Universal and Efficient Electroporation Protocol for Genetic Engineering of Gastrointestinal Organoids
Özet
This protocol describes an efficient electroporation method for the transfection of four different gastrointestinal organoid entities with larger plasmids (to the extent of 10 kB). It can be performed within one day and does not need extensive preparation or special, cost-intensive electroporation buffers.
Abstract
Electroporation is a common method for transfection with different kinds of molecules by electrical permeabilization of the plasma membrane. With the increasing use of organoids as a culturing method for primary patient material in the last years, efficient transfer methods of components for genetic engineering in this 3D culture system are in need. Especially for organoids, the efficiency of genetic manipulations depends on a successful transfection. Thus, this protocol was developed to facilitate the electroporation of organoids and to prove its universal functionality in different entities. Human colorectal, pancreatic, hepatic and gastric cancer organoids were successfully electroporated with small and large plasmids in comparison. Based on GFP encoding vectors, the transfection efficiency was determined by FACS. No extensive preparation of the cells or special, cost-intensive electroporation buffers are necessary, and the protocol can be performed within one day.
Introduction
In recent years, a novel 3D cell culture system, termed organoids, was developed for various normal and cancerous tissues. Organoids are functionally and morphologically very close to their tissue of origin. They can be generated from different species, are easily expandable, genomically stable and genetically modifiable, which makes them an ideal model system for genetic investigations1,2,3. Genetic engineering techniques like the CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats)/Cas9 system enable diverse manipulations. The selection of clones can be realized by defined media conditions, for example, by WNT ligand withdrawal for APC (Adenomatosis Polyposis Coli) knockout clones4,5. Alternatively, selection markers have to be introduced by homologous directed repair of a targeting vector6,7. Due to the fact that often more than one plasmid needs to be introduced, an efficient transfection becomes a crucial parameter. Additionally, to reduce unspecific off-target effects, a transient expression of the Cas9 endonuclease is desirable8.
Electroporation is a comparatively simple method to transfect cells with DNA, RNA, proteins or other macromolecules. By means of electric pulses, the cell membrane becomes more permeable and causes an increased uptake9. In a previously published electroporation protocol of colon organoids a 30 % efficiency with a piggy-bac GFP (green fluorescent protein) expressing vector (7.4 kB) was reached in a four days procedure10. The following protocol was developed to facilitate an efficient transfection of cancerous or healthy organoids with large plasmids encoding for single guide RNA (sgRNA) and the Cas9 endonuclease sequence (e.g. px458 as vector with 9.3 kb). The whole electroporation process can be performed within one day, without special electroporation buffers, and with at least comparable efficiencies between different gastrointestinal organoids, namely pancreatic ductal adenocarcinoma (PDAC), colorectal cancer (CRC), cholangiocarcinoma (CCC) and gastric cancer (GC) organoids.
Protocol
Ethics approval was obtained from the ethical committee of the TU Dresden (#EK451122014).
1. Organoid Culture and Preparations Before Electroporation
- Establish organoids by tissue digestion as described previously and expand them with their corresponding entity specific culture medium in a basement matrix (overview see Table 1 and Table of Materials)11,12,13,14,15,16,17.
NOTE: For human tissue samples informed consent and approval of the study by an ethical committee is necessary. - Prewarm 48-well plates at 37 °C for post-electroporation seeding.
- Prepare basal medium w/o antibiotics as well as entity specific organoid culture medium w/o antibiotics (see Table 1) including 10 µM Y-27632 and 3 µM CHIR99021.
NOTE: The withdrawal of antibiotics is important to reduce toxic effects. Y-27632 and CHIR99021 ameliorate cell recovery10. - Preparation of the organoids (see Figure 1)
- Cultivate 5 wells of organoids in a 48-well plate per electroporation sample in culture medium.
NOTE: Proliferative organoids should be used (around 2-3 days after last splitting). - Prepare 230 µL of dissociation reagent (see Table of Materials) including 10 µM Y-27632 per well.
- Remove the culture medium from each well and dissociate the organoids mechanically in 230 µL of the prepared dissociation mixture. Pool 5 wells per electroporation sample into one 15 mL tube.
- Mix by vortexing and incubate for 5-15 min at 37 °C until clusters of 10-15 cells occur. Therefore, check the dissociation microscopically. Stop the digestion by adding basal medium w/o antibiotics up to 10 mL.
NOTE: This step is very critical! The electroporation efficiency will be reduced, when incubation is too short, but long digestion will reduce survivability. - Centrifuge at 450 x g for 5 min at room temperature, discard the supernatant and wash twice with 4 mL of electroporation buffer (see Table of Materials).
- Cultivate 5 wells of organoids in a 48-well plate per electroporation sample in culture medium.
2. Electroporation
NOTE: The following protocol is developed for electroporators capable of square waves and separated poring and transfer pulse sequences (see Figure 2). Optionally, impedance values as well as the voltages, currents and energies transferred into the sample can be measured as control for reproducible experiments.
- Resuspend the organoid pellet in 100 µL of electroporation buffer (see Table of Materials) containing 30 µg of plasmid DNA.
NOTE: The concentration of the used plasmid DNA should exceed 5 µg/µL for an optimal salt concentration during the electroporation process. Therefore, endofree plasmid maxi kits (see Table of Materials) for the preparation of vectors are recommended. A total amount up to 45 µg of DNA can be used without cytotoxic effects. - Dispense the complete DNA-organoid mixture into an electroporation cuvette with 2 mm gap width without producing air bubbles.
- Set the electroporation parameters according to Fujii et al.10 (see Table 2, Figure 2).
- Mix the cells slightly without foaming by tapping the cuvette with a finger. Place the cuvette into the cuvette chamber.
- Press the Ω button of the electroporator and make a note of the impedance value.
NOTE: An impedance between 30-40 Ω showed the best results. In general, it should be in the range between 30-55 Ω. If this is not the case, please control the following aspects: gap width of the cuvette used, cable connections of the electroporator, possible air bubbles, correct volume and salt concentration of the electroporation mixture. - Press the Start button to execute the electroporation program and control the values of currents, voltages and energies displayed.
NOTE: The values of measured voltages, currents and energies should correspond to the set electroporation parameters. For the comparison of repeated experiments, it can be helpful to note these data. - After electroporation, immediately add 500 µL of culture medium w/o antibiotics (with CHIR99021 and Y-27632; see step 1.3). Mix by pipetting up and down to dissociate the white foam.
NOTE: The white foam appears after the electroporation process and a significant number of cells is attached to it. So, dissociation of it is very important for not losing cells. - Transfer the sample completely from the cuvette into a new 15 mL tube using the pipette belonging to the electroporation cuvettes (see Table of Materials). Rinsing the cuvette again with basal medium is recommended to obtain remaining cells.
- For regeneration of the cells, incubate them for 40 min at room temperature.
3. Seeding of Cells
- Centrifuge the cells at 450 x g for 5 min at room temperature and discard the supernatant.
- Resuspend the pellet in 100 µL of basement matrix and seed 20 µL drops in a prewarmed 48-well plate (see step 1.2). Incubate for 10 min at 37 °C for polymerization and add 250 µL of culture medium, which is supplemented with Y-27632 and CHIR99021 until the next splitting of the grown organoids (around 5-7 days).
4. Determination of Transfection Efficiency
NOTE: In general, it is recommended to electroporate a vector carrying a fluorescence marker as additional transfection control. Dependent on the chosen marker and its chromophore maturation the fluorescence will be visible within around 24-48 h post transfection18.
- Check the fluorescence microscopically after 24-48 h in the transfection control (see Figure 4B).
- Fluorescent activated cell scanning (FACS)
- Harvest the cells analogously to step 1.4.2-1.4.4 and digest around 10-20 min until there are single cells. Add up to 10 mL of phosphate-buffered saline (PBS).
- Centrifuge at 450 x g for 5 min at room temperature and discard the supernatant.
- Optionally for discrimination of living cells: Resuspend the pellet in 1 mL of phosphate-buffered saline (PBS) and add a suitable antibody (see Table of Materials) or propidium iodide (PI). Mix very carefully only by tapping and incubate for 30 min at room temperature in the dark. Wash with 10 mL of PBS, centrifuge and discard the supernatant.
- Resuspend the cell pellet in 200 µL of PBS and optionally filter the suspension through a 100 µm cell strainer into a FACS tube.
- Analyze the cells by a FACS machine using an appropriate gating strategy (see Figure 3; Figure 4A) and determine the transfection efficiency.
Representative Results
Organoids of four different cancer entities (CRC, CCC, PDAC, GC) were electroporated at least 3 times using 30 µg of a small plasmid (pCMV-EGFP, 4.2 kb) or a large plasmid (px458, 9.3 kb). Both vectors carry a GFP cassette allowing the determination of transfection efficiency 48 h after electroporation by flow cytometry. To analyze only living cells, staining with a life-death antibody before scanning was performed. The gating strategy is shown in Figure 3.
In all four organoid entities the 4.2 kB sized plasmid was transfected with higher efficiency compared to the larger one (see Figure 4). The most efficient transfection of the small plasmid was reached in PDAC organoids with 92.1 ± 5.2 % GFP positive cells, whereas the large plasmid was transfected with an efficiency of 46.7 ± 3.7 % (mean ± standard deviation, n = 3). Compared to pancreatic cancer organoids, the larger plasmid was more efficiently transfected into CRC organoids with a mean efficiency of 53.4 ± 11.7 %, while the small plasmid was transfected with a mean efficiency of 84.3 ± 5.8 %. The most difficult entity to transfect were gastric cancer organoids: for both the large and the small plasmid, the lowest transfection efficiency was reached in this entity (32.3 ± 12.7 % and 74.1 ± 5.5 %, respectively). CCC organoids showed a mean transfection efficiency of 83.0 ± 13.1 % for the small plasmid and for the large plasmid 39.5 ± 10.4 % were obtained.
As proof of concept, human normal stomach organoids were electroporated with a px458_Conc2 plasmid encoding for Cas9, GFP and two sgRNAs targeting TP53. The Cas9-induced double strand breaks on exon 8 were repaired by non-homologous end-joining (NHEJ), resulting in frameshifts and consequently a knockout of the gene (see Supplementary Figure 1).
Table 1: Composition of basal media, digestion mixtures and cultivation media. Please click here to view this file (Right click to download).

Table 2: Electroporation settings according to Fujii et al.10.
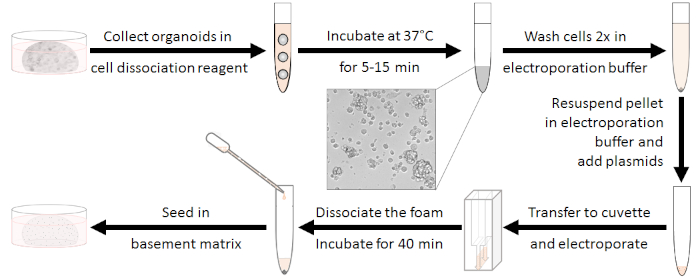
Figure 1: Electroporation preparation workflow. First, organoids ought to be dissociated to clusters of 10-15 cells and antibiotics should get washed out. After electroporation the white foam needs to be dissociated. Cells can be seeded after regenerating for 40 min at room temperature. Please click here to view a larger version of this figure.

Figure 2: Two-step electroporation. Two poring pulses with higher voltage und short duration (175 V and 157.5 V, each for 5 ms, pause for 50 ms, voltage decay 10%) lead to the formation of pores in cell membranes. The following transfer pulses deliver the DNA into the cells: five positive transfer pulses (with 20 V, 12 V, 7.2 V, 4.32 V and 2,592 V, each for 50 ms, pause for 50 ms, voltage decay 40%), followed by five polarity exchanged transfer pulses (with 20 V, 12 V, 7.2 V, 4.32 V and 2,592 V, each for 50 ms, pause for 50 ms, voltage decay 40%). Please click here to view a larger version of this figure.
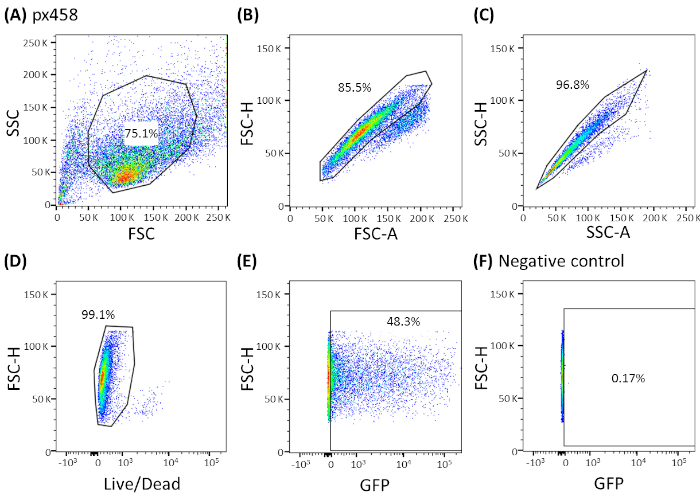
Figure 3: Representative gating strategy shown by CCC organoids. All electroporated organoids were analyzed by flow cytometry 48 h after electroporation. Cells electroporated without plasmid DNA were used as negative controls. The gates were set as following: (A) gating for cell shape, (B,C) gating for single cells (doublet discrimination), (D) gating for living cells (stained with an antibody for apoptotic cells) and (E,F) finally gating for eGFP expressing cells (FITC channel). FSC = forward scatter; SSC = side scatter. Please click here to view a larger version of this figure.
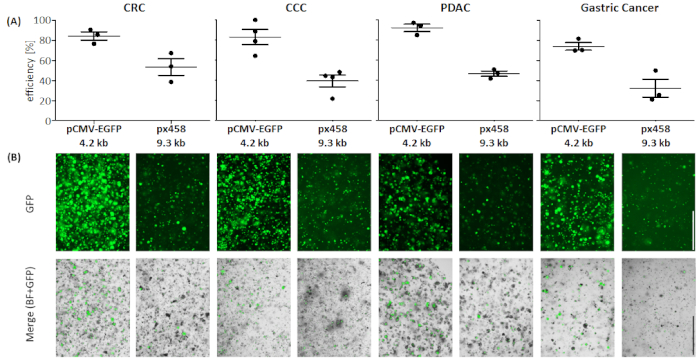
Figure 4: Electroporation efficiency of four organoid entities. (A) FACS analysis (n = 34, mean standard deviation and each single value are shown) and (B) visual comparison by fluorescence microscope. Scale bar = 1,000 µm. BF = bright field; CCC = cholangiocarcinoma; CRC = colorectal cancer; GC = gastric cancer; PDAC = pancreatic ductal adenocarcinoma. Please click here to view a larger version of this figure.

Supplementary Figure 1: Exemplary CRISPR/Cas9-based knockout of TP53 in normal human stomach organoids.The px458_Conc2 vector (see Table of Materials) was cloned by combining the 2 gRNA concatemer vector, a generous gift from Bon-Kyoung Koo19, with px45820, resulting in a plasmid encoding for 2 sgRNAs, Cas9 and GFP. Two sgRNAs targeting TP53 were introduced in px458_Conc2 vector by golden gate cloning (analogously to Andersson-Rolf et al.19). 10 µg of plasmid DNA were electroporated in human normal gastric organoids (A). Clones were selected by Nutlin3 administration (B) and the TP53 knockout was confirmed by TOPO TA cloning and sequencing of the alleles, here exemplary shown for one clone (C). The sgRNAs are underlined in the reference. Scale bar = 200 µm. BF = bright field. Please click here to view a larger version of this figure.
Discussion
This protocol gives detailed instructions for an efficient, quick and easy to perform electroporation of different organoid entities. Additional to the presented tumor organoids from PDAC, CRC, CCC and GC, it works successfully for organoids derived from healthy tissue as well. The protocol can be performed within one day. In published organoid transfection protocols the whole procedure lasted four days including two days of preparations with different types of cultivation media10,21. In our protocol no special pretreatment is required. By washing with electroporation buffer before electroporation the antibiotic components of the media were washed out and an adjustment of saline concentrations for optimal impedance values was achieved. Nevertheless, some critical aspects should be considered for a successful electroporation:
Cells
In the electroporation protocol by Fujii et al.10 it is recommended to dissociate organoids to single cells and to filter them through a 20 µm cell-strainer. In our hands digestion to single cells strongly decreases the survivability of cells. As suggested in Merenda et al.21, we also dissociated organoids to clusters of 10-15 cells and could not determine a decreased efficiency compared to single cell dissociation. After electroporation, it is a very important step to dissociate the white foam, so that no attached cells are getting lost.
For 2D cell culture, it has been shown that a regeneration time after electroporation of more than 10 min up to 40 min increases survivability and transfection efficiency especially of large plasmids22. In test experiments, the same could be documented for organoids, leading to an incubation step of 40 min after electroporation in this protocol. In order to increase recovery from the electroporation, we cultured them with Rho-associated protein kinase (ROCK) inhibitor Y-27632 for five to seven days23. Similarly, the additional supplementation of glycogen synthase kinase 3 (GSK3) inhibitor CHIR99021 is meant to help single cells to recover10.
Settings
One of the advantages of the used electroporator is that the impedance can be measured before electroporation for optimal conditions. According to the manufacturer, the impedance values should be 30-55 Ω. In our hands, impedance values of 30-40 Ω have shown optimal efficiencies. In a preliminary experiment, different voltages and pulse length values of the poring pulse were varied to find the optimal proportion of efficiency to survivability. In summary, we could confirm the described values of Fujii et al.10 in the different entities described here.
DNA
The effect of different DNA amounts was tested in preliminary experiments up to 45 µg of DNA per sample. No cytotoxic effects could be detected. Transfection efficiency was increased in a dose dependent way with saturation > 30 µg. So, we used 30 µg per sample in the final protocol, but of course it can be increased (e.g., for the electroporation of more plasmids in parallel). Additionally, the purity and concentration of the DNA seems to be very important. A concentration exceeding 5 µg/µL has shown optimal transfection efficiencies.
As expected, the 9.3 kB plasmid could be transfected with a lower efficiency than the smaller 4.2 kB plasmid (see Figure 4). The use of even larger plasmids than 10 kB is anticipated to further decrease the efficiency. For future applications, it might be interesting to test minicircle DNA as a vector, since these gene carriers lack the bacterial backbone of a plasmid which makes them smaller24. This should result in an enhanced transfection efficiency. Furthermore, for CRISPR-based manipulations of organoids a direct electroporation of sgRNAs bound to Cas9 as a ribonucleoprotein (RNP) complex could be an alternative or addition25.
Açıklamalar
The authors have nothing to disclose.
Acknowledgements
We thank Juliane Fohgrub, Ann-Christin Meinecke and Max Heiduk for excellent technical assistance. Funding was provided by Deutsche Krebshilfe (No 111350 and 70112925), Sander Stiftung (No 2014.104.1), Hector Stiftung (No M65.2) and the European Union (ERC No 639050).
Materials
| For establishment and culture medium | |||
| [Leu15] Gastrin | Sigma-Aldrich | G9145 | |
| A83-01 | Tocris Bioscience | 2939 | |
| Advanced DMEM/F-12 | Invitrogen | 12634010 | |
| B27 | Invitrogen | 17504044 | |
| B27 Supplement, minus vitamin A | Thermo Fisher Scientific | 12587010 | |
| CHIR99021 | Stemgent | 04-0004 | |
| Collagenase II | Life Technologies | 17101-015 | |
| Collagenase XI | Sigma-Aldrich | C9407-100MG | |
| Collagenase D | Roche | 11088866001 | |
| Dispase II | Roche | 4942078001 | |
| Dnase I | Sigma-Aldrich | D5319 | |
| D-sorbitol | Roth | 6213.1 | |
| Dithiothreitol | Thermo Scientific | 1859330 | |
| EDTA | Roth | 8040 | |
| Forskolin | Tocris Bioscience | 1099 | |
| Glutamax | Life Technologies | 35050061 | |
| Hepes | Thermo Fisher Scientific | 15630106 | |
| hFGF-10 | Preprotech | 100-26 | |
| KCl | Sigma-Aldrich | P9541 | |
| KH2PO4 | Roth | 3904.2 | |
| Matrigel | Corning | 356231 | basement matrix |
| mEGF | Invitrogen | PMG8043 | |
| N2 | Invitrogen | 17502048 | |
| NaCl | Roth | 3957.1 | |
| Na2HPO4 | Roth | K300.2 | |
| N-Acetyl-L-Cystein | Sigma-Aldrich | A9165 | |
| Nicotinamid | Sigma-Aldrich | N0636 | |
| Noggin | n.a. | n.a. | Conditioned medium produced from HEK293 cells (Hek293-mNoggin-Fc) |
| PBS | Gibco | 14190169 | |
| Penicillin Streptomycin | Life Technologies | 15140122 | |
| Primocin | InvivoGen | ant-pm-1 | |
| Prostaglandin E2 | Tocris Bioscience | 2296 | |
| Recombinant Human HGF | Preprotech | 100-39H | |
| Rspondin | n.a. | n.a. | Conditioned medium produced from HEK293 cells (HA-Rspo1-Fc-293T) |
| SB202190 | Sigma-Aldrich | S7067 | |
| Sucrose | VWR | 27,480,294 | |
| TrypLE Express | Gibco | 12604021 | Dissociation reagent |
| Wnt3A | n.a. | n.a. | Conditioned medium produced from L-Wnt3a cells (from Sylvia Boj) |
| Y-27632 | Sigma-Aldrich | Y0503 | |
| Consumables | |||
| Cell Strainer 100 µm | Falcon | 352360 | |
| 48-well plate | Corning | 3548 | |
| Nepa Electroporation Cuvettes 2mm gap w/pipettes | Nepa Gene Co., Ltd. | EC-002S | |
| Tubes 15 ml | Greiner | 188271 | |
| Tubes 15 ml low binding | Eppendorf | 30122208 | Tubes for FACS preparing |
| Equipment | |||
| Electroporator Nepa21 | Nepa Gene Co., Ltd. | n.a. | |
| EVOS FL Auto | Invitrogen | AMAFD1000 | Fluorescence microscope |
| LSRFortessa | BD Bioscience | 647800E6 | FACS |
| Reagents and plasmids | |||
| Live/dead fixable blue dead cell stain kit | Invitrogen | L34962 | Includes antibody for live/dead staining for FACS analysis |
| EndoFree Plasmid Maxi Kit | Qiagen | 12362 | |
| Nutlin-3 | Selleckchem | S1061/07 | |
| Opti-MEM | Gibco | 31985047 | Electroporation buffer |
| 2 gRNA concatemer vector | AddGene | 84879 | |
| pCMV-EGFP | Nepa Gene Co., Ltd. | n.a. | |
| px458 plasmid | AddGene | 48138 | coding for sgRNA and Cas9 |
| px458_Conc2 plasmid | AddGene | 134449 | px458 plasmid containing 2x U6 promotors for two different sgRNAs |
| sgRNA_hTP53_1a | Eurofins Genomics | n.a. |  |
| sgRNA_hTP53_1b | Eurofins Genomics | n.a. |  |
| sgRNA_hTP53_2a | Eurofins Genomics | n.a. |  |
| sgRNA_hTP53_2b | Eurofins Genomics | n.a. |  |
Referanslar
- Huch, M., et al. Long-term culture of genome-stable bipotent stem cells from adult human liver. Cell. 160 (1-2), 299-312 (2015).
- Werner, K., Weitz, J., Stange, D. E. Organoids as Model Systems for Gastrointestinal Diseases: Tissue Engineering Meets Genetic Engineering. Current Pathobiology Reports. 4 (1), 1-9 (2016).
- Olayanju, A., Jones, L., Greco, K., Goldring, C. E., Ansari, T. Application of porcine gastrointestinal organoid units as a potential in vitro tool for drug discovery and development. Journal of applied toxicology. 39 (1), 4-15 (2019).
- Drost, J., et al. Sequential cancer mutations in cultured human intestinal stem cells. Nature. 521 (7550), 43-47 (2015).
- Matano, M., et al. Modeling colorectal cancer using CRISPR-Cas9-mediated engineering of human intestinal organoids. Nature Medicine. 21 (3), 256-262 (2015).
- Schwank, G., et al. Functional repair of CFTR by CRISPR/Cas9 in intestinal stem cell organoids of cystic fibrosis patients. Cell Stem Cell. 13 (6), 653-658 (2013).
- Drost, J., et al. Use of CRISPR-modified human stem cell organoids to study the origin of mutational signatures in cancer. Science. 358 (6360), 234-238 (2017).
- Zhang, X. H., Tee, L. Y., Wang, X. G., Huang, Q. S., Yang, S. H. Off-target Effects in CRISPR/Cas9-mediated Genome Engineering. Molecular Therapy Nucleic Acids. 4, e264 (2015).
- Tsong, T. Y. Electroporation of cell membranes. Biophysical journal. 60 (2), 297-306 (1991).
- Fujii, M., Matano, M., Nanki, K., Sato, T. Efficient genetic engineering of human intestinal organoids using electroporation. Nature Protocols. 10 (10), 1474-1485 (2015).
- Sato, T., et al. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett’s epithelium. Gastroenterology. 141 (5), 1762-1772 (2011).
- Bartfeld, S., et al. In vitro expansion of human gastric epithelial stem cells and their responses to bacterial infection. Gastroenterology. 148 (1), 126-136 (2015).
- Boj, S. F., et al. Organoid models of human and mouse ductal pancreatic cancer. Cell. 160 (1-2), 324-338 (2015).
- Broutier, L., et al. Culture and establishment of self-renewing human and mouse adult liver and pancreas 3D organoids and their genetic manipulation. Nature Protocols. 11 (9), 1724-1743 (2016).
- Broutier, L., et al. Human primary liver cancer-derived organoid cultures for disease modeling and drug screening. Nature Medicine. 23 (12), 1424-1435 (2017).
- Hennig, A., et al. CFTR Expression Analysis for Subtyping of Human Pancreatic Cancer Organoids. Stem Cells Int. 2019, 1024614 (2019).
- Seidlitz, T., et al. Human gastric cancer modelling using organoids. Gut. 68 (2), 207-217 (2019).
- Stepanenko, O. V., Verkhusha, V. V., Kuznetsova, I. M., Uversky, V. N., Turoverov, K. K. Fluorescent proteins as biomarkers and biosensors: throwing color lights on molecular and cellular processes. Current protein & peptide science. 9 (4), 338-369 (2008).
- Andersson-Rolf, A., et al. Simultaneous paralogue knockout using a CRISPR-concatemer in mouse small intestinal organoids. Dev Biol. 420 (2), 271-277 (2016).
- Ran, F. A., et al. Genome engineering using the CRISPR-Cas9 system. Nature Protocols. 8, 2281 (2013).
- Merenda, A., et al. A Protocol for Multiple Gene Knockout in Mouse Small Intestinal Organoids Using a CRISPR-concatemer. Journal of Visualized Experiments. (125), (2017).
- Lesueur, L. L., Mir, L. M., Andre, F. M. Overcoming the Specific Toxicity of Large Plasmids Electrotransfer in Primary Cells In Vitro. Molecular Therapy Nucleic Acids. 5, e291 (2016).
- Watanabe, K., et al. A ROCK inhibitor permits survival of dissociated human embryonic stem cells. Nature Biotechnology. 25 (6), 681-686 (2007).
- Darquet, A. M., Cameron, B., Wils, P., Scherman, D., Crouzet, J. A new DNA vehicle for nonviral gene delivery: supercoiled minicircle. Gene Therapy. 4 (12), 1341-1349 (1997).
- Jacobi, A. M., et al. Simplified CRISPR tools for efficient genome editing and streamlined protocols for their delivery into mammalian cells and mouse zygotes. Methods. 121-122, 16-28 (2017).

