human Immune Monitoring Center for use of their Helios machines. This work was supported by NIH U19AI057229, NIH R21 AI135287, NIH R21 AI130532, NIH DP1 DA046089, and Burroughs Wellcome Fund Investigators in the Pathogenesis of Infectious Diseases #1016687 to CB, NIH Ruth L. Kirschstein Institutional National Research Service Award T32 AI007502, TL1 TR001084 and NIH/NIAID K08 AI138640 to EV, National Science Foundation Graduate Research Fellowship DGE-1656518 to JM and NIH training grant T32-AI-007290 (PI Olivia Martinez). The ACTG study received grant support from AI-68634 (Statistical and Data Management Center), UM1-A1-26617, AI-131798, and AI-68636 (ACTG). CB is the Tashia and John Morgridge Faculty Scholar in Pediatric Translational Medicine from the Stanford Maternal Child Health Research Institute and an Investigator of the Chan Zuckerberg Biohub.
Materials
| 89Y | Sigma-Aldrich | 204919 | |
| 102-Palladium nitrate | Trace Sciences International | Special Order | |
| 104-Palladium nitrate | Trace Sciences International | Special Order | |
| 106-Palladium nitrate | Trace Sciences International | Special Order | |
| 108-Palladium nitrate | Trace Sciences International | Special Order | |
| 115In | Trace Sciences International | Special Order | |
| 141Pr | Fluidigm | 201141A | |
| 142Nd | Fluidigm | 201142A | |
| 143Nd | Fluidigm | 201143A | |
| 144Nd | Fluidigm | 201144A | |
| 145Nd | Fluidigm | 201145A | |
| 146Nd | Fluidigm | 201146A | |
| 147Sm | Fluidigm | 201147A | |
| 148Nd | Fluidigm | 201148A | |
| 149Sm | Fluidigm | 201149A | |
| 150Nd | Fluidigm | 201150A | |
| 151Eu | Fluidigm | 201151A | |
| 152Sm | Fluidigm | 201152A | |
| 153Eu | Fluidigm | 201153A | |
| 154Sm | Fluidigm | 201154A | |
| 155Gd | Fluidigm | 201155A | |
| 156Gd | Fluidigm | 201156A | |
| 157Gd | Trace Sciences International | N/A | |
| 158Gd | Fluidigm | 201158A | |
| 159Tb | Fluidigm | 201159A | |
| 160Gd | Fluidigm | 201160A | |
| 161Dy | Fluidigm | 201161A | |
| 162Dy | Fluidigm | 201162A | |
| 163Dy | Fluidigm | 201163A | |
| 164Dy | Fluidigm | 201164A | |
| 165Ho | Fluidigm | 201165A | |
| 166Er | Fluidigm | 201166A | |
| 167Er | Fluidigm | 201167A | |
| 168Er | Fluidigm | 201168A | |
| 169Tm | Fluidigm | 201169A | |
| 170Er | Fluidigm | 201170A | |
| 171Yb | Fluidigm | 201171A | |
| 172Yb | Fluidigm | 201172A | |
| 173Yb | Fluidigm | 201173A | |
| 174Yb | Fluidigm | 201174A | |
| 175Lu | Fluidigm | 201175A | |
| 176Yb | Fluidigm | 201176A | |
| 209Bi anti-CD16 | Fluidigm | 3209002B | Clone 3G8. Used at a 1:50 dilution. |
| 697 cells | Creative Bioarray | CSC-C0217 | |
| Amicon Ultra Centrifugal Filter Units 0.5 with Ultracel-30 Membrane, 30 kDa | Millipore | UFC503096 | |
| Anhydrous acetonitrile | Fisher Scientific | BP1165-50 | |
| anti-2B4 | Biolegend | 329502 | Clone C1.7. |
| anti-B7-H6 | R&D Systems | MAB7144 | Clone 875001. |
| anti-CCR2 | Biolegend | 357202 | Clone K036C2. |
| anti-CD2 | Biolegend | 300202 | Clone RPA-2.10. |
| anti-CD3 | Biolegend | 300402 | Clone UCHT1. |
| anti-CD4 | Biolegend | 317402 | Clone OKT4. |
| anti-CD4 | Biolegend | 344602 | Clone SK3. |
| anti-CD7 | Biolegend | 343102 | Clone CD7-6B7. |
| anti-CD8 | Biolegend | 344702 | Clone SK1. |
| anti-CD11b | Biolegend | 301302 | Clone ICRF44. |
| anti-CD14 | Biolegend | 301802 | Clone M5E2. |
| anti-CD19 | Biolegend | 302202 | Clone HIB19. |
| anti-CD33 | Biolegend | 303402 | Clone WM53. |
| anti-CD38 | Biolegend | 303502 | Clone HIT2. |
| anti-CD48 | Biolegend | 336702 | Clone BJ40. |
| anti-CD56 | BD Pharmingen | 559043 | Clone NCAM16.2. |
| anti-CD57 | Biolegend | 322302 | Clone HCD57. |
| anti-CD62L | Biolegend | 304802 | Clone DREG-56. |
| anti-CD69 | Biolegend | 310902 | Clone FN50. |
| anti-CD94 | Biolegend | 305502 | Clone DX22. |
| anti-CD95 | Biolegend | 305602 | Clone DX2. |
| anti-CD155 | Biolegend | 337602 | Clone SKII.4. |
| anti-CXCR6 | Biolegend | 356002 | Clone K041E5. |
| anti-DNAM-1 | BD Biosciences | 559787 | Clone DX11. |
| anti-DR4 | Biolegend | 307202 | Clone DJR1. |
| anti-DR5 | Biolegend | 307302 | Clone DJR2-2. |
| anti-FAS-L | Biolegend | 306402 | Clone NOK-1. |
| anti-FcRg | Millipore | 06-727 | Polyclonal antibody. |
| anti-HLA-C,E | Millipore | MABF233 | Clone DT9. |
| anti-HLA-Bw4 | Miltenyi Biotec | Special Order | Clone REA274. |
| anti-HLA-Bw6 | Miltenyi Biotec | 130-124-530 | Clone REA143. |
| anti-HLA-DR | Biolegend | 307602 | Clone L243. |
| anti-HLA-E | Biolegend | 342602 | Clone 3D12. |
| anti-ICAM-1 | Biolegend | 353102 | Clone HA58. |
| anti-Ki-67 | Biolegend | 350502 | Clone Ki-67. |
| anti-KIR2DL1/KIR2DS5 | R&D Systems | MAB1844 | Clone 143211. |
| anti-KIR2DL3 | R&D Systems | MAB2014 | Clone 180701. |
| anti-KIR2DL5 | Miltenyi Biotec | 130-096-200 | Clone UP-R1. |
| anti-KIR2DS4 | R&D Systems | MAB1847 | Clone 179315. |
| anti-KIR3DL1 | BD Biosciences | 555964 | Clone DX-9. |
| anti-LFA-3 | Biolegend | 330902 | Clone TS2/9. |
| anti-LILRB1 | R&D Systems | 292319 | Clone MAB20172. |
| anti-LLT-1 | R&D Systems | AF3480 | Clone 402659. |
| anti-MICA | R&D Systems | MAB1300-100 | Clone 159227. |
| anti-MICB | R&D Systems | MAB1599-100 | Clone 236511. |
| anti-Nectin-1 | Biolegend | 340402 | Clone R1.302. |
| anti-Nectin-2 | Biolegend | 337402 | Clone TX31. |
| anti-NKG2A | R&D Systems | MAB1059 | Clone 131411. |
| anti-NKG2C | R&D Systems | MAB1381 | Clone 134522. |
| anti-NKG2D | Biolegend | 320802 | Clone 1D11. |
| anti-NKp30 | Biolegend | 325202 | Clone P30-15. |
| anti-NKp44 | Biolegend | 325102 | Clone P44-8. |
| anti-NKp46 | Biolegend | 331902 | Clone 9E2. |
| anti-NTB-A | Biolegend | 317202 | Clone NT-7. |
| anti-Pan HLA class I | Biolegend | 311402 | Clone W6/32. |
| anti-PD1 | Biolegend | 329902 | Clone EH12.2H7. |
| anti-Perforin | Abcam | ab47225 | Clone B-D48. |
| anti-Siglec-7 | Biolegend | 347702 | Clone S7.7. |
| anti-Syk | Biolegend | 644302 | Clone 4D10.2. |
| anti-TACTILE | Biolegend | 338402 | Clone NK92.39. |
| anti-TIGIT | R&D Systems | MAB7898 | Clone 741182. |
| anti-ULBP-1 | R&D Systems | MAB1380-100 | Clone 170818. |
| anti-ULBP-2, 5, 6 | R&D Systems | MAB1298-100 | Clone 165903. |
| Antibody Stabilizer | Candor Bioscience | 131 050 | |
| Benzonase Nuclease | Millipore | 70664 | |
| Bond-Breaker TCEP Solution | Thermo Fisher Scientific | 77720 | |
| Bovine Serum Albumin solution | Sigma-Aldrich | A9576 | |
| Calcium chloride dihydrate (CaCl2+2H2O) | Sigma-Aldrich | 223506-25G | |
| Cis-Platinum(II)diamine dichloride (cisplatin) | Enzo Life Sciences | ALX-400-040-M250 | A 100 mM stock solution was prepared in DMSO and divided into 25 µL aliquots. Used at a 25 µM dilution for live/dead stain. Signal appears in 194Pt and 195Pt channels. |
| DMSO | Sigma-Aldrich | D2650 | |
| eBioscience Permeabilization Buffer | Thermo Fisher Scientific | 00-8333-56 | |
| EDTA (0.5 M) | Hoefer | GR123-100 | A double-concentrated HEPES buffer with EDTA was made according to the following recipe: 1.3 g NaCl (Thermo Fisher Scientific), 27 mg CaCl2+2H2O (Sigma-Aldrich), 23 mg MgCl2 (Sigma-Aldrich), 83.6 mg KH2PO4 (Thermo Fisher Scientific), 4 mL of 1M HEPES (Thermo Fisher Scientific), 2 mL of 0.5M EDTA (Hoefer, Holliston, MA, USA), and 100mL H2O. The pH of this double-concentrated HEPES buffer was adjusted to a pH of 7.3 using 1M HCl and 1M NaOH. |
| EQ Four Element Calibration Beads | Fluidigm | 201078 | |
| Fetal Bovine Serum | Thermo Fisher Scientific | N/A | |
| Helios mass cytometer | Fluidigm | N/A | |
| HEPES (1M) | Thermo Fisher Scientific | 15630080 | |
| HyClone Antibiotic/Antimycotic Solution (Pen/Strep/Fungiezone) solution | Fisher Scientific | SV3007901 | |
| Iridium – 191Ir/193Ir intercalator | DVS Sciences (Fluidigm) | 201192B | Used at a 1:10000 dilution. |
| Isothiocyanobenzyl-EDTA (ITCB-EDTA) | Dojindo Molecular Technologies, Inc. | M030-10 | Diluted to 1.25 mg/mL in anhydrous acetonitrile. |
| K562 cells | American Type Culture Collection (ATCC) | ATCC CCL-243 | |
| L-Glutamine (200 mM) | Thermo Fisher Scientific | SH30034 | |
| Magnesium chloride (MgCl2) | Sigma-Aldrich | 208337-100G | |
| Maxpar X8 Antibody Labeling Kits | Fluidigm | N/A | No catalog number as kits come with metals. |
| Millex-VV Syringe Filter Unit, 0.1 µm | Millipore | SLVV033RS | |
| Milli-Q Advantage A10 Water Purification System | Millipore | Z00Q0V0WW | |
| MS Columns | Miltenyi Biotec | ||
| NALM6 cells | American Type Culture Collection (ATCC) | ATCC CRL-3273 | |
| Nanosep Centrifugal Devices with Omega Membrane 3K | Pall Corporation | OD003C35 | |
| NK Cell Isolation Kit, human | Miltenyi Biotec | 130-092-657 | |
| Paraformaldehyde (16%) | Electron Microscopy Sciences | 15710 | |
| PBS | Thermo Fisher Scientific | 10010023 | |
| Potassium Phosphate Monobasic (KH2PO4) | Fisher Scientific | MP021954531 | |
| Qdot 655 anti-CD19 | Thermo Fisher Scientific | Q10179 | Clone SJ25-C1. Used at a 1:50 dilution. Signal appears in 112Cd-114Cd channels. |
| Qdot 655 anti-HLA-DR | Thermo Fisher Scientific | Q22158 | Clone Tü36. Used at a 1:200 dilution. |
| Rockland PBS | Rockland Immunochemicals, Inc. | MB-008 | Used to make CyPBS (10X Rockland PBS diluted to 1X in Milli-Q water) and CyFACS buffers (10X Rockland PBS diluted to 1X in Milli-Q water with 0.1% BSA and 0.05% sodium azide). Buffers were sterile-filtered through a 0.22 µM filter and sotred at 4°C in Stericup bottles. |
| RPMI 1640 | Thermo Fisher Scientific | 21870092 | |
| Sodium azide (NaN3) | Sigma-Aldrich | S2002 | |
| Sodium chloride (NaCl) | Fisher Scientific | S271-500 | |
| Stericup Quick Release-GP Sterile Vacuum Filtration System | Millipore Sigma | S2GPU10RE | |
| Tuning solution | Fluidigm | 201072 | |
| Washing solution | Fluidigm | 201070 |
References
- Shimasaki, N., Jain, A., Campana, D. NK cells for cancer immunotherapy. Nature Reviews. Drug Discovery. 19 (3), 200-218 (2020).
- Vivier, E., Tomasello, E., Baratin, M., Walzer, T., Ugolini, S. Functions of natural killer cells. Nature Immunology. 9 (5), 503-510 (2008).
- Eller, M. A., Currier, J. R. OMIP-007: phenotypic analysis of human natural killer cells. Cytometry. Part A: The Journal of the International Society for Analytical Cytology. 81 (6), 447-449 (2012).
- Mahnke, Y. D., Beddall, M. H., Roederer, M. OMIP-029: Human NK-cell phenotypization. Cytometry. Part A: The Journal of the International Society for Analytical Cytology. 87 (11), 986-988 (2015).
- Hammer, Q., Romagnani, C. OMIP-039: Detection and analysis of human adaptive NKG2C + natural killer cells : Detection of Human Adaptive NKG2C + NK Cells. Cytometry. 91 (10), 997-1000 (2017).
- Liechti, T., Roederer, M. OMIP-058: 30-Parameter flow cytometry panel to characterize iNKT, NK, unconventional and conventional T cells. Cytometry. 95 (9), 946-951 (2019).
- Béziat, V., et al. NK cell responses to cytomegalovirus infection lead to stable imprints in the human KIR repertoire and involve activating KIRs. Blood. 121 (14), 2678-2688 (2013).
- Pfefferle, A., et al. Intra-lineage plasticity and functional reprogramming maintain natural killer cell repertoire diversity. Cell Reports. 29 (8), 2284-2294 (2019).
- Barcenilla, H., Åkerman, L., Pihl, M., Ludvigsson, J., Casas, R. Mass cytometry identifies distinct subsets of regulatory T cells and Natural killer cells associated with high risk for Type 1 diabetes. Frontiers in Immunology. 10, 982 (2019).
- Kurioka, A., et al. CD161 defines a functionally distinct subset of pro-inflammatory Natural killer cells. Frontiers in Immunology. 9, 486 (2018).
- Romee, R., et al. Cytokine-induced memory-like natural killer cells exhibit enhanced responses against myeloid leukemia. Science Translational Medicine. 8 (357), (2016).
- Shinko, D., et al. Mass cytometry reveals a sustained reduction in CD16+ Natural killer cells following chemotherapy in colorectal cancer patients. Frontiers in Immunology. 10, 2584 (2019).
- Pohlmeyer, C. W., et al. Identification of NK cell subpopulations that differentiate HIV-infected subject cohorts with diverse levels of virus control. Journal of Virology. 93 (7), 01790 (2019).
- Palgen, J. -. L., et al. NK cell immune responses differ after prime and boost vaccination. Journal of Leukocyte Biology. 105 (5), 1055-1073 (2019).
- Mei, H. E., Leipold, M. D., Schulz, A. R. Barcoding of live human peripheral blood mononuclear cells for multiplexed mass cytometry. The Journal of Immunology. 194 (4), 2022-2031 (2015).
- Takahashi, C., et al. Mass cytometry panel optimization through the designed distribution of signal interference. Cytometry. Part A: The Journal of the International Society for Analytical Cytology. 91 (1), 39-47 (2017).
- Baumgart, S., Peddinghaus, A., Schulte-Wrede, U., Mei, H. E., Grützkau, A. OMIP-034: Comprehensive immune phenotyping of human peripheral leukocytes by mass cytometry for monitoring immunomodulatory therapies. Cytometry. Part A: The Journal of the International Society for Analytical Cytology. 91 (1), 34-38 (2017).
- Leipold, M. D. Another step on the path to mass cytometry standardization. Cytometry. Part A: The Journal of the International Society for Analytical Cytology. 87 (5), 380-382 (2015).
- Bendall, S. C., et al. Single-cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum. Science. 332 (6030), 687-696 (2011).
- Leipold, M. D., Newell, E. W., Maecker, H. T. Multiparameter phenotyping of human PBMCs using mass cytometry. Methods in Molecular Biology. 1343, 81-95 (2015).
- Fienberg, H. G., Simonds, E. F., Fantl, W. J., Nolan, G. P., Bodenmiller, B. A platinum-based covalent viability reagent for single-cell mass cytometry. Cytometry. Part A: The Journal of the International Society for Analytical Cytology. 81 (6), 467-475 (2012).
- Zivanovic, N., Jacobs, A., Bodenmiller, B. A practical guide to multiplexed mass cytometry. Current Topics in Microbiology and Immunology. 377, 95-109 (2014).
- Béziat, V., Hilton, H. G., Norman, P. J., Traherne, J. A. Deciphering the killer-cell immunoglobulin-like receptor system at super-resolution for natural killer and T-cell biology. Immunology. 150 (3), 248-264 (2017).
- Wilk, A. J., et al. Charge-altering releasable transporters enable specific phenotypic manipulation of resting primary natural killer cells. BioRxiv. , 970491 (2020).
- Vendrame, E., et al. TIGIT is upregulated by HIV-1 infection and marks a highly functional adaptive and mature subset of natural killer cells. AIDS. 34 (6), 801-813 (2020).
- Zhao, N. Q., et al. Natural killer cell phenotype is altered in HIV-exposed seronegative women. PloS One. 15 (9), 0238347 (2020).
- McKechnie, J. L., et al. HLA upregulation during dengue virus infection suppresses the natural killer cell response. Frontiers in Cellular and Infection Microbiology. 9, 268 (2019).
- McKechnie, J. L., et al. Mass cytometry analysis of the NK cell receptor-ligand repertoire reveals unique differences between dengue-infected children and adults. ImmunoHorizons. 4 (10), 634-647 (2020).
- Ranganath, T., et al. Characterization of the impact of daclizumab beta on circulating natural killer cells by mass cytometry. Frontiers in immunology. 11, 714 (2020).
- Fernandez, I. Z., et al. A novel human IL2RB mutation results in T and NK cell–driven immune dysregulation. The Journal of Experimental Medicine. 216 (6), 1255-1267 (2019).
- Herndler-Brandstetter, D., et al. Humanized mouse model supports development, function, and tissue residency of human natural killer cells. Proceedings of the National Academy of Sciences. 114 (45), 9626-9634 (2017).
- Nikzad, R., et al. Human natural killer cells mediate adaptive immunity to viral antigens. Science Immunology. 4 (35), (2019).

.
