Competing-Risk Nomogram for Predicting Cancer-Specific Survival in Multiple Primary Colorectal Cancer Patients after Surgery
Özet
The study found that male gender, poor tumor grade, and advanced Tumor Node Metastasis stage were associated with poorer cancer-specific survival (CSS) in multiple primary colorectal cancer (MPCC) patients after surgery. We developed a nomogram to predict the CSS of MPCC patients and contribute to clinical treatment decision-making.
Abstract
Cancer-specific survival (CSS) in multiple primary colorectal cancer (MPCC) patients is competitively affected by death from other causes. This study aimed to investigate CSS and associated risk factors by competing risk analysis in MPCC patients. The data of this study is from the SEER database. Using univariable and multivariable analysis of the competitive risk model to weaken the impact of competitive events, it explores the risk factors of CSS and develops a nomogram model. Then, the performance of the model is verified by the ROC curve, calibration curve, and DCA. The study encompasses a total of 8931 patients, with 6255 in the training cohort and 2676 in the validation cohort. Univariable and multivariable analyses showed that sex, Tumor Node Metastasis (TNM) stage, and tumor grade are independent risk factors for cancer-specific survival in MPCC patients. Based on the risk factors, we developed a diagram model to predict CSS. ROC curve, calibration curve, and DCA also show good results. In conclusion, a nomogram is developed that serves as a valuable tool for predicting CSS in MPCC patients, providing clinicians with crucial insights for personalized treatment planning.
Introduction
Colorectal cancer, as one of the most common digestive system tumors globally, has seen a continuous increase in its incidence over the past few decades. The persistently high incidence and mortality rates have drawn significant attention. According to the latest statistics, colorectal cancer ranks third among common malignant tumors worldwide, with its mortality rate ranking second globally1. Multiple primary colorectal cancer (MPCC) is a special subtype of colorectal cancer that has garnered increasing attention from researchers in recent years. It is defined as the diagnosis of two or more independent colorectal cancer lesions in the same patient, with a distance of more than 5 cm between the edges of the lesions. In MPCC, when multiple tumors are diagnosed simultaneously or within less than 6 months, it is defined as synchronous colorectal cancer (SCRC), while if the interval between diagnoses is greater than 6 months, it is defined as metachronous colorectal cancer (MCRC)2,3,4,5.
The proportion of MPCC within all colorectal cancers is relatively low, and reports on its incidence vary across different regions and studies. Recent studies have reported that MPCC accounts for between 2%-10% of colorectal cancers2,3,4,5. Compared to solitary colorectal cancer, MPCC has a worse prognosis3,6. Currently, clinical assessment of MPCC prognosis mainly relies on the tumor lymph node metastasis (TNM) staging system of the American Joint Committee of Cancer, which uses the most advanced stage of multiple lesions as the actual stage of MPCC. However, basing prognosis predictions solely on TNM staging is insufficient. There is still a lack of an effective tool to predict the prognosis of MPCC post-surgery. In the current era of precision medicine, clinical prediction models for quantifying risk are widely used in clinical decision-making and patient prognosis assessment7,8. In recent years, nomograms based on independent prognostic risk factors have been widely accepted for predicting tumor prognosis9. Nomograms can visualize complex statistical models, making them easier to apply in clinical settings. In the assessment of tumor prognosis, overall survival (OS) and cancer-specific survival (CSS) are commonly used outcome indicators9. OS refers to the time from the confirmation of a cancer in a patient to death from any cause. CSS refers to the time from the diagnosis of a tumor to death caused specifically by the cancer, offering a more precise reflection of the risk of death due to the cancer. When CSS is used as the outcome indicator, deaths caused by other factors can affect the probability of cancer-specific mortality, introducing a competing risk relationship between the two events10,11. Therefore, studies of tumor-specific survival should use competing risk models to eliminate the impact of competing events. Previous studies have constructed models to predict the prognosis of MPCC, but these have been limited to synchronous colorectal cancer and have not utilized competing risk models to account for the impact of competing events on CSS12,13,14.
In this study, we explored the competing independent risk factors affecting cancer-specific survival for MPCC after surgery. The rationale for employing a competing risks model stems from its ability to account for the possibility that patients may die from causes other than cancer, which is critical for obtaining unbiased survival estimates15,16. Traditional survival analysis techniques, such as the Cox model, may overestimate survival probabilities in the presence of competing events, making the use of competing risk models more appropriate in these scenarios17.
Based on the identified independent risk factors, we constructed a nomogram to predict survival probability and validated its performance. Nomograms have gained popularity in clinical settings because they provide a user-friendly graphical representation of complex statistical models, allowing clinicians to easily calculate individualized risk scores18,19. Unlike other predictive tools, nomograms incorporate multiple risk factors and provide more accurate, personalized prognosis estimates. This approach has been increasingly validated across various cancers, demonstrating superior performance over traditional staging systems20,21,22. Our tool aims to assist clinicians in making personalized and more accurate prognosis estimates during diagnosis and treatment, thereby enhancing decision-making in the management of MPCC.
Protocol
This study includes two steps. First, clinical and survival data of MPCC were obtained from the SEER database. Then, the R software (version 4.3.3) was used to analyze and construct a competing risk model. The workflow diagram of the study is presented in Figure 1. This study does not require ethical approval and consent to participate. The data used in this study was obtained from databases.
1. Data acquisition
- Download the SEER.Stat 8.4.3 software from the SEER database website (http://seer.cancer.gov/about/overview.html). Register and log in to SEER.Stat 8.4.3 to obtain relevant patient data.
- After logging in to SEER.Stat 8.4.3, click on Case Listing Session > Data and select the Incidence SEER Research Data, 17 Registries, Nov 2022 Sub (2000-2020) database.
- Click on Selection > Edit and choose {Race, Sex, Year Dx. Year of diagnosis} = '2004', '2005', '2006', '2007', '2008', '2009', '2010', '2011', '2012', '2013', '2014', '2015' AND {Site and Morphology. Site recode ICD-O-3/WHO 2008} = 'Colon and Rectum' AND {Site and Morphology. Diagnostic Confirmation} = 'Positive histology' AND {Multiple Primary Fields. Sequence number}! = 'One primary only'.
- Click on OK and save the selection. Click on Table, and in the Available variables interface, select Age recode with single ages and 85+, Sex, Site recode ICD-O-3/WHO 2008, CS tumor size, Grade Recode (thru 2017), Derived AJCC T, 6th ed (2004-2015), Derived AJCC N, 6th ed (2004-2015), Derived AJCC M, 6th ed (2004-2015), Radiation recode, Chemotherapy recode (yes, no/unk), SEER cause-specific death classification, SEER other cause of death classification, Survival months, Patient ID, RX Summ-Surg Prim Site (1998+), and click on Column.
- Click on Output, name the data, and click on Execute to output and save the data.
- The SEER database does not directly provide information on whether a patient has MPCC. After completing the data download as described above, use the patient ID to filter out patients with MPCC, that is, those diagnosed with two or more occurrences. After identifying MPCC patients, calculate the interval between tumor diagnoses based on their survival times. Using 6 months as the cutoff, classify patients into SMPCC and MMPCC.
- Based on their age at the time of initial diagnosis, categorize patients' ages as = 65 years and> 65 years. For tumor grade, grade I corresponds to good differentiation, grade II corresponds to moderate differentiation, grade III corresponds to low differentiation, and grade IV corresponds to undifferentiation. Classify tumor location based on the distribution of multiple tumors as right colon, left colorectum, and entire colorectum.
- There are obvious differences in embryonic development and biological characteristics between the proximal and distal colorectum, divide the tumor location based on the splenic flexure23. The right colon is defined as proximal to the splenic flexure, including the cecum, ascending colon, hepatic flexure, and transverse colon, while the left colorectum includes the splenic flexure, descending colon, sigmoid colon, rectosigmoid junction, and rectum. If a patient's tumors are entirely located in the right colon, define the tumor location as right colon; if all tumors are located in the left colorectum, define the location as left colorectum; if there are tumors in both the right colon and left colorectum, define the location as entire colorectum.
- Determine tumor size by selecting the largest tumor diameter among multiple tumors in the same patient. Based on the tumor diameter, categorize size as = 5 cm or >5 cm. A total of 8,931 patients were included in the study. To create a training cohort and a validation cohort, randomly divide all cases in a 7:3 ratio.
2. Model construction
- Download RStudio (2023.12.1+402) and R software (4.3.3). Open RStudio to run R software. Click on New File and select R Script to create a new R programming interface. Enter the relevant code in the code editor and click on Run to execute the code.
NOTE: The R language and its functions offer a wide variety of parameters. Adding or modifying these parameters can enhance data analysis and visualization. - Use the following code to perform univariate analysis and plot the CIF curve.
library(tidycmprsk)
library(gtsummary)
library(ggsurvfit)
library(ggprism)
data <-read.csv('data.csv')"
data$status<-factor(data$status, levels=c(0,1,2),labels=c("0","CSS","OCS"))
CIF <- tidycmprsk::cuminc(Surv(time, status) ~ Sex, data = data)
ggcuminc(CIF,outcome= c("CSS", "OCS"),size=1.5)
where data.csv is data obtained from the SEER database. - After running the above code, click on Export, then click Save as Image, and finally click Save to save the image. The method for saving subsequent images will be the same as in this step. Replace Sex in the above code one by one with other factors to perform univariate analysis for all factors.
- Use the following code to perform BSR and multivariate analysis and visualization.
library(leaps)
library(riskRegression)
library(prodlim)
library(forestplot)
leaps<-regsubsets(status==0~Sex+Size+Grade+T+N+M,data = data)
plot(leaps,scale="adjr2")
multi <- FGR(Hist(time,status)~Sex + T + N + M +Grade+ Size,cause=1,data=data)
summary(multi)
Multi <- read.csv("multi.csv",header = T)
forestplot(labeltext=as.matrix(Multi [,1:4]), mean= Multi $HR_mean, ower= Multi $HR_1, upper= Multi $HR_2)
where the data in multi.csv comes from the results of the previous code. - Follow step 2.3. to save the image.
- Use the following code to plot the nomogram, ROC curve, calibration curve, and DCA curve.
library(QHScrnomo)
library(rms)
library(riskRegression)
library(prodlim)
data <- read.csv("data.csv")
d <- datadist(data)
options(datadist = "d")
e <-cph(Surv(time,status==1)~Sex + T + N + M +Grade,data = data,
x=T, y=T, surv=T,time.inc=60)
nomo <- crr.fit(e,failcode=1,cencode = 0)
nomogram.crr(fit =nomo, lp = F, xfrac = 0.5, fun.at =seq(from=0, to=1, by= 0.1) , failtime =c(12,36,60),funlabel = c("1-year CSS Cumulative Incidence", "3-year CSS Cumulative Incidence","5-year CSS Cumulative Incidence"))
set.seed(123)
data$pro <- tenf.crr(m3,time = 60)
groupci(x=data$pro, ftime = data$time, fstatus = data$status, failcode = 1, cencode = 0, ci = TRUE)
f <- CSC(Hist(time,status)~Sex + T + N + M +Grade,data = data)
x <- Score(list(model1=f), Hist(time,status)~1, data=data, cause=1, times=c(12,36,60), se.fit=1L, plots="roc", metrics="auc")
g <- as.data.frame(x$AUC$score)
h <- g[g$times %in% c(12,36,60),]
col = c("darkcyan","tomato","purple")
plotROC(x, xlab="1-Specificity", ylab="Sensitivity",col=col[1], cex=1.5, legend="", auc.in.legend = F, times = 12)
plotROC(x,col=col[2],legend = '', cex=1.5,times =36,auc.in.legend = F,add=T)
plotROC(x,col=col[3], times =60, add=T, cex=1.5, legend = '',auc.in.legend = F)
leg <- paste0(c("1-Year: ","3-Year: ","5-Year: "),substr(a$AUC,1,5)) - Follow step 2.3. to save the image.
- Use the ggscidca package to plot the DCA curve.
library(ggscidca)
df_surv<-read.csv("data.csv",header = T)
cox_model <- coxph(Surv(time,status==1)~Sex + T + N+ M +Grade, data = df_surv)
cox_model1<-newcrr(cox_model)
scidca(cox_model1, newdata= df_surv,threshold.line = T,threshold.text = T)
where data.csv is data obtained from the SEER database. - Follow step 2.3. to save the image.
Representative Results
Patient characteristics
A total of 8,931 patients were included in the study. In terms of sex distribution, there was a higher proportion of male patients (56%) compared to female patients (44%). Regarding tumor location distribution, the majority of tumors were distributed across the total colorectum, while the fewest tumors were located in the right colon (21%). In terms of tumor grade, the most common grade was grade II, with 6,251 patients, accounting for 70% of the total; this was followed by grade III, with 2,026 patients (23%). Grades I and IV had 355 and 299 patients, respectively, accounting for 4% and 3.3% of the total. In terms of tumor invasion, T3 and T4 stages were the most common. More than half of the patients did not have lymph node metastasis, while 17% of the patients experienced distant metastasis. Radiation therapy was received by 1,446 patients (16%), and chemotherapy was administered to 3,843 patients (43%). SCRC was more common, with 5,327 patients accounting for 60% of the total; there were 3,604 patients with metachronous colorectal cancer, accounting for 40%. The patients were randomly divided into training and validation sets in a 7:3 ratio. There were no statistically significant differences in baseline data between the training and validation cohorts, as shown in Table 1.
Univariable analysis
After controlling the influence of competitive events, the results of the univariate analysis showed sex, tumor grade and size, TNM stage, radiation, chemotherapy, synchronous or metachronous status, and tumor location were the prognostic factors affecting CSS in MPCC patients. Only age is not a prognostic factor for CSS in MPCC patients. We note that there is a significant intersection of CIF in radiation, chemotherapy, location, and synchronous or metachronous status, indicating that the short-term and long-term prognostic effects of radiation on MPCC patients were different, as did chemotherapy, location, and synchronous or metachronous status. The cumulative risk curve of each subgroup is shown in Figure 2.
Multivariable analysis
The prognostic factors obtained by univariable analysis were incorporated into the best subsets regression (BSR) and multivariable analysis of the competitive risk model. Among them, radiation, chemotherapy, location, and synchronous or metachronous status are excluded because of their dual effects on prognosis. The results of the BSR and multivariable analysis both showed that sex, TNM stage, and tumor grade were independent risk factors for CSS in MPCC patients. (Figure 2)
Construction and verification of Nomogram
Based on the independent risk factors obtained by multivariable analysis, we construct a line nomogram to predict CSS and verify the performance of the prediction model (Figure 3). Then, we use the ROC curve, calibration curve, and DCA to evaluate the model. The ROC curve showed that the AUCs of 1-year, 3-year, and 5-year of the training cohort were 0.762, 0.742, and 0.734, and the AUCs of 1-year, 3-year, and 5-year of the verification cohort were 0.801, 0.740, and 0.743. In the training cohort and verification cohort, the calibration curve showed a high agreement between the projected probability and the actual data (Figure 4). In order to verify the performance of the model in clinical application, we used DCA to evaluate the clinical value of the model. The results show that the model shows a good net benefit (Figure 5).
Figure 1: Workflow diagram of the study. This study consists of two steps: first, data was obtained using SEER.Stat, and then data analysis and visualization were performed using R. Please click here to view a larger version of this figure.
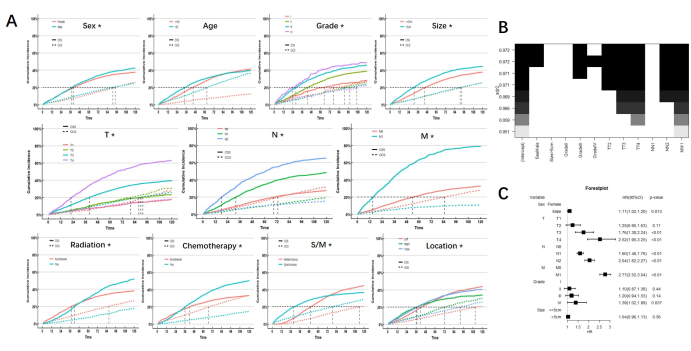
Figure 2: Analysis results. (A) CIF of subgroups. * indicates p <0.05. (B) Best subsets regression. Under the best goodness of fit, sex, grade, and TNM stage were considered for inclusion (shown as black blocks at the top). (C) Multivariate analysis also showed that sex, grade, and TNM stage are independent risk factors. Please click here to view a larger version of this figure.
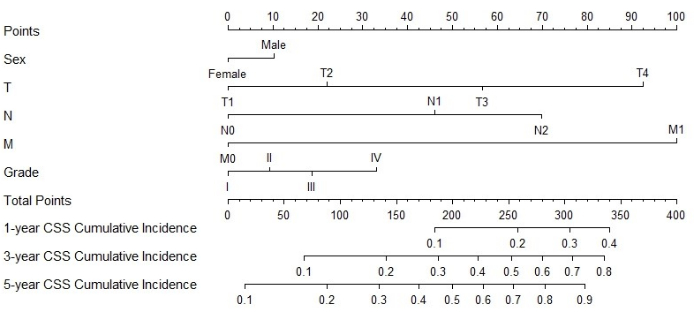
Figure 3: Nomogram for CSS in MPCC patients. The patient's total score can be calculated by adding the scores corresponding to each factor. Based on the total score, the probability of cancer-specific death at 1, 3, and 5 years can be predicted Please click here to view a larger version of this figure.
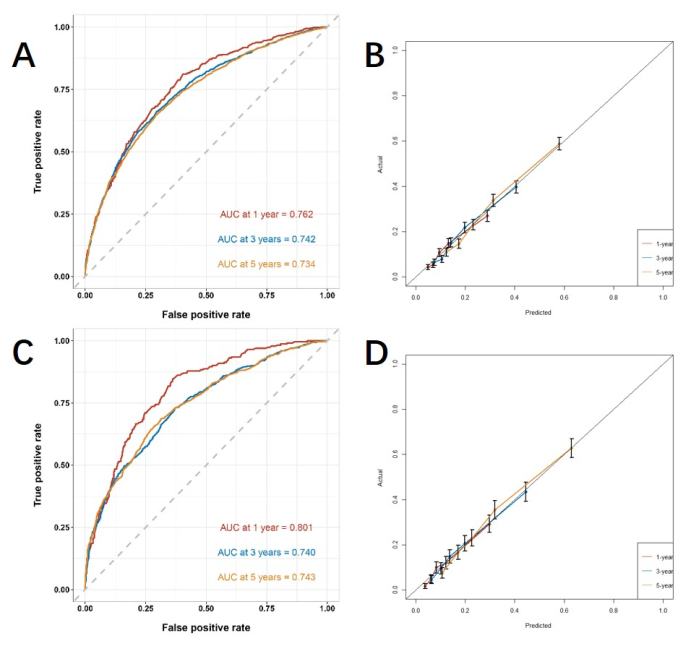
Figure 4: ROC curve and calibration curve. (A, B) The curves in training cohort (n=6255) and (C, D) validation cohort (n=2676). The closer the AUC value is to 1, the better the model's classification performance. The error bars show the 95% confidence interval for the probability of the actual event occurring. Please click here to view a larger version of this figure.
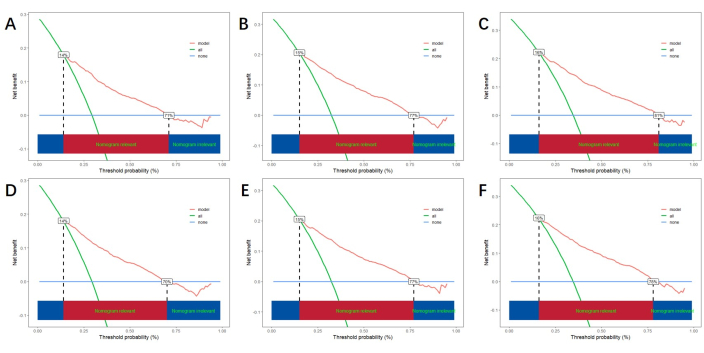
Figure 5: DCA for 1-year, 3-year, and 5-year. (A, B, C) DCA in training cohort and (D, E, F) validation cohort. The green line represents the net benefit of all positives, the blue line represents the net benefit of none positives, and the red line indicates the net benefit of the model. The red area below represents the model's benefit exceeding that of all positives and none positives, indicating the actual benefit range of the model. Please click here to view a larger version of this figure.
Table 1: Clinicopathologic and baseline characteristics of patients. Please click here to download this Table.
Discussion
As a common digestive system tumor, factors influencing the prognosis of solitary colorectal cancer have been studied and confirmed in previous research24. However, there has been limited research on prognostic factors for MPCC patients. This study included 8,931 MPCC patients who underwent surgery from the SEER database between 2004 and 2015. We used a competing risk model to investigate the risk factors affecting CSS and constructed a predictive model. In this study, 39.1% of the deceased patients died due to causes other than cancer, emphasizing the necessity of using a competing risk model to analyze risk factors for CSS.
Previous study has found that advanced age is a risk factor affecting the OS of MPCC patients12. However, in this study, we found that age was not a risk factor for CSS. Elderly patients often exhibit poorer baseline health conditions and more comorbidities, which can lead to a shorter OS compared to younger patients. The elderly are also more likely to die from other causes, such as cardiovascular events and severe infections, making them more susceptible to these factors than younger patients. In this study, we focused on the competing relationship between deaths due to other causes and deaths due to cancer itself. By using a competing risk model and excluding the interference of deaths caused by other reasons, we found that under these conditions, advanced age was no longer a risk factor for CSS of MPCC. This indicates that MPCC patients across different age groups may face similar tumor burdens. Overall, young and elderly patients with this disease have similar CSS, which provides valuable guidance for developing personalized treatment strategies.
This study also found that the incidence and prognosis of MPCC exhibit gender-related tendencies, with a higher proportion and poorer CSS of male patients. This is consistent with the situation observed in solitary colorectal cancer. Previous studies have shown that solitary colorectal cancer is more common in males, and male patients have a worse prognosis than female patients. This may be due to the influence of estrogen on the occurrence and progression of colorectal cancer25. Additionally, other research has suggested that the gut microbiota and gut metabolites in male patients may be one of the reasons for the gender differences observed in colorectal cancer patients26. A study in Germany found that type 2 diabetes has a greater impact on colorectal cancer in female27. What's more, vitamin D has a protective effect against colorectal neoplasia in females, but no similar findings were found in males28.
Competing univariate analysis found that MPCC patients with larger tumor sizes had poorer CSS. In previous studies, tumor size has often been considered an indicator of tumor aggressiveness 29,30,31. However, the results from competing multivariate analyses did not support tumor size as an independent risk factor. This suggests that in hollow organs like the colorectum, tumor size may have limited ability to reflect tumor aggressiveness. This phenomenon may stem from various factors, including the complexity of tumor biology and the differing patterns of tumor growth and spread across different organs.
Poor tumor grade indicates stronger invasive and migratory abilities of tumor cells, which aligns with the conclusions of this study: a worse tumor grade suggests a poorer CSS for MPCC patients. TNM stage is the most commonly used clinical method for guiding patient treatment and predicting prognosis32,33. This study found that the deeper the tumor infiltration, the greater the number of metastatic lymph nodes, and the presence of organ metastasis, the worse the patient's cancer-specific survival. This is entirely consistent with clinical consensus.
In colorectal cancer, prognosis varies depending on the location of the tumor. Many studies have found that the right colon has a poorer prognosis compared to the left colorectum34,35,36,37. However, some research suggests that in resectable colorectal cancer, the side of the tumor location does not impact the long-term prognosis38. In this study, MPCC located in the right colon had worse CSS in the short term but a better long-term prognosis. Similarly, MPCC patients who received radiation and chemotherapy had better short-term CSS but worse long-term CSS. SCRC had worse short-term CSS compared to MCRC but better long-term CSS. The reasons for these dual effects on prognosis are still unclear.
This study has some limitations. Due to the inherent constraints of the SEER database, we could not obtain or analyze some known prognostic indicators, such as carcinoembryonic antigen (CEA)levels and microsatellite status. Moreover, we could not exclude patients with inflammatory bowel disease, hereditary nonpolyposis colorectal cancer, and familial adenomatous polyposis who have higher risks of MPCC39. Additionally, immunotherapy has become an increasingly important aspect of colorectal cancer treatment, but we were unable to obtain data on this. The retrospective nature of our analysis and reliance on a single data set may introduce inherent biases.
The study based on the SEER database found that male gender, poor tumor grade, and advanced TNM stage were associated with poorer CSS in MPCC patients after surgery. It is important to pay close attention to patients with these risk factors during diagnosis and treatment. Furthermore, this study developed a nomogram to predict the CSS of MPCC patients, which can accurately predict the prognosis and contribute to clinical treatment decision-making.
Açıklamalar
The authors have nothing to disclose.
Acknowledgements
None.
Materials
| forestplot package | Comprehensive R Archive Network (CRAN) | forest plot 3.1.3 | A forest plot that allows for multiple confidence intervals per row, custom fonts for each text element, custom confidence intervals, text mixed with expressions, and more. |
| ggprism package | Comprehensive R Archive Network (CRAN) | ggprism 1.0.5 | The ggprism package provides various themes, palettes, and other useful functions to customise ggplots and give them the ‘GraphPad Prism’ look. |
| ggscidca package | Comprehensive R Archive Network (CRAN) | ggscidca package | The 'ggscidca' package adds coloured bars of discriminant relevance to the traditional decision curve. Improved practicality and aesthetics. |
| ggsurvfit package | Comprehensive R Archive Network (CRAN) | ggsurvfit 1.1.0 | The ggsurvfit package eases the creation of time-to-event (aka survival) summary figures with ggplot2. The concise and modular code creates images that are ready for publication or sharing. |
| gtsummary package | Comprehensive R Archive Network (CRAN) | gtsummary 2.0.0 | The gtsummary package provides an elegant and flexible way to create publication-ready analytical and summary tables using the R programming language. The {gtsummary} package summarizes data sets, regression models, and more, using sensible defaults with highly customizable capabilities. |
| QHScrnomo package | Comprehensive R Archive Network (CRAN) | QHScrnomo 3.0.1 | The goal of QHScrnomo is to provide functionality to construct nomograms in the context of time-to-event (survival) analysis in the presence of competing risks. It also contains functions to build, validate, and summarize these models. |
| R Software | R Core Team | R 4.3.3 | Free software environment for statistical computing and graphics |
| riskRegression package | Comprehensive R Archive Network (CRAN) | riskRegression 1.3.7 | Risk Regression Models and Prediction Scores for Survival Analysis with Competing Risks |
| rms package | Comprehensive R Archive Network (CRAN) | rms 6.8-1 | rms does regression modeling, testing, estimation, validation, graphics, prediction, and typesetting by storing enhanced model design attributes in the fit. |
| RStudio | RStudio, Public Benefit Corporation(PBC) | Rstudio 2023.12.1+402 | Integrated Development Environment (IDE) used for running R scripts, data analysis, and model development. Provides a user-friendly interface for R programming with advanced features like script editing, debugging, and version control. |
| SEERstat | National Cancer Institute (NCI) | SEERstat 8.4.3 | Software for statistical analysis of SEER and other cancer-related databases |
| tidycmprsk package | Comprehensive R Archive Network (CRAN) | tidycmprsk 1.0.0 | The tidycmprsk package provides an intuitive interface for working with the competing risk endpoints |
Referanslar
- Bray, F., et al. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 74 (3), 229-263 (2024).
- Derwinger, K., Gustavsson, B. A study of aspects on gender and prognosis in synchronous colorectal cancer. Clin Med Insights Oncol. 5, 259-264 (2011).
- Chin, C. C., Kuo, Y. H., Chiang, J. M. Synchronous colorectal carcinoma: predisposing factors and characteristics. Colorectal Dis. 21 (4), 432-440 (2019).
- Nikoloudis, N., et al. Synchronous colorectal cancer. Tech Coloproctol. 8 (Suppl 1), s177-s179 (2004).
- Huang, C. S., et al. Synchronous and metachronous colorectal cancers: Distinct disease entities or different disease courses. Hepatogastroenterology. 62 (140), 838-842 (2015).
- He, W., et al. Prognosis of synchronous colorectal carcinoma compared to solitary colorectal carcinoma: a matched pair analysis. Eur J Gastroenterol Hepatol. 31 (12), 1489-1495 (2019).
- Ranstam, J., Cook, J. A., Collins, G. S. Clinical prediction models. Br J Surg. 103 (13), 1886 (2016).
- Smith, T., et al. Comparison of prognostic models to predict the occurrence of colorectal cancer in asymptomatic individuals: a systematic literature review and external validation in the EPIC and UK Biobank prospective cohort studies. Gut. 68 (4), 672-683 (2019).
- Iasonos, A., Schrag, D., Raj, G. V., Panageas, K. S. How to build and interpret a nomogram for cancer prognosis. J Clin Oncol. 26 (8), 1364-1370 (2008).
- Conroy, T., et al. Five-year outcomes of FOLFIRINOX vs Gemcitabine as adjuvant therapy for pancreatic cancer: A randomized clinical trial. JAMA Oncol. 8 (11), 1571-1578 (2022).
- Zhong, M., et al. Impact of lung metastasis versus metastasis of bone, brain, or liver on overall survival and thyroid cancer-specific survival of thyroid cancer patients: A population-based study. Cancers. 14 (13), 3133 (2022).
- Xu, Y., Wang, X., Huang, Y., Ye, D., Chi, P. A LASSO-based survival prediction model for patients with synchronous colorectal carcinomas based on SEER. Transl Cancer Res. 11 (8), 2795-2809 (2022).
- Zhang, X., Zhao, L., Hu, Y., Deng, K., Ren, W. A novel risk prediction nomogram for early death in patients with resected synchronous multiple primary colorectal cancer based on the SEER database. Int J Colorectal Dis. 38 (1), 130 (2023).
- Zhang, X., et al. Developing prognostic nomograms to predict overall survival and cancer-specific survival in synchronous multiple primary colorectal cancer based on the SEER database. J Cancer Res Clin Oncol. 149 (15), 14057-14070 (2023).
- Austin, P. C., Lee, D. S., Fine, J. P. Introduction to the analysis of survival data in the presence of competing risks. Circulation. 133 (6), 601-609 (2016).
- Lau, B., Cole, S. R., Gange, S. J. Competing risk regression models for epidemiologic data. Am J Epidemiol. 170 (2), 244-256 (2009).
- Nolan, E. K., Chen, H. Y. A comparison of the Cox model to the Fine-Gray model for survival analyses of re-fracture rates. Arch Osteoporos. 15 (1), 86 (2020).
- Park, S. Y. Nomogram: An analogue tool to deliver digital knowledge. J Thorac Cardiovasc Surg. 155 (4), 1793 (2018).
- Balachandran, V. P., Gonen, M., Smith, J. J., DeMatteo, R. P. Nomograms in oncology: more than meets the eye. Lancet Oncol. 16 (4), e173-e180 (2015).
- Liang, W., et al. Development and validation of a nomogram for predicting survival in patients with resected non-small-cell lung cancer. J Clin Oncol. 33 (8), 861-869 (2015).
- Chen, D., et al. Predicting postoperative peritoneal metastasis in gastric cancer with serosal invasion using a collagen nomogram. Nat Commun. 12 (1), 179 (2021).
- Niu, X., et al. A prognostic nomogram for patients with newly diagnosed adult thalamic glioma in a surgical cohort. Neuro Oncol. 23 (2), 337-338 (2021).
- Luo, S., et al. Comparison of left- and right-sided colorectal cancer to explore prognostic signatures related to pyroptosis. Heliyon. 10 (7), e28091 (2024).
- Keum, N., Giovannucci, E. Global burden of colorectal cancer: emerging trends, risk factors and prevention strategies. Nat Rev Gastroenterol Hepatol. 16 (12), 713-732 (2019).
- Foster, P. A. Oestrogen and colorectal cancer: mechanisms and controversies. Int J Colorectal Dis. 28 (6), 737-749 (2013).
- Wang, L., et al. Male-biased gut microbiome and metabolites aggravate colorectal cancer development. Adv Sci. 10 (25), e2206238 (2023).
- Krämer, H. U., et al. Type 2 diabetes mellitus and gender-specific risk for colorectal neoplasia. Eur J Epidemiol. 27 (5), 341-347 (2012).
- Aigner, E., et al. Gender- and site-specific differences of colorectal neoplasia relate to vitamin D. Aliment Pharmacol Ther. 40 (11-12), 1341-1348 (2014).
- Dai, W., et al. Does tumor size have its prognostic role in colorectal cancer? Re-evaluating its value in colorectal adenocarcinoma with different macroscopic growth pattern. Int J Surg. 45, 105-112 (2017).
- Rössler, O., et al. Tumor size, tumor location, and antitumor inflammatory response are associated with lymph node size in colorectal cancer patients. Mod Pathol. 30 (6), 897-904 (2017).
- Zhang, Q., et al. Prognostic impact of tumor size on patients with metastatic colorectal cancer: a large SEER-based retrospective cohort study. Updates Surg. 75 (5), 1135-1147 (2023).
- Greene, F. L. TNM: our language of cancer. CA Cancer J Clin. 54 (3), 129-130 (2004).
- Ueno, H., et al. Optimal colorectal cancer staging criteria in TNM classification. J Clin Oncol. 30 (13), 1519-1526 (2012).
- Yahagi, M., Okabayashi, K., Hasegawa, H., Tsuruta, M., Kitagawa, Y. The worse prognosis of right-sided compared with left-sided colon cancers: a systematic review and meta-analysis. J Gastrointest Surg. 20 (3), 648-655 (2016).
- Holch, J. W., Ricard, I., Stintzing, S., Modest, D. P., Heinemann, V. The relevance of primary tumour location in patients with metastatic colorectal cancer: A meta-analysis of first-line clinical trials. Eur J Cancer. 70, 87-98 (2017).
- Petrelli, F., et al. Prognostic survival associated with left-sided vs right-sided colon cancer: A systematic review and meta-analysis. JAMA Oncol. 3 (2), 211-219 (2017).
- Zheng, C., Jiang, F., Lin, H., Li, S. Clinical characteristics and prognosis of different primary tumor location in colorectal cancer: a population-based cohort study. Clin Transl Oncol. 21 (11), 1524-1531 (2019).
- Karim, S., Brennan, K., Nanji, S., Berry, S. R., Booth, C. M. Association between prognosis and tumor laterality in early-stage colon cancer. JAMA Oncol. 3 (10), 1386-1392 (2017).
- Lindberg, L. J., et al. Risk of multiple colorectal cancer development depends on age and subgroup in individuals with hereditary predisposition. Fam Cancer. 18 (2), 183-191 (2019).

.
