Stepwise Dosing Protocol for Increased Throughput in Label-Free Impedance-Based GPCR Assays
Özet
This protocol demonstrates real-time recording of full dose-response relationships for agonist-induced GPCR activation from a single cell layer grown on a single microelectrode using label-free impedance measurements. The new dosing scheme significantly increases throughput without loss in time resolution.
Abstract
Label-free impedance-based assays are increasingly used to non-invasively study ligand-induced GPCR activation in cell culture experiments. The approach provides real-time cell monitoring with a device-dependent time resolution down to several tens of milliseconds and it is highly automated. However, when sample numbers get high (e.g., dose-response studies for various different ligands), the cost for the disposable electrode arrays as well as the available time resolution for sequential well-by-well recordings may become limiting. Therefore, we here present a serial agonist addition protocol which has the potential to significantly increase the output of label-free GPCR assays. Using the serial agonist addition protocol, a GPCR agonist is added sequentially in increasing concentrations to a single cell layer while continuously monitoring the sample's impedance (agonist mode). With this serial approach, it is now possible to establish a full dose-response curve for a GPCR agonist from just one single cell layer. The serial agonist addition protocol is applicable to different GPCR coupling types, Gq Gi/0 or Gs and it is compatible with recombinant and endogenous expression levels of the receptor under study. Receptor blocking by GPCR antagonists is assessable as well (antagonist mode).
Introduction
This report presents a detailed description of a serial addition protocol developed for the quantification of ligand-induced G protein-coupled receptor (GPCR) activation in adherently grown cells by label-free impedance measurements. G protein-coupled receptors (GPCRs) are involved in a multitude of physiological functions and human diseases1. Because of this and their good accessibility at the cell surface, GPCRs are one of the most important drug targets. This assessment is reflected in an estimated number of ~700 approved drugs targeting GPCRs, equivalent to a ~35% share on all marketed drugs2.
The development of new drugs comprises two central processes: (i) the identification and functional characterization of biological target molecules, and (ii) the discovery of new lead substances and their development into administrable drugs. In both processes, efficient methods are required to quantitatively evaluate drug-target interactions and the subsequent biological downstream response. Different stages of the pre-clinical drug development process make use of different methods of analysis ranging from biomolecular interaction studies between drug and target, over functional studies on cells in culture, to experiments on excised organ material or whole animals. Both, physiological significance and biological complexity increase from the former to the latter3. Although it is the overall goal to minimize animal experiments, pharmacological studies using isolated organs from laboratory animals or even whole animals are regarded inevitable to comprehensively characterize new drug candidates. In terms of analytical readout, organ pharmacology studies provide a distal, integrative "holistic" functional response of highest physiological relevance. A drawback of such experiments is that they are not compatible with high throughput screening for technical as well as ethical reasons and have been largely substituted by studies based on in vitro cell culture4.
Methods to quantify GPCR activation in cell cultures include different label-based chemical assays, which specifically detect second messengers, phosphorylation state of downstream signaling proteins, transcriptional activation via certain transcription factors or ligand-induced intracellular receptor trafficking4,5. A drawback of such label-based assays is the necessity to label the cells with potentially harmful dyes or radioactive markers. This often requires running the assay as an endpoint-determination for an exposure time that has to be specified a priori. Using label-based endpoint assays suffers from this very limited and biased timing of the experiment and the risk that chemical labels and probes may interfere with the regular cell physiology – potentially unnoticed by the experimenter.
In recent years, label-free assays for monitoring GPCR activation have emerged, like impedance-based techniques or optical methods applying resonant waveguide gratings5,6. Labelling of the cells is experimentally not required with these approaches. As these physical readouts operate on low amplitude signals, such methods are considered non-invasive, they allow for continuous cell monitoring potentially in real time and the observation time is only limited by the cell culture not by the readout. Similar to readouts from whole organs, label-free approaches commonly report on holistic cell responses, far downstream of receptor activation when integration along the entire signaling network leads to time-dependent but rather unspecific changes in cell morphology or mass redistribution. Whereas impedance-based assays measure the dielectric signature of changes in cell shape7,8, measurements using resonant waveguide gratings are sensitive to changes in refractive index at the cell-substrate interface arising from dynamic mass redistribution (DMR)9. The integrative character makes label-free methods extremely sensitive to receptor-mediated events regardless of the type(s) of G protein (Gq, Gi/0, Gs, G12/13) or β-arrestin involved in the signaling cascade6 and well-suited for endogenous expression levels of the receptor.
In a standard label-free impedance-based assay the cells are adherently grown in multi-well plates with coplanar gold-film electrodes deposited on the bottom of each well10. These electrode arrays are connected to an impedance analyzer and the cell responses to an experimental stimulus are recorded from individual wells by time-resolved impedance readings. In a typical GPCR assay a ligand is added in individually different concentrations to each individual well. The ligand-induced changes in the impedance time course are then analyzed with respect to characteristic curve features such as maximum signal change, area under the curve, signal change within a given time interval or slope of the curve at a specified time point, in order to quantify the ligand's potency and efficacy11.
The cost of the electrode arrays may limit the application of this technique in high throughput screening (HTS) campaigns. Moreover, with increasing numbers of samples to be followed in parallel, the number of individual measurements increases and thereby reduces the available time resolution for each well gradually – even for state of the art multichannel recordings. Under such conditions fast and transient cell responses may escape the measurement. In addition, the conventional one well – one concentration approach imposes a significant time and cost factor on perfused organ-on-chip or body-on-chip developments with respect to their suitability in ligand-GPCR interaction analysis.
For this reason, we developed a stepwise dosing protocol that enables recording of full dose-response curves of ligand-induced GPCR activation in cultured cell monolayers by continuously monitoring the impedance of a single well while the agonist concentration is increased stepwise. The serial agonist addition protocol significantly increases throughput per well from one concentration to 10 or more concentrations, as shown on the current example of human U-373 MG cells, which endogenously express the histamine 1 receptor (H1R). Thus, the method has the potential to significantly improve throughput in label-free dose-response studies, while the time resolution is retained at the instrumental maximum.
Protocol
1. Cell seeding on electrode arrays
NOTE: Selection of electrode layout is a trade-off between sensitivity and number of cells under study. The smaller the electrode, the more sensitive is the measurement but the smaller is the number of cells under study. For cells showing strong impedance fluctuations over time under baseline conditions, bigger or interdigitated electrodes are preferable.
- Pre-warm all solutions needed for standard cell passaging and seeding in a 37 °C water bath. For assays with human U-373 MG cells it takes: phosphate buffered saline (PBS) without calcium and magnesium, 0.05% (w/v) trypsin, cell culture medium (Eagle's Minimum Essential Medium (EMEM) supplemented with 5% (v/v) fetal calf serum (FCS), 2 mM L-glutamine and 100 µg/mL penicillin/streptomycin).
- Rinse the cell layer of the stock culture grown on the bottom of conventional cell culture flasks or dishes with PBS twice.
- Remove the PBS, add the trypsin solution (1 mL for 25 cm2) and let the cells incubate at 37 °C for 5 min (applies to U-373 MG cells).
- Control for complete detachment of the cells from the bottom of the growth substrate by microscopy.
- Stop the trypsin reaction as soon as the cells are completely detached by adding 9 mL of cell culture medium per 1 mL of trypsin to the cell suspension. Carefully rinse remaining cells from the bottom of the cell culture substrate by pipetting the suspension over the substrate.
- Collect the cell suspension with a pipette and transfer it to a centrifugation tube (15 mL or 50 mL tube).
- Spin down the cells by centrifugation at 110 x g for 10 min at room temperature.
- Carefully remove the supernatant and thoroughly re-suspend the cell pellet in culture medium before counting the cells (e.g. by using a hemacytometer for phase contrast microscopes and manual counting).
- Adjust the cell suspension to the desired cell density. For experiments with U-373 MG cells, use 100 000 cells/cm2 to grow a confluent cell layer within 48 h. This translates to a cell density of 200 000 cells/mL for 8-well electrode arrays with a growth area of ~0.8 cm2 and a well volume of 400 µL.
NOTE: For reproducible GPCR activation experiments, cells should be grown to confluent monolayers on the electrode arrays. To ensure proper receptor expression on the cell surface, cells should be seeded at least 36 h before performing the experiment. Testing different cell densities for cell seeding is often meaningful to identify the best experimental conditions. - Add the cell suspension to the wells of the electrode array and let the sells settle down at room temperature for 10 – 15 min in order to ensure homogenous distribution of cells on the bottom of the wells.
- Grow cells for at least 36 h in a standard cell culture incubator with 5% CO2 (dependent on medium type) at 37 °C and humidified atmosphere. Change cell culture medium 24 h prior to the experiment.
- On the day of the experiment, inspect the cell layers on the electrode arrays by (phase contrast) microscopy to ensure complete coverage of electrodes with cells.
2. Equilibration of cells in serum-free medium
- Pre-warm serum-free medium, in this study: Leibovitz's L15 medium.
- Remove cell culture medium from cells grown on electrode arrays and replace by pre-warmed serum-free medium. Use 200 µL for 8-well format electrode arrays, and 150 µL for 96-well electrode arrays.
- Let the cells equilibrate in serum-free medium at 37 °C for at least 2 h. The equilibration time strongly depends on the cell type. For example, U-373 MG cells need 2 h, CHO cells need 4 h, BAEC may require overnight equilibration in L-15 medium.
NOTE: L-15 medium is CO2 independent and requires a CO2-free atmosphere. For equilibration in L-15 set the incubator to 0 % CO2. Equilibration can be monitored by impedance readings, which we recommend to do for the first experiments.
3. Monitoring cell equilibration with impedance readings
- Place the electrode array into the connecting array holder of the impedance analyzer.
- Ensure proper low impedance contact between electrodes and impedance analyzer. This check is individually different for different instruments.
NOTE: If the instrument fails to make connection to the electrodes, open the contact clamp again, readjust the electrode array for proper positioning inside the holder and retry. - Select electrode type and/or multi-well format from the user interface of the software.
- Set up the measurement parameters. Different options are available.
NOTE: To select the most sensitive AC frequency, we refer to the literature and instrument manuals as it depends on electrode layout and cell type under study. Typically, the sensing frequency is in the range of 4 kHz – 50 kHz. Here, U-373 MG cells were grown on circular working electrodes of 250 µm diameter and they were monitored at an AC frequency of 12 kHz.- If single and multiple frequency data acquisition modes are available, select the single frequency mode to ensure maximum time resolution. The measurement will be performed at this single frequency. For the most widely spread instruments, there is a number of pre-set frequencies along the available frequency window.
- If the number of wells under study is low or time resolution is not critical, select multiple frequency recordings instead. Impedance readings at a specified number of frequencies will be recorded for every well for later in-depth analysis.
NOTE: The time resolution decreases with increasing number of frequencies recorded per well and increasing number of wells. The options for frequency and data acquisition mode selection vary with instrument type and version.
- Start acquisition of time course data.
- Follow the cell layer impedance (at least 2 h) until the impedance has stabilized. In the meantime, prepare the agonist solutions.
- When the cell layers have reached a stable impedance level, either (i) proceed to the serial agonist addition within the same experiment or (ii) terminate data acquisition and start a new dataset for monitoring agonist-induced receptor activation.
4. Preparation of agonist solutions for experiments in agonist mode
- Calculate the concentration of agonist solutions as needed for each step of serial dosing according to equation (1). n ranges from 1 to the total number of serial additions i. x denotes concentration and volume in the well at step n. y denotes concentration and volume of the "solution-to-be-added" in step n.

NOTE: Consider the number of replicates and calculate the total volume of "solution-to-be-added" for each concentration step. Results of a typical calculation are given in Tables 1-4. It takes a general idea about the agonist concentration range to be studied as the range defines the concentrations and number of portions to be administered during serial addition. Using the serial agonist addition protocol the agonist concentration is increased stepwise. Therefore, the amount of agonist already in the well when the next dose is added has to be taken into account. When the number of agonist molecules already present in the well is nx = cx ∙Vx (with the current concentration cx and volume Vx) and the number of molecules in the well after the next addition is nx+y, the number of molecules to be added ny is determined by the concentration cy and volume Vy of the solution to be applied to the well (ny = cy ∙ Vy). After adding a portion of agonist, the new amount of agonist molecules in the well is: cx+y ∙ Vx+y = cx ∙ Vx + cy ∙ Vy. This calculation applies for each subsequent step. Because of the interdependence of agonist concentration in the well and the amount of agonist in the portions to be added with each step, it is important to define final concentrations after each step in advance.
Mode 1: The volume in the well will increase with each step as liquid is continuously added.
Using this mode and an 8-well format, use Vx1 = 200 µL and Vy1 …. Vyi = 30 µL.
Mode 2: The volume in the well is constant as the volume added with each step is removed just before the subsequent addition.
Using this mode and a 96-well format, use Vx1 = 150 µL and Vy1 …. Vyi = 75 µL.
- Print the data sheet with the total volume per concentration and detailed instructions for pipetting.
- Prepare all solutions in the required amount. Make all agonist solutions in the same serum-free medium as used for equilibration of the cells.
CAUTION: Histamine dihydrochloride is considered hazardous by the 2012 OSHA Hazard Communication Standard (29 CFR 1910.1200). Histamine causes skin irritation, serious eye irritation, may cause an allergic skin reaction, allergy, asthma symptoms or breathing difficulties if inhaled and may cause respiratory irritation. Please consider the safety data sheet.
NOTE: Make agonist solutions as fresh as possible. Stability of agonists in solution may vary considerably. Store solutions at 4 °C or below until use in the experiment. For some molecules additional stabilizing additives, like BSA when using peptide-based or lipid-based molecules, may be considered to prevent adsorption to the walls of wells and tubes. - If performing the experiment in 96-well format, transfer solutions to a conventional 96-well plate (no electrodes) and use an 8- (or 12-) channel pipet for quick liquid transfer to the electrode array.
5. Preparation of agonist solutions for experiment in antagonist mode
NOTE: Prepare the antagonist solution(s) with the concentration to be applied throughout. Volume and concentration of antagonist solution depend on the mode of agonist addition (1 or 2). Examples for experiments in 8-well or 96-well format in addition mode 2: (A) 8-well format (Vx1 = 200 µL, VAntagonist = 200 µL); (B) 96-well format (Vx1 = 150 µL, VAntagonist = 75 µL).
- Calculate the concentration of each agonist solution needed for each step of serial dosing as described in step 4.1.
- Make all agonist solutions in the same serum-free medium as used for equilibration of the cells and add the antagonist at the same final concentration as planned for the respective wells in the experiment.
NOTE: In this case the histamine stock solution (10 mM) is prepared in L-15 medium. When agonists are dissolved in other solvents (e.g., dimethyl sulfoxid (DMSO), ethanol) a solvent control should be included to account for the increasing solvent load with each step of addition.
6. Performing the serial addition protocol in agonist mode
- Start data acquisition as described in steps 3.1 – 3.5.
- Pre-warm the agonist solutions prior to use by placing them in the incubator about 10 – 15 min before addition.
NOTE: When using thermo-labile substances, the solutions should not be kept at 37 °C for too long. If pre-warming for 10 – 15 min is considered critical, bring solutions to 37 °C shortly before addition in a water bath. - Run the agonist serial dosing dependent on the selected addition mode. Following mode 1 the total volume in the well increases with each addition of the next dose. In mode 2 the same volume which is added with each step is also removed again just before adding the next higher dose.
NOTE: Time needed for equilibration of the cell layer in between two subsequent agonist doses depends on the cells' response time. An initial experiment in parallel mode (one well – one concentration) reveals (i) cell response times for different agonist concentrations and (ii) the most sensitive curve parameter (e.g., impedance maximum, impedance after time x).
A: Mode 1 / 8-well format
- Add 30 µL of the first solution with the lowest concentration of agonist to the cells that were equilibrated in 200 µL of serum-free medium.
- Let the cells respond and equilibrate for the pre-defined period of time (e.g., 15 min).
- Add 30 µL of the second solution with the next higher concentration.
- Repeat steps 6.3.1-6.3.3 with the third, fourth, and so on, agonist solution.
NOTE: Working with 10 concentration steps will result in a total volume of 500 µL in the end of the experiment, which is just below the maximum applicable volume of these wells of ~ 550 µL.
B: Mode 2 / 96-well format
NOTE: Pause data acquisition during each liquid handling step (addition/removal) via the impedance instrument's software when running experiments in 96-well format. The more elaborate liquid handling may interfere with data acquisition. Use a multi-channel pipette.
- Pause data acquisition.
- Add 75 µL of the first solution with the lowest concentration of agonist to the cells that were equilibrated in 150 µL of serum-free medium.
- Resume data acquisition.
- Let the cells respond and equilibrate for the pre-defined period of time (e.g., 15 min).
- About 1 – 2 min before the regular equilibration time ends, pause the measurement and remove 75 µL from each well.
NOTE: The time point at which solution should be removed depends on the number of wells monitored in parallel and the pipetting speed. Time needed for removal of the solutions should not exceed the time between subsequent steps. - Add 75 µL of the second solution with the next higher concentration and resume the measurement.
- Repeat steps 6.3.8-6.3.10 with the third, fourth, and so on, agonist solution.
7. Performing the serial addition protocol in antagonist mode
- Start the measurement as described in steps 3.1 – 3.5.
- During equilibration of the cell layers, prepare antagonist solution (e.g., 200 µL of 1.5 µM diphenhydramine hydrochloride in L15 medium).
CAUTION: Diphenhydramine hydrochloride has potential acute health effects. It is harmful if swallowed or inhaled, may cause eye and skin irritation. It may cause respiratory and digestive tract irritation. Please consider the safety data sheet. - Pre-warm the antagonist and agonist solutions by placing them in the incubator about 10 – 15 min before addition to the cell cultures (cf. 6.2). If antagonist-free wells are included in the assay as well, also pre-warm serum-free medium.
- Add the antagonist solution to the designated wells. Let the cells equilibrate with antagonist for 15 – 20 min. If antagonist-free wells are included, add the same volume of serum-free medium to these wells.
- According to antagonist addition in mode 2, remove solution from the wells
(A) 8-well format (200 µL)
(B) 96-well format (75 µL) - Run the agonist addition sequence as described in step 6.3.
8. Data export and analysis
- Export data using the impedance instrument's software in order to convert all recorded data from proprietary into common data formats (e.g., csv). This step allows for data re-organization and presentation with other software packages.
- Load the csv-formatted data to scientific data analysis software.
- Normalize impedance values by subtracting the impedance of the last data point before the first addition of agonist solution and by setting the time of addition to t = 0. Plot the time course of normalized impedance.
- Plot the individual time courses and identify the maxima in impedance after each addition step. Compose a data sheet with these values.
- Plot the values of maximum (or minimum, if applicable) impedance changes as a function of agonist concentration. This can be done for individual wells or for the averages (mean ± SD).
- Use a data fitting routine to determine half-maximal effective concentrations (EC50) and maximum response (EMax) by using the four parameter logistic model (equation 2):

NOTE: c indicates the agonist concentration, A1 is the minimum and A2 is the maximum asymptote of the sigmoidal dose-response curve (A2 = EMax). EC50 is the concentration at the inflection point of the curve, and n corresponds to the Hill slope.
Representative Results
A typical scheme for preparing the various agonist solutions is shown for an experiment using 8-well electrode arrays with histamine as agonist in Tables 1-4. Table 1 and Table 2 present volumes and concentrations for an experiment using addition mode 1 (cf., Figure 1), while Table 3 and Table 4 present volumes and concentrations for an experiment following addition mode 2 (cf., Figure 1). Data have been calculated using equation (1).
A representative experiment is shown in Figure 2 for confluent layers of U-373 MG cells, which endogenously express the histamine 1 receptor, grown on an 8-well electrode array with a working electrode of 250 µm diameter using histamine as agonist. The measurement was performed at a single frequency of 12 kHz. Addition of the agonist concentration series was performed following mode 1. In the selected experiment 10 solutions with increasing histamine concentration (prepared according to step 4) were added sequentially every 15 min (Figure 2A). The serial dosing scheme was applied to seven of eight wells. In one well agonist-free medium (L-15) was added sequentially as a negative control in nine of ten steps. The last addition to this control was a histamine solution providing a final concentration of 100 µM histamine in the well. Data has been plotted with data analysis software and is shown as impedance change Δ|Z|12 kHz / kΩ along the experiment, with the impedance change and the time point of the first addition being set to zero. The initial equilibration time in the experimental buffer of 2 h is not shown in this plot. The time points of agonist addition are indicated by arrows.
The change in impedance relative to the baseline after each concentration step was extracted from the curves using a data cursor. At steps with no clear maximum, the last data point before the next addition was used for analysis. Impedance changes Δ|Z|12 kHz were plotted as a function of histamine concentration (cx+y) (Figure 2B, symbols). The transfer function of a four parameter logistic model (cf. step 8.6) was fitted to the experimental data points from seven wells (Figure 2B, lines). The fit results are shown in Table 5. Figure 2C and Figure 2D show the result of averaging the data in Figure 2A and Figure 2B, presenting mean ± SD for seven wells. Average time course data is summarized in Figure 2C, whereas the average dose-response curve is provided in Figure 2D.
In dose-response relationships as shown in Figure 2B,D, the fitting procedure was applied to the entire concentration range returning EC50 = (0.86 ± 0.13) µM. The impedance response increases monotonically with increasing histamine concentration except for the last two concentration steps (30 µM, 100 µM final concentration). This response is presumably explained by downregulation of the histamine receptor, desensitization or over-stimulation of the cells (see discussion). To account for this effect, the data range for analysis has been reduced, as shown for one single serial dosing experiment (Figure 3; well 1) selected from the data shown in Figure 2. Moreover, setting the lower asymptote (A1) to zero during least square optimization assuming no impedance change in response to 0 µM histamine is well justified. Applying this three parameter optimization provides an EC50 of (0.75 ± 0.12) µM (cf., Figure 3B). EC50 values were found to be insensitive for either of the data analysis modes and were not significantly different.
Typical results for a 96-well plate experiment are summarized in Figure 4. U-373 MG cells were grown on 96-well electrode arrays as described above. 32 wells were included in the measurement. 8 wells served as agonist-free controls, exposed to medium only. 24 wells were subjected to a series of increasing histamine concentrations calculated and prepared for addition mode 2. The time course of impedance change is shown for all 32 individual wells in Figure 4A. One of the curves (highlighted in red) shows an unusual time course and was excluded from further analysis. Accordingly, data from 23 wells were averaged and plotted as time course of average impedance change (mean ± SD) in Figure 4B. Each individual curve was analyzed as described above for the dose-response relationship. Data points were averaged and plotted as function of the final histamine concentration (Figure 4C). Dose-response data were analyzed by means of the four parameter logistic model, which returned an average EC50 value of (1.0 ± 0.3) µM.
A typical result for the serial agonist addition protocol in antagonist mode is shown in Figure 5. Here, one well was pre-treated with 0.75 µM of the histamine receptor antagonist diphenhydramine (red curve) 20 min before the first histamine was added (Figure 5A). The control received antagonist-free medium (blue curve). The antagonist was added following mode 2, which means that 200 µL of the finally 400 µL were removed before the agonist series was applied. The agonist (histamine) was added following serial dosing mode 1 as described in the examples above. It is important for experiments in antagonist mode that the antagonist concentration is kept constant over the entire time course of the experiment. Accordingly, all agonist solutions added to the well must also contain the pre-defined antagonist concentration (in this case, 0.75 µM), while the control remains antagonist-free. An antagonistic effect becomes apparent by a "delayed" increase of impedance within the serial dosing scheme corresponding to higher agonist concentrations needed to induce a cell response. In the dose-response relationship the effect of the antagonist is expressed as a rightward shift of the curves, which corresponds to an increase in EC50, provided the antagonist is a competitive ligand relative to the agonist (Figure 5B). The EC50 was determined to (0.92 ± 0.05) µM in absence of the antagonist and (14.7 ± 0.6) µM in its presence.
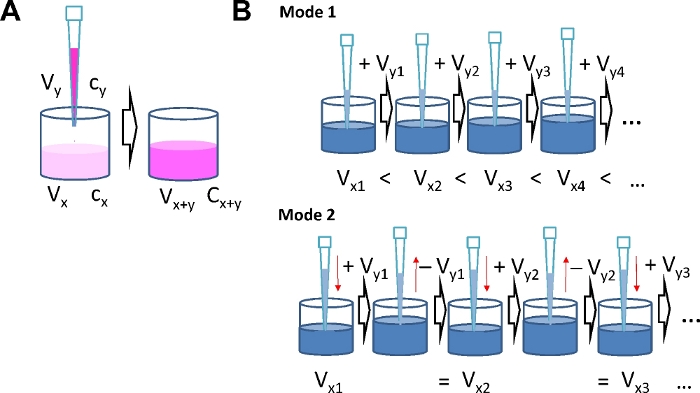
Figure 1: Serial agonist addition modes. (A) Schematic illustrating concentration calculations. By adding agonist solution of concentration cy and volume Vy to a well with volume Vx and agonist concentration cx, the volume increases to Vx+y and the concentration increases to cx+y. (B) Two different modes for serial agonist addition. Mode 1: The total volume in the well Vx+y increases with each addition. Mode 2: The volume added at each concentration step is removed again before the next dose is added, which keeps the total volume in the well constant. Please click here to view a larger version of this figure.

Figure 2: Results of a typical experiment in 8-well format. (A) Time course of impedance change at 12 kHz for confluent U-373 MG cell layers grown on circular working electrodes of 250 µm diameter. In seven of eight wells serial addition of histamine was applied. One well (grey curve) was sequentially loaded with histamine-free buffer at each step (mode 1), except for the last step when 100 µM histamine were added as a positive control. (B) Dose-response relationships generated from the maximum impedance change relative to the baseline for seven individual wells after each concentration step. (C) Average time course of impedance change (mean ± SD) generated from seven individual wells as shown in (A). (D) Average dose-response relationships generated from individual data sets as shown in (B). Please click here to view a larger version of this figure.
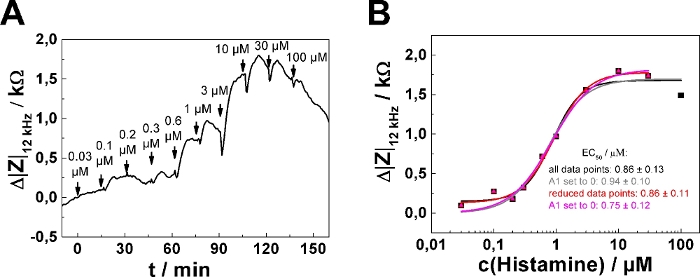
Figure 3: Adaptation of data analysis modes. (A) Typical time course of impedance change at 12 kHz for a single confluent U-373 MG cell layer grown on circular working electrodes of 250 µm diameter. (B) Experimental dose-response relationship (filled symbols) generated from the maximum impedance change relative to baseline after each concentration increase. Either the full data set was subjected to the fitting procedure (black and grey curve) or the last data point – indicating the onset of receptor desensitization and/or internalization – was omitted from quantitative analysis (red and magenta curve). All four parameters were adjusted during least square optimization (black and red curve) or the parameter A1, representing the lower asymptote, was set and fixed to zero (grey and magenta curve). EC50 values were not significantly different for the two data analysis modes. Please click here to view a larger version of this figure.
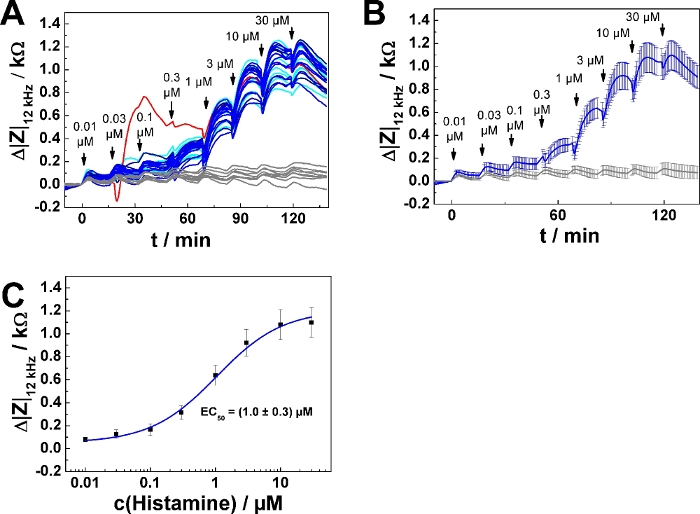
Figure 4: Typical experiment in 96-well format. (A) Time course of impedance change measured at 12 kHz for confluent U-373 MG cell layers grown on a 96-well electrode array. Data are shown for 32 wells. 24 wells were subjected to a series of increasing histamine concentrations added with an equilibration time of 15 min between each step. The curve highlighted in red was excluded from averaging. Eight wells (control) were treated with histamine-free medium at each time point of addition. (B) Time course of average impedance change for 23 individual wells in response to histamine serial addition and eight control wells as shown in (A). (C) Average dose-response relationship generated from the maximum impedance change relative to the baseline after each concentration increase (mean ± SD). Data were fitted with a four parameter logistic function. Please click here to view a larger version of this figure.
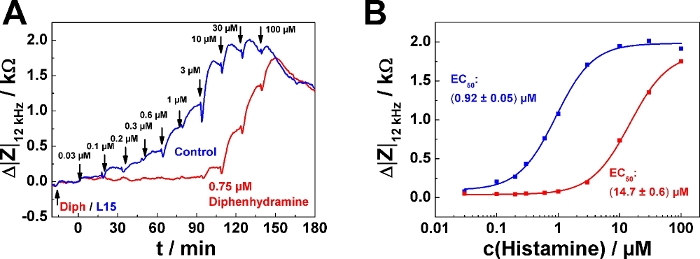
Figure 5: Typical experiment in antagonist mode. (A) Time course of impedance change at 12 kHz for confluent U-373 MG cell layers grown on circular working electrodes of 250 µm diameter upon addition of increasing concentrations of histamine with or without presence of histamine receptor antagonist diphenhydramine. Diphenhydramine (0.75 µM) or antagonist-free medium (L15) were added 20 min before the agonist serial dosing started. (B) Dose-response relationships generated from the maximum impedance change relative to the baseline after each concentration increase from data shown in (A). Presence of 0.75 µM Diphenhydramine increased the EC50 from (0.92 ± 0.05) µM to (14.7 ± 0.6) µM. Please click here to view a larger version of this figure.
| Solution # | cx+y (µM) | cx (µM) | Vx (µL) | Vx+y (µL) | Vy (µL) | cy (µM) |
| 1 | 0.01 | 0 | 200 | 230 | 30 | 0.08 |
| 2 | 0.1 | 0.01 | 230 | 260 | 30 | 0.79 |
| 3 | 0.3 | 0.1 | 260 | 290 | 30 | 2.03 |
| 4 | 1 | 0.3 | 290 | 320 | 30 | 7.77 |
| 5 | 3 | 1 | 320 | 350 | 30 | 24.33 |
| 6 | 10 | 3 | 350 | 380 | 30 | 91.67 |
| 7 | 30 | 10 | 380 | 410 | 30 | 283.33 |
| 8 | 100 | 30 | 410 | 440 | 30 | 1056.67 |
Table 1: Concentration calculation scheme for experiments using 8-well electrode arrays following addition mode 1.
| Solution # | cy (µM) | Vy (µL) | make from | c (µM) | dilution factor | make total V (µl) | use (µl) | add V (µl) |
| 1 | 0.08 | 30 | 3 | 2.03 | 26.52 | 600 | 22.6 | 577.4 |
| 2 | 0.79 | 30 | 4 | 7.77 | 9.83 | 600 | 61 | 539 |
| 3 | 2.03 | 30 | 5 | 24.33 | 11.97 | 600 | 50.1 | 549.9 |
| 4 | 7.77 | 30 | 6 | 91.67 | 11.8 | 600 | 50.8 | 549.2 |
| 5 | 24.33 | 30 | 7 | 283.33 | 11.64 | 600 | 51.5 | 548.5 |
| 6 | 91.67 | 30 | 8 | 1056.67 | 11.53 | 600 | 52.1 | 547.9 |
| 7 | 283.33 | 30 | 8 | 1056.67 | 3.73 | 600 | 160.9 | 439.1 |
| 8 | 1056.67 | 30 | 10 mM Stock | 10000 | 9.46 | 800 | 84.5 | 715.5 |
Table 2: Pipetting scheme for experiments using 8-well electrode arrays following addition mode 1.
| Solution # | cx+y (µM) | cx (µM) | Vx (µL) | Vx+y (µL) | Vy (µL) | cy (µM) |
| 1 | 0.01 | 0 | 200 | 400 | 200 | 0.02 |
| 2 | 0.1 | 0.01 | 200 | 400 | 200 | 0.19 |
| 3 | 0.3 | 0.1 | 200 | 400 | 200 | 0.5 |
| 4 | 1 | 0.3 | 200 | 400 | 200 | 1.7 |
| 5 | 3 | 1 | 200 | 400 | 200 | 5 |
| 6 | 10 | 3 | 200 | 400 | 200 | 17 |
| 7 | 30 | 10 | 200 | 400 | 200 | 50 |
| 8 | 100 | 30 | 200 | 400 | 200 | 170 |
Table 3: Concentration calculation scheme for experiments using 8-well electrode arrays following addition mode 2.
| Solution # | cy (µM) | Vy (µL) | make from | c (µM) | dilution factor | make total V (µl) | use (µl) | add V (µl) |
| 1 | 0.02 | 200 | 3 | 0.5 | 25 | 1000 | 40 | 960 |
| 2 | 0.19 | 200 | 4 | 1.7 | 8.95 | 1000 | 111.8 | 888.2 |
| 3 | 0.5 | 200 | 5 | 5 | 10 | 1000 | 100 | 900 |
| 4 | 1.7 | 200 | 6 | 17 | 10 | 1000 | 100 | 900 |
| 5 | 5 | 200 | 7 | 50 | 10 | 1000 | 100 | 900 |
| 6 | 17 | 200 | 8 | 170 | 10 | 1000 | 100 | 900 |
| 7 | 50 | 200 | 8 | 170 | 3.4 | 1000 | 294.1 | 705.9 |
| 8 | 170 | 200 | 200 µM Stock | 200 | 1.18 | 1300 | 1105 | 195 |
Table 4: Pipetting scheme for experiments using 8-well electrode arrays following addition mode 2.
| Well | EC50 | Emax (=A2) | A1 | n |
| (µM) | (kΩ) | (Ω) | ||
| 1 | 0.9 ± 0.1 | 1680 ± 70 | 150 ± 80 | 1.9 ± 0.5 |
| 2 | 0.7 ± 0.2 | 1770 ± 90 | 200 ±140 | 1.3 ± 0.4 |
| 3 | 0.6 ± 0.2 | 1700 ± 100 | 60 ± 250 | 1.1 ± 0.5 |
| 4 | 0.6 ± 0.2 | 2000 ± 100 | 140 ± 180 | 1.3 ± 0.4 |
| 5 | 0.9 ± 0.1 | 1810 ± 60 | 160 ± 80 | 1.4 ± 0.3 |
| 6 | 0.8 ± 0.1 | 1950 ± 70 | 200 ± 110 | 1.3 ± 0.3 |
| 7 | 0.6 ± 0.3 | 1700 ± 200 | 260 ± 250 | 1.6 ± 1.0 |
| 1 – 7 | 0.7 ± 0.2 | 1900 ± 100 | 100 ± 100 | 1.1 ± 0.2 |
Table 5: Results obtained from four parameter logistic fits applied to typical dose-response data. The dose-response data used for analysis is presented in Figure 2. The full concentration range was applied to analysis for the individual wells one through seven (Figure 2B) and for the average data obtained from wells one to seven (Figure 2D). None of the parameters of the logistic function was fixed during least square optimization. Fit results were rounded to the decade of the error, except if the value was smaller than the error.
Discussion
This protocol describes a method for label-free impedance measurements to determine the dose-response relationship of agonist-induced GPCR activation in absence or presence of specific antagonists for the same receptor. The proof of concept of this method was presented in a recent publication12. To our knowledge it is the first study describing the establishment of a full dose-response curve of agonist-mediated GPCR activation using a single cell layer in vitro. The approach inevitably requires the use of label-free cell monitoring approaches and will not be applicable to endpoint assays.
The protocol has been demonstrated on the example of U-373 MG cells, which endogenously express the human histamine 1 receptor (hHR1) that is stimulated with a series of increasing concentrations of its endogenous agonist histamine while the impedance of the cells is continuously recorded. To demonstrate the applicability of the protocol to antagonist studies, the experiments were repeated in presence of one constant concentration of diphenhydramine, a competitive antagonist to the hH1R. Impedance recordings have also been used successfully to study other H1R agonists (e.g., UR-KUM53011, individual and stepwise dosing) and antagonists (e.g., Mepyramine)12 in addition to the examples shown here. Moreover, the protocol has been applied to other cell lines and receptors as well, e.g. CHO-D2R expressing the dopamine D2 receptor (D2R) or BAEC expressing the ß2-adrenergic receptor (ß2AR)12 indicating that it presents a general scheme for very efficient generation of dose-response data. While U373-MG and BAEC cells express the respective receptors H1R and ß2AR at endogenous levels, the CHO-D2R is an example for a recombinant cell model. Taken together, the serial protocol is in general applicable to cell types with endogenous or ectopic GPCR expression, GPCRs with different coupling types Gq (e.g., H1R), Gs (e.g., ß2AR), Gi (e.g., D2R) as well as different ligand types (agonist/antagonist). In all those cell/receptor types mentioned above, the impedance increases with increasing agonist concentration. However, some cells respond to GPCR activation by an impedance decrease. We tested the protocol for one such case (HaCaT/histamine) and found also a concentration-dependent stepwise decrease, supporting the notion of a generally applicable approach that is also compatible with inverse impedance changes. It has been proposed that the detailed time course of impedance data indicates the type of G-protein coupling of a given receptor. Even though the concept is attractive, the pool of our data does not support a generally applicable rule that correlates the impedance time profile to G-protein coupling of a given GPCR6.
The serial addition protocol is adaptable to different types of electrode layouts and multi-well formats. It is applicable to on-chip perfusion systems, which is a significant advantage over many label-based approaches that often require one cell layer for one concentration to be tested. Accordingly, the serial dosing scheme may pave the way to dose-response studies in more complex biological models like organ-on-a chip or even human-on-a-chip approaches. Impedance measurements detect an integrated, distal response of the cells to receptor activation that ultimately induces cell morphology changes. This rather general and unspecific but also unbiased readout is the basis for its broad applicability that is not restricted to GPCRs but might also work for other classes of cell surface receptors, cytoplasmic receptors or even intracellular proteins as targets.
It is critical for the stepwise agonist addition protocol to work with confluent cell layers on the electrodes, as sparse cultures will interfere with sensitivity, reproducibility and data interpretation. An additional prerequisite for successful monitoring of receptor activation even in confluent cell monolayers is a change in cell shape along the signal transduction cascade. If the cells under study do not change their morphology along the experiment, impedance-based readouts are blind and will not report on any change in receptor activity. To properly layout the serial addition protocol, initial pre-testing in conventional one-well-one-concentration mode is recommended to determine the concentration range of the agonist roughly and the time interval between subsequent agonist additions. Time intervals between sequential doses of the agonist should be tailored such that the system adopts a new equilibrium before the next, higher dose is added. Accordingly, the method may be less useful for receptors that trigger a very quick and only transient response of the cells or a very slow response that may take hours to complete. The latter situation would increase the total time of the experiment and call for parallel addition protocols to cut short on lab time. In the examples presented above, the total runtime of the experiment was 2.5 – 3.5 h, depending on the number of concentrations (8 or 10) and a potential pre-incubation with antagonist. The same experiment in conventional parallel agonist addition mode would only take ~ 1 h for the same cell-receptor model but with much smaller throughput. A clear advantage of the serial approach is that the number of concentrations that need to be tested for a full dose-response relationship is not limited by the number of available wells. If higher agonist concentrations are needed, the experiment is easily extended and completed without any additional cell culture work. Agonist addition itself should be carried out carefully, as some cells are sensitive to shear stress and respond with considerable impedance changes simply due to mechanical stimulation imposed by liquid handling. The cells might even come off the electrode. This problem is, however, not unique to impedance-based cell monitoring but applies to other techniques as well. For sensitive cells addition mode 1 is recommended due to reduced mechanical load on the cells. However, this mode operates with lower liquid volumes during addition which raises the risk of incomplete mixing since intensive agitation is not recommended to avoid unspecific cell responses.
Addition mode 1 goes along with a stepwise increase of the total volume in the well whereas mode 2 keeps the total volume constant since equal amounts of solution are sequentially removed. These are just the two extreme strategies and might be adapted for special cell types, multi-well formats or electrode layouts in any intermediate way. In the case of the U373-MG/H1R model slight differences in EC50 and EMax values have been observed for the serial addition mode compared to the conventional parallel addition mode (serial, N = 30: EC50: (0.8 ± 0.3) µM, EMax: (1.9 ± 0.2) kΩ; parallel, N = 15: EC50: (0.5 ± 0.2) µM, EMax: (1.3 ± 0.3) kΩ), while other cell/receptor models did not show any differences. Shifts in EC50 and EMax values could originate from receptor desensitization that is more likely to occur when the receptors are exposed to the agonist for longer times. The desensitization influence will strongly depend on the receptor and the ligand under study. It remains to be elucidated whether a detailed analysis of serial and parallel agonist addition may provide a new means to study receptor desensitization.
We recommend performing a control experiment with conventional parallel mode agonist addition in order to test for potential shifts in dose-response curves. Further controls should include a sample treated only with medium/solvent in appropriate doses to unravel any effects due to solvent load or mechanical stress upon medium addition. This becomes particularly important if the stock solution of the ligand is prepared in other solvents than medium (e.g., DMSO, ethanol). When investigating the effect of different ligands a standard agonist should be included as reference.
Future directions should involve automatic liquid handling units, which might allow for completely automated dosing or titration. Applying the serial addition mode in perfusion systems holds great potential for lab-on-a chip and organ-on a-chip approaches, as well as for experiments performed under liquid shear stress.
Açıklamalar
The authors have nothing to disclose.
Acknowledgements
We thank Barbara Goricnick and Nadja Hinterreiter for their help with cell culturing and preparation of experimental solutions. The authors gratefully acknowledge financial support by the Research Training Group 1910 "Medicinal chemistry of selective GPCR ligands" funded by the German Research Foundation (DFG) under grant number 222125149. JAS is particularly grateful for a scholarship granted by the Bavarian Gender Equality Program.
Materials
| Bürker counting chamber | Marienfeld (Lauda-Königshofen, Germany) | 640210 | |
| cell culture flasks 25 cm2 | Greiner bio-one (Frickenhausen, Germany) | 690175 | |
| Cell incubator (Heraeus Function Line BB15) | Thermo Scientific (Darmstadt, Germany) | ||
| Centrifuge (Heraeus 1S-R) | Thermo Scientific (Darmstadt, Germany) | ||
| Diphenhydramine hydrochloride | Sigma Aldrich (Taufkirchen, Germany) | D3630 | |
| Eagle's Minimum Essential Medium with 4.5 g/L D-Glucose and 2.2 g/L NaHCO3 | Sigma Aldrich (Taufkirchen, Germany) | D5671 | |
| Impedance Instrument (ECIS Zθ) | Applied BioPhysics Inc. (Troy, NY, USA) | ||
| 8-well electrode Arrays (8W1E PET) | Applied BioPhysics Inc. (Troy, NY, USA) | PET base with 0.049 mm2 working electrode and ~50 mm2 counter electrode (gold) | |
| 96-well electrode arrays (96W1E+ PET) | Applied BioPhysics Inc. (Troy, NY, USA) | PET base with two electrodes (gold) with 0.256 mm2 total electrode area | |
| Fetal calf serum (FCS) | Biochrom (Berlin, Germany) | S0615 | |
| Histamine dihydrochloride | Carl Roth (Karlsruhe, Germany) | 4017.1 | |
| Laminar flow hood (Herasafe, KS 12) | Thermo Scientific (Darmstadt, Germany) | 51022515 | class II safety cabinet |
| Leibovitz' L-15 medium | Thermo Scientific (Darmstadt, Germany) | 21083-027 | |
| L-glutamine | Sigma Aldrich (Taufkirchen, Germany) | G7513 | |
| Micropipette large (100 – 1000 µL) | Brandt (Wertheim, Germany) | 704780 | |
| Micropipette large (20 – 200 µL) | Brandt (Wertheim, Germany) | 704778 | |
| Microscope (phase contrast, Nikon Diaphot) | Nikon (Düsseldorf, Germany) | ||
| Penicillin/streptomycin | Sigma Aldrich (Taufkirchen, Germany) | P0781 | |
| Phosphate buffered saline (PBS) | Sigma Aldrich (Taufkirchen, Germany) | D8537 | |
| Pipette, serological | Greiner bio-one (Frickenhausen, Germany) | 607 180 | |
| Pipettor (accu-jet pro) | Brandt (Wertheim, Germany) | 26300 | |
| Trypsin | Sigma Aldrich (Taufkirchen, Germany) | T4174 | in PBS with 1 mM EDTA |
| Tube, 15 mL | Greiner bio-one (Frickenhausen, Germany) | 188 271 | |
| Tube, 50 mL | Greiner bio-one (Frickenhausen, Germany) | 210 261 | |
| U-373 MG cells | ATCC (Rockville, MD, USA) | ATCC HTB-17 | |
| water bath (TW21) | Julabo (Seelbach, Germany) |
Referanslar
- Rosenbaum, D. M., Rasmussen, S. G., Kobilka, B. K. The structure and function of G-protein-coupled receptors. Nature. 459, 356-363 (2009).
- Sriram, K., Insel, P. A. G Protein-Coupled Receptors as Targets for Approved Drugs: How Many Targets and How Many Drugs. Molecular Pharmacology. 93, 251-258 (2018).
- Kaitin, K. I. Deconstructing the drug development process: The new face of innovation. Clinical Pharmacoly and Therapeutics. 87, 6 (2010).
- Kenakin, T. P. Cellular assays as portals to seven-transmembrane receptor-based drug discovery. Nature reviews. Drug discovery. 8, 617-626 (2009).
- Zhang, R., Xie, X. Tools for GPCR drug discovery. Acta Pharmacologica Sinica. 33, 372-384 (2012).
- Scott, C. W., Peters, M. F. Label-free whole-cell assays: expanding the scope of GPCR screening. Drug Discovery Today. 15, 704-716 (2010).
- Giaever, I., Keese, C. R. Monitoring fibroblast behavior in tissue culture with an applied electric field. Proceedings of the National Academy of Sciences of the United States of America. 81, 3761-3764 (1984).
- Giaever, I., Keese, C. R. A morphological biosensor for mammalian cells. Nature. 366, 591-592 (1993).
- Fang, Y., Ferrie, A. M., Fontaine, N. H., Mauro, J., Balakrishnan, J. Resonant waveguide grating biosensor for living cell sensing. Biophysical Journal. 91, 16 (2006).
- Stolwijk, J. A., Matrougui, K., Renken, C. W., Trebak, M. Impedance analysis of GPCR-mediated changes in endothelial barrier function: overview and fundamental considerations for stable and reproducible measurements. Pflugers Archiv : European Journal of Physiology. 467, 2193-2218 (2015).
- Lieb, S., et al. Label-free versus conventional cellular assays: Functional investigations on the human histamine H1 receptor. Pharmacological Research. 114, 13-26 (2016).
- Stolwijk, J. A., et al. Increasing the throughput of label-free cell assays to study the activation of G-protein-coupled receptors by using a serial agonist exposure protocol. Integrative Biology. 11, 10 (2019).

