High-Throughput Optical Controlling and Recording Calcium Signal in iPSC-Derived Cardiomyocytes for Toxicity Testing and Phenotypic Drug Screening
Özet
The present protocol describes all-optical control and observation of triggered cellular activity in iPSC-derived cardiomyocytes (iPSC-CMs) for high throughput drug screening and toxicity testing. Multi-parametric quantification of phenotypic patterns in time, and space, are shown. Long-term effects of drugs over hours, or sequential measurements over days, are demonstrated.
Abstract
Understanding how excitable cells work in health and disease and how that behavior can be altered by small molecules or genetic manipulation is important. Genetically encoded calcium indicators (GECIs) with multiple emission windows can be combined (e.g., for simultaneous observation of distinct subcellular events) or used in extended applications with other light-dependent actuators in excitable cells (e.g., combining genetically encoded optogenetic control with spectrally compatible calcium indicators). Such approaches have been used in primary or stem cell-derived neurons, cardiomyocytes, and pancreatic beta-cells. However, it has been challenging to increase the throughput, or duration of observation, of such approaches due to limitations of the instruments, analysis software, indicator performance, and gene delivery efficiency. Here, a high-performance green GECI, mNeonGreen-GECO (mNG-GECO), and red-shifted GECI, K-GECO, is combined with optogenetic control to achieve all-optical control and visualization of cellular activity in a high-throughput imaging format using a High-Content Imaging System. Applications demonstrating cardiotoxicity testing and phenotypic drug screening with healthy and patient-derived iPSC-CMs are shown. In addition, multi-parametric assessments using combinations of spectral and calcium affinity indicator variants (NIR-GECO, LAR-GECO, and mtGCEPIA or Orai1-G-GECO) are restricted to different cellular compartments are also demonstrated in the iPSC-CM model.
Introduction
Human-based induced pluripotent stem cells (iPSC)-derived models are considered a promising solution to the 3Rs aims (Replacement, Reduction, and Refinement) set out for animal studies1,2. iPSC-derived cardiomyocytes have been used for disease modeling and drug discovery as they recapitulate essential aspects of human biology3. Calcium imaging, typically with chemical dyes, has been used to observe cellular activity before and after drug treatment4,5. However, chemical dye-based calcium-sensing probes directly inhibit the Na, K-ATPase and disrupt cellular function6. Thus, tracking the same cells over hours or days has been problematic when chemical dyes are used. Here, a range of genetically encoded calcium indicators (GECIs)7,8,9,10 are utilized across a broad excitation/emission and calcium affinity spectrum to form a cardiac toxicity testing platform offering real-time multi-organelle measurements over extended periods of time11.
To further develop the high-throughput calcium-based screening concept in the iPSC-CM model beyond the previously demonstrated academic context using genetically encoded tools for optical control and calcium imaging by co-expression of a red-shifted calcium indicator, R-GECO or K-GECO with an optogenetic tool, ChR212,13, off-the-shelf viral kits have been generated. By transitioning imaging into a high-content instrument equipped with an auto-microfluidic system, single-channel pipetting of compound addition is layered on top of live-cell imaging. Finally, both crucial aspects of the experimental pathway, image processing, and analysis are improved and automated.
Protocol
Left ventricular non-compaction cardiomyopathy (LVNC) patient iPSC-derived cardiomyocytes (iPSC-CM) were provided by the National Taiwan University Hospital. The collection of patient samples for reprogramming to iPSC and the maintenance protocols14 for research purposes followed the WMA Declaration of Helsinki (ethical principles for medical research involving human subjects) and were approved by Institutional Review Board: Research Ethics Committee Office at NTUH (#201612099RINC). Other iPSC-CM's were obtained from commercial vendors. The use of iPSC derivatives does not require specific permissions in our institution.
1. iPSC derived cardiomyocytes preparation
- Prepare the multi-well plate before iPSC-CM is removed from cold storage for thawing. Coat each well of the 96-well microplate with 100 µL of 50 µg/mL fibronectin/0.2% gelatin solution (see Table of Materials) to cover all surfaces thoroughly.
- Incubate the plate at 37 °C for 1 h in a humidified incubator.
- Warm the medium (see Table of Materials) to room temperature for 30 min before cell thawing.
NOTE: The present protocol describes room temperature as 25 °C. - Transfer the iPSC-CM from the liquid nitrogen storage tank to a 37 °C water bath.
NOTE: iPS-CMs are typically shipped on dry ice by academic or commercial sources. They are transferred to liquid nitrogen storage on receipt until use. - Remove the vial before the ice melts entirely and spray the vial with 75% ethanol.
- Take the vial to a Class II biosafety cabinet and gently transfer the contents to a new 15 mL conical tube containing 9 mL of room temperature medium.
- Centrifuge the cells at 200 x g for 3 min at room temperature.
- Discard the supernatant by pipette and gently resuspend the cells in 1 mL of medium with 10 µM of ROCK inhibitor (Y27632) (see Table of Materials).
NOTE: Avoid repeated pipetting of thawed cells to ensure maximum cell recovery. - Remove the coating solution from the well and add 50 µL of medium with 10 µM of ROCK inhibitor quickly.
- Confirm the number of viable cells using the trypan blue exclusion method with a hemocytometer15.
- Plate cells in coated 96-well plate at a density of 100,000-150,000 cell/cm2,as per manufacturer's instructions16.
- After 24 h, ensure that the cells attached to the surface of the plate are in contact with other cells.
NOTE: Single cells survive poorly in long-term culture and typically behave more variably. - Replace pre-warmed maintenance medium every 2 days.
NOTE: For long-term drug treatment, the LVNC iPSC-CM were changed with half medium replacements with, or without, small molecule drugs every 2 days.
2. Expression of genetically encoded calcium indicators
- After plating the cells for 72 h, thaw the GECIs viral kits (see Table of Materials) on ice.
- Prepare a 2x viral kit stock solution with the maintenance medium.
- Prepare the cells for infection by adjusting the medium volume to 100 µL per well. Add 100 µL of the pre-mixed viral solution and incubate for 8-16 h at 37 °C.
- Replace with 200 µL pre-warmed medium after incubation.
- Visualize the GECIs expression 24 h post-transduction using an imaging system (see Table of Materials).
NOTE: The expression level reaches the maximum at 48-72 h post-transduction. Cells can be imaged from the 24 h time-point. However, the signals obtained from GECIs sustain for more than one month. - Maintain cells in the humidified incubator before functional assays are performed.
- Infect the iPSC-CMs (LVNC patient-derived iPSC-CMs) with ChR2-K-GECO viral kit (see Table of Materials) at day 25 post differentiation using the infection protocol (step 2.1-2.6).
3. Dye-based calcium imaging using Fluo-4
- Dissolve Fluo-4 powder (see Table of Materials) in DMSO as a stock solution (5 mM).
- Dilute the stock solution to a concentration of 10 µM Fluo-4 in Tyrode's buffer.
NOTE: Tyrode's buffer consists of 1.0 g/L of sodium bicarbonate, 0.2 g/L of calcium chloride, 0.1 g/L of magnesium chloride, 0.2 g/L of potassium chloride, 8.0 g/L of sodium chloride, 0.05 g/L of sodium phosphate monobasic, and 1.0 g/L of D-glucose. - Replace half medium with 10 µM of Fluo-4 solution (giving a final concentration of 5 µM).
- Incubate the cells at 37 °C for 30 min.
- Wash the cells with pre-warmed Tyrode's buffer and replace them with the medium before image acquisition.
- Start recording with the stream acquisition protocol.
NOTE: Stream acquisition is continuously acquiring images without interval time. The beating frequency of iPSC-CM is about 1 Hz, and this stream acquisition protocol is used for collecting beating patterns.
4. Image acquisition and functional assays for toxicity test and phenotypic screening
- Switch on all the devices of the High-Content Imaging System (see Table of Materials).
- Verify that the chamber temperature reaches 37 °C, with 5% CO2 supplementation. Keep the system at equilibrium for 2 h before image acquisition.
NOTE: Thermal drift is a significant problem for long-term live-cell observations, 1 °C can change the focal plane by ~1 µm, so allowing the lens, and stage, to reach the target temperature is essential before image acquisition. - Open the imaging software (see Table of Materials) and choose the plate setting for a 96-well plate.
- Place an unlidded 96-well plate into the chamber and load with the live-cell sealing ring on top.
NOTE: The live-cell sealing ring is an adapter for reaching environmental equilibrium. - Choose the 20x (Figure 1 and Figure 2), 60x (Figure 3 and Figure 4), and 20x (Figure 5) water immersion objective lens according to the spatial resolution required.
- Adjust the focus to take clear images before experimental acquisition.
- Select 3-5 regions randomly per well for calcium activity recording.
NOTE: Every region must include at least 20 cells (n ≥ 20). - Choose appropriate filters for different indicators. FITC 475/536 for mNG-GECO, mtGCEPIA3, and Oria1-G-GECO; Cy5 631/692 for NIR-GECO2; ER 530/593 for ER-LAR-GECO; Texas Red 560/624 for K-GECO.
- Set the appropriate light-emitting diode (LED) power for each imaging channel used.
NOTE: When optimizing LED power for imaging acquisition by signal (detected intensity of object) to noise (detected intensity of background) ratio, the S/N ratio should be higher than 2. In this protocol, 10% LED power for TRITC and FITC; 20% for Cy5 was typical. - Set the camera exposure time to a maximum of 40 ms (25 Hz) and a recording duration of 30 sfor stream acquisition to observe calcium transients reported by mNG-GECO and K-GECO probes expressed in cardiomyocytes (Figure 1-3 and Figure 5). Increase the LED power if the signal/noise is <2 at higher frame rates.
- For optogenetic stimulation (Figure 2 and Figure 3C,D), optimize the power (typically 5%-10%) and frequency of blue light at 1 Hz.
NOTE: Generally, human iPSC-CM beats spontaneously around ~ 1 Hz, and the operator needs to pace faster than the spontaneous rate. - If sequential (Figure 3) or extended (Figure 4) measurements are planned, avoid phototoxicity at all costs. This may mean reducing illumination power intensity and compensating for this by increasing camera exposure time, for example, to 100 ms with the time-lapse protocol used for three-channel real-time observation of subcellular Ca2+ signals (Figure 4).
- Use the asynchronous dispense protocol for 10 mM caffeine addition via the auto microfluidics system (see Table of Materials) (Figure 4).
NOTE: The asynchronous dispense protocol allows image acquisition to occur while drugs such as caffeine are added, without gaps between images. - Start acquisition.
5. Small molecule preparation
- Resuspend the E4031, dofetilide, and verapamil in DMSO and dilute them with Tyrode's buffer. The final concentration of DMSO in the medium was 0.1% (v/v).
- Prepare the compound solutions at the double (i.e., 2x) the desired working concentration and equilibrate at 37 °C before addition.
NOTE: Minimizing temperature fluctuations following medium addition is vital as contractility is temperature-dependent, and the effect of changing temperature is rapid. - Arrange compounds to the corresponding target well.
- Add 100 µL of the double strength (2x) compounds via the auto microfluidics system (see Table of Materials) into the wells and start acquisition after the desired (typically 15 min) incubation period.
6. Imaging processing and analysis
- Isolate the signal area from the background by detecting the feature of the pulse over time-revolution automatically with the software.
- Use the Calcium peak analysis software (see Table of Materials) to analyze Beating frequency (BPM), Peak signal (Amplitude), Calcium transient duration 50% (CTD50), Calcium transient duration 90% (CTD90), rise time, and decay time (Figure 1C).
NOTE: In these settings, photobleaching was apparent over the first 10 s, and the signals are stabilized after that. Thus, the calcium peak was analyzed from 11-30 s. In three-channel recordings (Figure 4), the cell contour is bordered manually for measuring intensity change.
Representative Results
The observation of spontaneous calcium oscillation in mNG-GECO10 expressed by iPSC-derived cardiomyocytes (iPSC-CMs) with or without drug treatments is demonstrated in Figure 1 and Animated Video 1. Kinetic traces obtained using mNG-GECO show that small molecular ion channel inhibitors, verapamil, dofetilide, and E4031, affected calcium transients (Figure 1B) as expected. The typical calcium transient shown in Figure 1C can be analyzed by Calcium peak analysis software to derive Peak signal (Amplitude), Calcium transient duration 50% (CTD50), Calcium transient duration 90% (CTD90), rise time, and decay time. Verapamil, an L-type calcium blocker17, completely inhibits the calcium influx in the cells as expected (Figure 1B). Dofetilide and E4031 are hERG channel inhibitors18,19,20, listed as experimental class III antiarrhythmic drugs. The decay time is prolonged with both Dofetilide (p < 0.05) and E4031 treatment (p < 0.001) when compared with the vehicle group. Prolongation of CTD50 (p < 0.01) and CTD90 (p < 0.001) with E4031 treatment was observed compared with vehicle only (Table 1), these results are consistent with reported effects on the QT interval, a clinically relevant measurement of repolarization determined from the surface electrocardiogram from a long time between the start of the Q-wave and the end of T-wave, in previous studies12,21.
A difficulty of CTD assessment in spontaneously beating cells is the variability beat rate imposes on the CTD. This is a particular problem for the iPSC-CM model, where each well of a 96 well plate can have its own beat rate even though all cells come from the same parent vial. It is possible to impose a beat rate by optical or electrical pacing. To evaluate the dose-response of E4031 under standardized paced conditions, ChR2 and K-GECO7 expressing iPSC-CMs were optical controlled using 1 Hz 470 nm pulses of light and observed using the red K-GECO signal (Figure 2A and Supplementary Figure 1). Progressive reductions of peak amplitude (p < 0.01) and increase in decay time (p < 0.05) of the calcium transients occur with increasing concentrations of E4031 (Figure 2B1). A dose-dependent effect of E4031 was most apparent for the reductions in peak amplitude (Figure 2B1,B2).
Although short-term studies lasting minutes are typically used for cardiotoxicity assessments, there is a blind spot for chronic toxicity assessments which parallel patient exposures to drugs over weeks, months, or years. It would be advantageous to study disease, or toxicity, effects over the intervals reflecting the treatment durations relevant to clinical practice. We utilized our all-optical control and detection system to study the iPSC-derived cardiomyocytes from a patient with left ventricular non-compaction (LVNC). iPSC-CMs were transduced with a K-GECO viral kit (step 2.2-2.6) post differentiation Day 25. The K-GECO signal was then tracked every 1-2 weeks in the same wells, with or without additional drug treatments. Both spontaneous calcium activity (Figure 3A,3B) and calcium dynamics under optical pacing (Figure 3C,D, step 4.11) were obtained using the high-Content Imaging System (step 4.1-4.12) and processed by calcium peak analysis software (step 6). As demonstrated in Figure 3A,C, clear calcium transients remain visible a month after viral transduction, with or without the additional light used for optical pacing. This work identified a drug that appears to increase beat rate, regularise contraction interval, and increase the transient calcium amplitude in the LVNC iPSC-CM model (Figure 3A,B). There are two important observations to make from this data. Firstly, from the therapeutic perspective, the drug effect appears durable at Day 63 and Day 70-time points. Secondly, and of relevance to a field which in general assays compound effects during brief windows (<1 min) at earlier time-points (typically <50 days); the disease phenotype modifying effects of the compound are only apparent after Day 60, suggesting it may be easy to discount drugs which have slow mechanisms of action in standard screening protocols.
The variability of beat frequency, and the subsequent impact on calcium transient duration, is not just a well-to-well problem at any given time point. It also changes as iPSC-CM are maintained in culture, which varies within a well over time (Figure 3B). To impose consistency within sequential experiments, 1 Hz optical pacing can be applied with or without drug treatment (Figure 3C,D). Here although the Day 63 results show the encouraging trends in the calcium transient with a significantly higher amplitude, shorter CTD90, and shorter decay time; by the 70 day time-point – and indicative of the need for chronic assessments during the drug screening process for rare disease – early after depolarizations (EAD) are detected. The value of adding a pacing protocol to disease phenotyping, or small molecule evaluation, can be qualitatively assessed by comparison of the results presented in Figure 3B,D for beat rate, calcium transient amplitude, CTD90, and decay time.
An advantage of a GECI, compared to chemical dye, based methodology to visualize the calcium transient is the ability to restrict the probe to a specific compartment within a cell. This can be further expanded by multiplexing multiple color and/or affinity variants in single cells. Hence, several GECIs including ER-LAR-GECO9, mtGCEPIA22,23 (or Oria1-G-GECO), and NIR-GECO28,24 have been developed for expression singly, or in combination, in cell models for real-time calcium activity measurement arising in the endoplasmic reticulum / sarcoplasmic reticulum (ER/SR), mitochondria (or CRAC channel) and cytosol, respectively. They can be combined in the iPSC-CM model (Figure 4A,C) to study the interaction between different intracellular calcium stores. For example, 10 mM caffeine treatment can be used to empty the SR calcium store. Here a decrease of ER-LAR-GECO signal (indicative of loss of calcium from the SR) paralleled a decline of the NIR-GECO signal (indicative of an increase in cytosolic calcium concentration) as expected. How other intracellular compartments are affected by these fluctuations is not well studied. Still, the inclusion of a green mitochondrial-targeted mtGCEPIA3 shows that calcium uptake occurs in mitochondria in these conditions (Figure 4A,B).
Similarly, visualizing the interplay between different calcium currents can be seen by including a probe, Orai1-G-GECO25, to reveal the calcium release-activated channel (CRAC) Ca2+ current (Figure 4C,D). In the iPSC-CM model, this signal increases with the spontaneous cytosolic calcium transient (Figure 4D1,D2). In keeping with this, treatment with caffeine evokes a large calcium transient in both the cytosol and from the Orai1 channel.
It is acknowledged that most calcium transient imaging in cardiomyocyte models has been determined using calcium dyes26. A genetically encoded calcium indicator was compared to a chemical dye in the iPSC-derived cardiomyocyte model to enable a side-by-side comparison of different calcium imaging approaches. K-GECO expressing iPSC-CMs were imaged alongside Fluo-4 loaded cells. K-GECO expressing cardiomyocytes showed consistent beating behavior over time (Figure 5A,C). However, Fluo-4 loading impacted both the beat frequency and CTD in this model (Figure 5B,C), suggesting Fluo-4 itself might influence the results of such experiments. This will be especially true after long-term exposure to the imaging probe, which can occur in the multiwall imaging format if well-by-well imaging occurs with a minute dwell time per well.
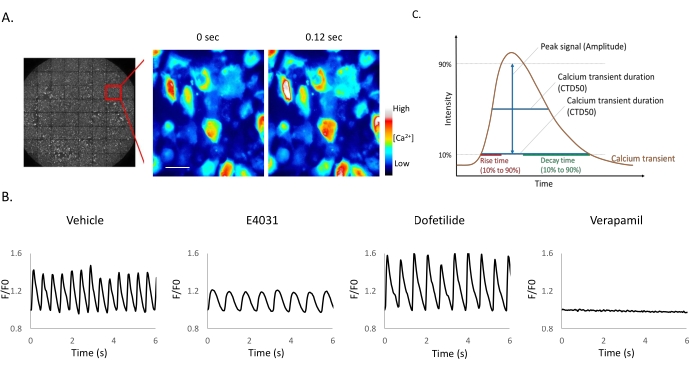
Figure 1: Small molecule ion channel inhibitors applied to iPSC-derived cardiomyocytes. (A-B) Time-lapse images of iPSC-CM expressing mNG-GECO taken at 25 Hz. Scale bar = 50 µm. (B) Representative Ca2+ oscillations after compound addition. Fluorescent signals obtained from mNG-GECO expressing cells are presented as a fluorescence ratio F/F0, where F0 is defined as basal intensity and F the intensity detected at each time point. (C) Calcium peak analysis software can extract parameters including half-maximal width (CTD50), 90% transient duration (CTD 90), rise time, and decay time. Please click here to view a larger version of this figure.
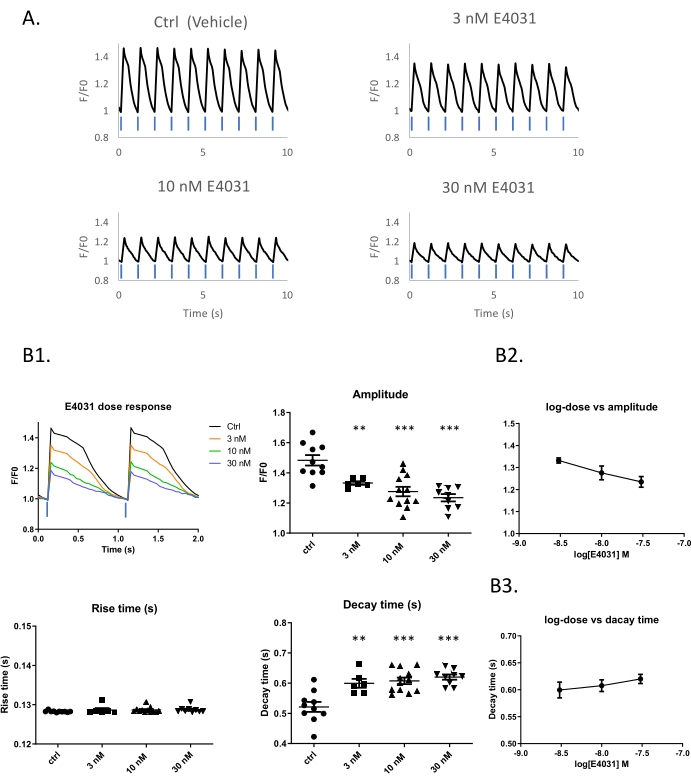
Figure 2: Dose-response example using the hERG inhibitor E4031 within an optical pacing system. (A) Traces of optical stimulation with 10% blue light in different concentrations of E4031. Time-lapse images of iPSC-derived cardiomyocytes expressing K-GECO taken at 25 Hz. (B1) Representative traces of calcium transients obtained with different compound doses. Peak analysis and dose-response of E4031 in amplitude (B2) and Decay time (B3). Blue bars indicate 1 Hz 470 nm pulse light stimulation. Please click here to view a larger version of this figure.
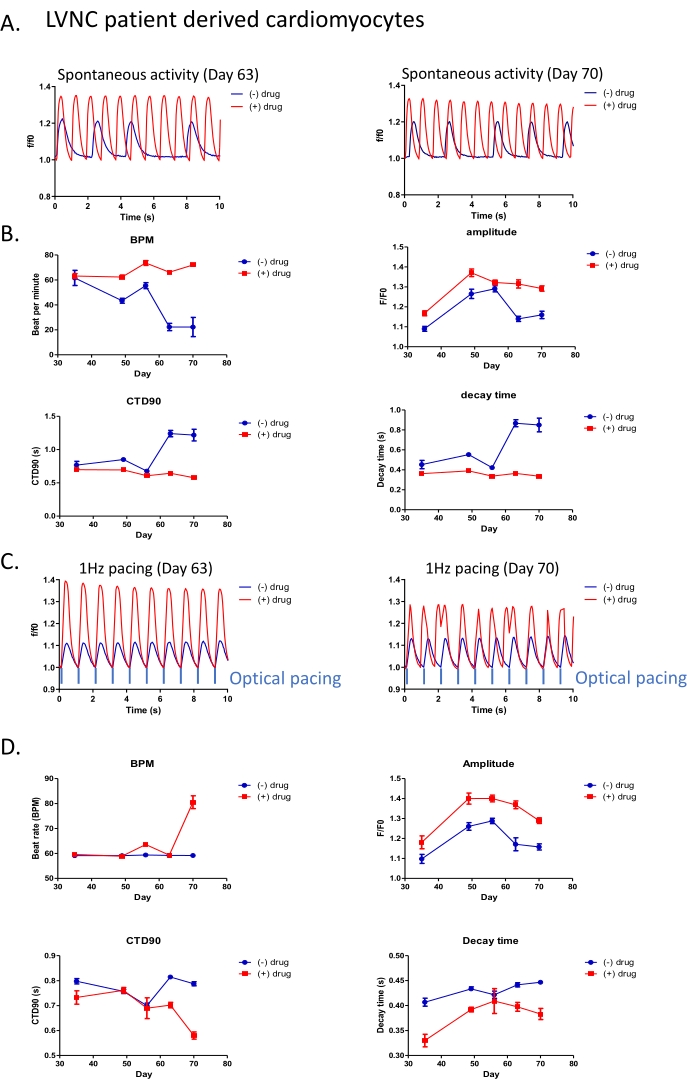
Figure 3: All-optical platform applied for long-term drug phenotypic screening in LVNC patient-derived cardiomyocytes. (A) Representative trace of spontaneous beating activity in patient-derived cardiomyocytes (post differentiation Day 70) with (red) or without (blue) 5 µM drug treatment. (B) Peak analysis of spontaneous beating activity with or without 5 µM drug treatment. (C) Intensity traces of drug response in patient-derived iPSC-CM under 1 Hz optical stimulation. (D) Peak analysis of patient-derived cardiomyocytes with 1 Hz optical stimulation. Please click here to view a larger version of this figure.
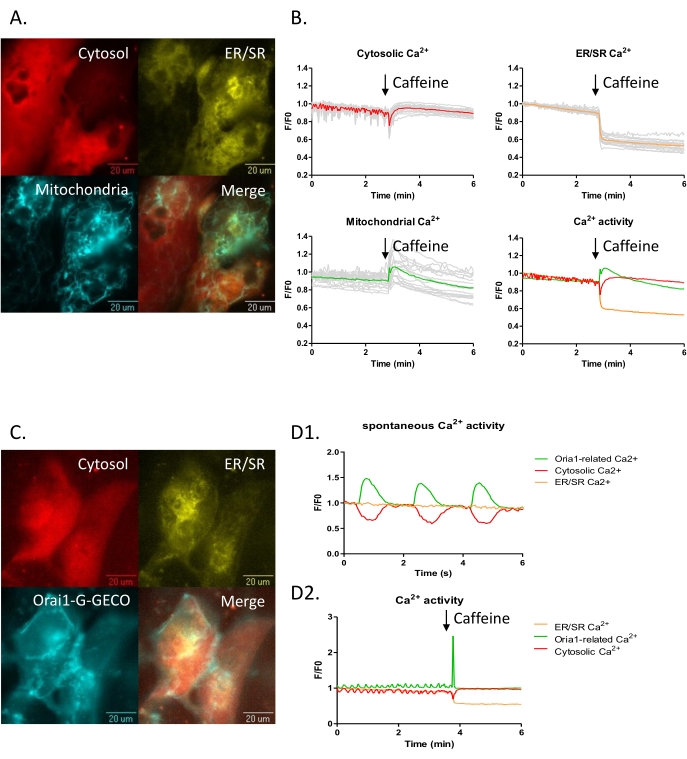
Figure 4: Three channels subcellular Ca2+ imaging in the iPSC-CM model. (A-B) iPSC-CMs were transduced with subcellular calcium probes for the cytoplasm, NIR-GECO2 (red), the endoplasmic reticulum ER-LAR-GECO (yellow), and mtGCEPIA3 (cyan), a mitochondrial Ca2+ indicator. (B) Three-channel time-lapse calcium imaging before and after 10 mM caffeine treatment. (C-D) iPSC-CMs transduced with cytoplasmic, NIR-GECO2 (red), endoplasmic reticulum ER-LAR-GECO (yellow), and CRAC channel indicators Orai1-G-GECO (cyan) were visualized. (D) Three-channel time-lapse calcium imaging was captured. (D1) Spontaneous beat-to-beat activity was observed with NIR-GECO2 and Orai1-G-GECO. (D2) Calcium efflux from the ER (decrease of ER-LAR-GECO signal) accompanied an increase in the cytosolic calcium signal upon caffeine treatment. Please click here to view a larger version of this figure.
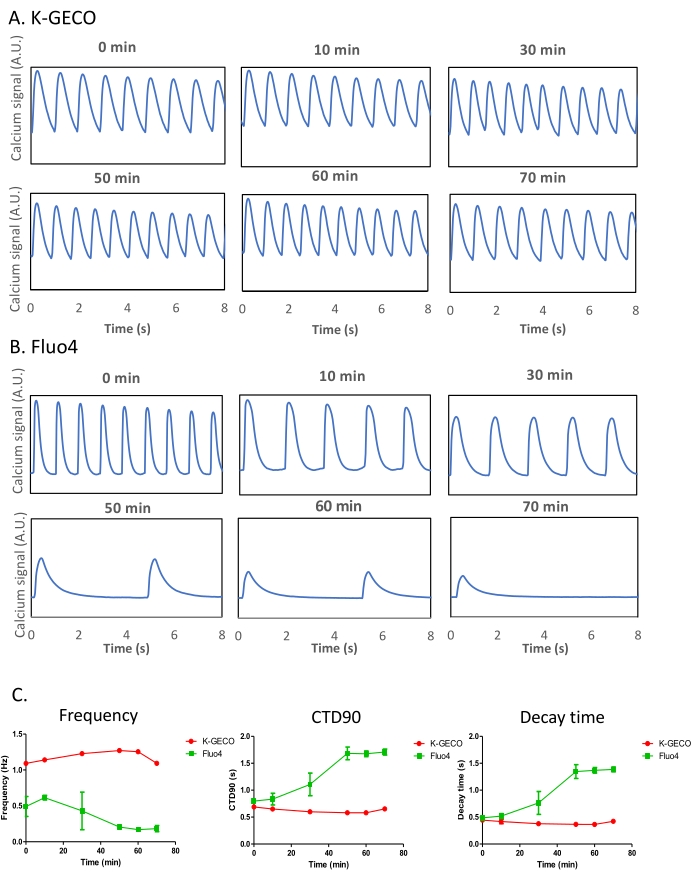
Figure 5: Comparison of the genetically encoded calcium indicator (K-GECO) to the chemical calcium-sensitive dye (Fluo-4) in iPSC-CMs. (A-B) Representative calcium traces of K-GECO (A) and Fluo-4 (B) are presented over time. (C) Calcium transient analysis of K-GECO transduced, or Fluo-4 loaded iPSC-CM. Please click here to view a larger version of this figure.
| BPM | CTD50 (s) | CTD90 (s) | Amplitude (F/F0) | Rise time | Decay time (s) | |
| Vehicle (0.1% DMSO) | 112.3657+/-10.95 | 0.25+/-0.04 | 0.41+/-0.01 | 1.99+/-0.02 | 0.07+/-0.01 | 0.28+/-0.01 |
| Verapamil (1 uM) | 0 | – | – | – | – | – |
| Dofetilide (5 nM) | 103.42+/-9.87** | 0.23+/-0.03 | 0.46+/-0.03 | 1.91+/-0.03 | 0.07+/-0.01 | 0.35+/-0.02* |
| E4031 (30 nM) | 75.73+/-12.08*** | 0.37+/-0.04** | 0.58+/-0.08*** | 1.55+/-0.02** | 0.11+/-0.04*** | 0.35+/-0.07** |
Table 1: Effects of compound addition to transient calcium parameters in the iPSC-CM model. Significance values are indicated by *(p ≤ 0.05), **(p < 0.01) and ***(p < 0.001).
Animated Video 1: Spontaneous calcium activity was observed by mNG-GECO in iPSC-derived cardiomyocytes with vehicle only. A Pseudo-coloured movie of a grey-scale image is provided. Please click here to download this Video.
Supplementary Figure 1: Schematic representation of the adenoviral-vector. This includes channel rhodopsin ChR2 as an actuator, with a red fluorescent calcium indicator K-GECO as a calcium reporter for an all-optical assay. The light-sensitive ion channel allows excitable cells to be depolarized by light in the blue-green range of the visible spectrum. 470 nm light was used in these experiments. Calcium transients in excitable cells can simultaneously be imaged by K-GECO, which requires green excitation light, producing red emissions. Please click here to download this File.
Discussion
To successfully perform high-throughput screening, signals must be evenly distributed across the culture vessel, guaranteeing that randomly selected regions of interest (ROIs) can be applied automatically during imaging acquisition. Although chemical calcium indicators easily meet this requirement, genetically encoded probes have historically produced patches, rather than sheets, of signal limiting their use. The use of a viral transduction system delivers high27,28 and equivalent expression levels of multiple indicators among the target cells compared to conventional transfection methods. Different cell types may require different promoters, and the choice of gene delivery system may need to change accordingly29,30,31. In heterogeneous cell models, transduction and expression efficiency may be different in specific cell types. On the one hand, this may limit the application to particular cells, but the phenomenon can be exploited to distinguish particular cells in a co-culture system32.
The main impact of this approach is the liberation from the one-to-one relationship between the experimenter and the sample by automation. In a standard multi-well experiment, for collecting data at four positions within a single well, we estimate it takes about 15 h at the microscope to gather 30 s videos from a 96 well plate. In the automated system, this takes ~4 h. Furthermore, analysis of the data manually takes far longer than data acquisition. The analysis system used here is about 200 times faster than the manual equivalent. The net result is an improved workflow and high cost and time savings.
In contrast to primary cardiomyocytes loaded with calcium dyes that survive in the order of hours, GECIs expressed in iPSC-CM's via viral delivery systems can yield a signal in an experimental model that lives for a few months. These models can be maintained in a multi-well format and tracked simultaneously for serial analysis in high-content imaging systems developed for small molecule screening. This produces a simple human experimental platform to test drug effects over durations closer to those experienced by patients (weeks or months) rather than the minutes typically used in toxicology assays.
The system can be expanded to deliver multi-parameter measurements by including GECIs with distinct emission properties from green to near infra-red. Such experiments require proper excitation light and filter sets in the experimental design to keep signals separate. Broad application in a drug screening context requires robust and efficient analysis platforms in addition to drug dispensing. Although the model described here focuses on the use of multiple dynamic calcium indicators, this could be modified to structural markers of cell form or function. These are typically easier to visualize as they show little beat-to-beat variability compared to the hundred-fold variation in intracellular calcium concentration. This approach may be particularly valuable for rare-disease where the iPSC intermediate derived from patients may contain all the genetic drivers and modifiers of a particular condition which can be preserved in a scaleable cell model, facilitating drug discovery in various diseases based on cellular phenotypes that can be visualized independently of the underpinning biological mechanism which may be challenging to elucidate.
Açıklamalar
The authors have nothing to disclose.
Acknowledgements
We thank Prof. Robert Campbell at the University of Tokyo for sharing the material and valuable discussion; Dr. Chia-Lin Ho in Molecular Device for technical support.
Materials
| 96-well plate | Perkin Elmer | 6055302 | |
| auto microfluidic system | Molecular Device | A device has a single channel pipetter that can be used to add compound automatically | |
| Caffeine | Sigma-Aldrich | C0750 | |
| Cardiosight-S l iPSC derived cardiomyocytes | NEXEL | C-002 | |
| Dimethyl sulfoxide | Millipore Sigma | 1096780100 | |
| Dofetilide | Sigma-Aldrich | PZ0016 | |
| E4031 | Tocris | 1808 | |
| ER-LAR-GECO viral kit | LumiSTAR | AA001a | Red-shifted GECI packed into adeno-viral vector |
| Fibronectin | Sigma-Aldrich | F1141 | |
| Fluo-4 AM | Invitrogen | F14201 | Chmical calcium sensitive dye |
| Gelatin | Sigma-Aldrich | G1890 | |
| ImageXpress Micro Confocal High-Content Imaging System | Molecular Device | ||
| iPSC-CMs Maintenance Medium | NEXEL | CMS-002 | iPSC-CMs Medium (Cardiosight-S medium) + Cardiosight-S Supplement |
| iPSC-CMs Medium (Cardiosight-S medium) | NEXEL | CMS-002 | |
| K-GECO viral kit | LumiSTAR | AA005a | Red-shifted GECI packed into adeno-viral vector |
| LumiCAL software | LumiSTAR | LUCS01a | Software for analysis of calcium peak in cardiomyocytes |
| mNG-GECO viral kit | LumiSTAR | AL008a | Brighter green GECI packed into lenti-viral vector |
| mt-GCEPIA3 viral kit | LumiSTAR | AL011a | GECI targgeting on mitochondria packed into lenti-viral vector |
| NIR-GECO viral kit | LumiSTAR | AV004a | Near infrared GECI packed into viral vector |
| Orai1-GGECO viral kit | LumiSTAR | AL010a | GECI targgeting on Orai1 packed into lenti-viral vector |
| Tyrode's salts | Sigma-Aldrich | T2145 | |
| Verapamil hydrochloride | Sigma-Aldrich | V4629 | |
| Y-27632 dihydrochloride | Tocris | 1254 |
Referanslar
- O’Connor, M. D. The 3R principle: Advancing clinical application of human pluripotent stem cells. Stem Cell Research & Therapy. 4 (2), 21 (2013).
- Ebert, A. D., Liang, P., Wu, J. C. Induced pluripotent stem cells as a disease modeling and drug screening platform. Journal of Cardiovascular Pharmacology. 60 (4), 408-416 (2012).
- Ovics, P., et al. Drug development and the use of induced pluripotent stem cell-derived cardiomyocytes for disease modeling and drug toxicity screening. International Journal of Molecular Sciences. 21 (19), 7320 (2020).
- Paredes, R. M., Etzler, J. C., Watts, L. T., Zheng, W., Lechleiter, J. D. Chemical calcium indicators. Methods. 46 (3), 143-151 (2008).
- Takahashi, A., Camacho, P., Lechleiter, J. D., Herman, B. Measurement of intracellular calcium. Physiological Reviews. 79 (4), 1089-1125 (1999).
- Smith, N. A., et al. Fluorescent Ca(2+) indicators directly inhibit the Na,K-ATPase and disrupt cellular functions. Science Signaling. 11 (515), (2018).
- Shen, Y., et al. A genetically encoded Ca2+ indicator based on circularly permutated sea anemone red fluorescent protein eqFP578. BMC Biology. 16 (1), 9 (2018).
- Qian, Y., et al. A genetically encoded near-infrared fluorescent calcium ion indicator. Nature Methods. 16 (2), 171-174 (2019).
- Wu, J., et al. Red fluorescent genetically encoded Ca2+ indicators for use in mitochondria and endoplasmic reticulum. Biochemical Journal. 464 (1), 13-22 (2014).
- Zarowny, L., et al. and high-performance genetically encoded Ca2+ indicator based on mneongreen fluorescent protein. ACS Sensors. 5 (7), 1959-1968 (2020).
- Potekhina, E. S., et al. Drug screening with genetically encoded fluorescent sensors: today and tomorrow. International Journal of Molecular Sciences. 22 (1), 148 (2021).
- Chang, Y. -. F., et al. Non-invasive phenotyping and drug testing in single cardiomyocytes or beta-cells by calcium imaging and optogenetics. PLoS One. 12 (4), 0174181 (2017).
- Chang, Y. -. F., Arai, Y., Nagai, T. Optogenetic activation during detector "dead time" enables compatible real-time fluorescence imaging. Neuroscience Research. 73 (4), 341-347 (2012).
- Chen, G., et al. Chemically defined conditions for human iPSC derivation and culture. Nature Methods. 8 (5), 424-429 (2011).
- Strober, W. Trypan blue exclusion test of cell viability. Current Protocols in Immunology. 111, 1-3 (2015).
- Jang, Y., et al. Modulating cardiomyocyte and fibroblast interaction using layer-by-layer deposition facilitates synchronisation of cardiac macro tissues. Soft Matter. 16 (2), 428-434 (2020).
- Leonard, R. G., Talbert, R. L. Calcium-channel blocking agents. Clinical Pharmacology. 1 (1), 17-33 (1982).
- Sanguinetti, M. C., Jurkiewicz, N. K. Two components of cardiac delayed rectifier K+ current. Differential sensitivity to block by class III antiarrhythmic agents. The Journal of General Physiology. 96 (1), 195-215 (1990).
- Lees-Miller, J. P., Duan, Y., Teng, G. Q., Duff, H. J. Molecular determinant of high-affinity dofetilide binding to HERG1 expressed in Xenopus oocytes: Involvement of S6 sites. Molecular Pharmacology. 57 (2), 367-374 (2000).
- Kamiya, K., Niwa, R., Mitcheson, J. S., Sanguinetti, M. C. Molecular determinants of HERG channel block. Molecular Pharmacology. 69 (5), 1709-1716 (2006).
- Fukushima, H., et al. Specific induction and long-term maintenance of high purity ventricular cardiomyocytes from human induced pluripotent stem cells. PLoS One. 15 (11), 0241287 (2020).
- Suzuki, J., Kanemaru, K., Iino, M. Genetically encoded fluorescent indicators for organellar calcium imaging. Biophysical Journal. 111 (6), 1119-1131 (2016).
- Suzuki, J., et al. Imaging intraorganellar Ca2+ at subcellular resolution using CEPIA. Nature Communications. 5, 4153 (2014).
- Qian, Y., et al. Improved genetically encoded near-infrared fluorescent calcium ion indicators for in vivo imaging. PLoS Biology. 18 (11), 3000965 (2020).
- Dynes, J. L., Amcheslavsky, A., Cahalan, M. D. Genetically targeted single-channel optical recording reveals multiple Orai1 gating states and oscillations in calcium influx. Proceedings of the National Academy of Sciences of the United States of America. 113 (2), 440-445 (2016).
- Broyles, C. N., Robinson, P., Daniels, M. J. Fluorescent, bioluminescent, and optogenetic approaches to study excitable physiology in the single cardiomyocyte. Cells. 7 (6), 51 (2018).
- Cao, F., et al. Comparison of gene-transfer efficiency in human embryonic stem cells. Molecular Imaging and Biology. 12 (1), 15-24 (2010).
- Ma, Y., Ramezani, A., Lewis, R., Hawley, R. G., Thomson, J. A. High-level sustained transgene expression in human embryonic stem cells using lentiviral vectors. Stem Cells. 21 (1), 111-117 (2003).
- Haery, L., et al. Adeno-associated virus technologies and methods for targeted neuronal manipulation. Frontiers in Neuroanatomy. 13, 93 (2019).
- Nieuwenhuis, B., et al. Optimization of adeno-associated viral vector-mediated transduction of the corticospinal tract: Comparison of four promoters. Gene Therapy. 28 (1), 56-74 (2021).
- Smith, R. L., et al. Characterization of promoter function and cell-type-specific expression from viral vectors in the nervous system. Journal of Virology. 74 (23), 11254-11261 (2000).
- Fang, Y., et al. Safety and efficacy of an immune cell-specific chimeric promoter in regulating anti-PD-1 antibody expression in. CAR T cells. Molecular Therapy – Methods & Clinical Development. 19, 14-23 (2020).

