Native Polyacrylamide Gel Electrophoresis Immunoblot Analysis of Endogenous IRF5 Dimerization
Özet
A native Western blot method for analyzing endogenous interferon regulatory factor 5 dimerization in the CAL-1 plasmacytoid dendritic cell line is described. This protocol can be applied to other cell lines as well.
Abstract
Interferon regulatory factor 5 (IRF5) is a key transcription factor for regulating the immune response. It is activated downstream of the Toll-like receptor myeloid differentiation primary response gene 88 (TLR-MyD88) signaling pathway. IRF5 activation involves phosphorylation, dimerization, and subsequent translocation from the cytoplasm into the nucleus, which in turn induces the gene expression of various pro-inflammatory cytokines. A detection assay for IRF5 activation is essential to studying IRF5 functions and its relevant pathways. This article describes a robust assay to detect endogenous IRF5 activation in the CAL-1 human plasmacytoid dendritic cell (pDC) line. The protocol consists of a modified nondenaturing electrophoresis assay that can distinguish IRF5 in its monomer and dimer forms, thus providing an affordable and sensitive approach to analyze IRF5 activation.
Introduction
Interferon regulatory factor 5 (IRF5) is an important transcription regulator that plays a prominent role in regulating the immune response, particularly in the release of pro-inflammatory cytokines and type I interferons (IFNs)1,2,3. Misregulation of IRF5 is a contributing factor in numerous autoimmune diseases, as evident by various polymorphisms in the IRF5 locus that are associated with systemic lupus erythematosus, multiple sclerosis, rheumatoid arthritis, etc.4,5,6,7,8,9,10. Therefore, a robust detection assay for endogenous IRF5 activation state is crucial for understanding the regulatory pathways and downstream effects of IRF5 in a physiologically relevant cellular context.
IRF5 is constitutively expressed in monocytes, dendritic cells (DCs), B cells, and macrophages1,11. As with other IRF family transcription factors, IRF5 resides in the cytoplasm in its latent state. Upon activation, IRF5 is phosphorylated and forms homodimers, which then translocate into the nucleus and bind to specific regulatory elements of genes encoding type I IFNs and pro-inflammatory cytokines, eventually inducing the expression of these genes1,2,11,12,13. IRF5 regulates the innate immune responses downstream of various Toll-like receptors (TLRs), such as TLR7, TLR 8, and TLR 9, which are localized in endosomes and use MyD88 for signaling1,11,14. These TLRs primarily recognize foreign nucleic acid species such as single-stranded RNA (ssRNA) and unmethylated CpG DNA that are symptomatic of an infection15,16,17,18. IRF5 has been shown to regulate immune responses against bacterial, viral, and fungal infections19,20,21. Considering IRF5’s influential and diverse role in the immune system, enhancing or dampening IRF5 activity could serve as a novel avenue for the development of therapeutic agents22. Hence, it is critical to develop a protocol to monitor the activation status of endogenous IRF5 to allow thorough investigation of the pathways and mechanisms regulating IRF5 activity in different cell types.
To the best of our knowledge, no biochemical or gel electrophoretic assay for endogenous IRF5 activation has been published prior to the development of this protocol. Phosphorylation has been shown to be an important first step of IRF5 activation, and a phosphospecific IRF5 antibody was developed that led to the discovery and confirmation of a serine residue important for IRF5 activity13. However, while the antibody clearly detects phosphorylated IRF5 when immunoprecipitated or overexpressed23, it fails to detect IRF5 phosphorylation in a whole cell lysate in our hands (data not shown). Dimerization is the next step of IRF5 activation, and many important studies to date investigating this step relied on overexpression of epitope-tagged IRF5, often in irrelevant cell types that do not normally express IRF511,12,24,25. Previous studies have shown that dimerized IRF5 may not always translocate into the nucleus and hence is not necessarily fully activated25,26. An assay for endogenous IRF5 nuclear localization was developed to assess IRF5 activation by imaging flow cytometry27. This assay has been applied in studies that were crucial to understanding IRF5 activity, especially in primary or rare cell types28,29 and greatly advanced the knowledge in the field. However, this assay relies on a specialized instrument that is not widely available to researchers. Further, it is often necessary to investigate the initial steps of activation while dissecting IRF5 regulatory pathways and identifying upstream regulators and pathway components. This study provides a robust and reliable biochemical assay for the early activation events of IRF5 that can be performed in labs equipped with molecular biology tools. The protocol described here will be very useful in investigating the pathways and mechanisms of IRF5 actions, especially when combined with orthogonal assays such as the imaging flow cytometric analysis of IRF5 nuclear localization23,27,28,30.
Native polyacrylamide gel electrophoresis (native PAGE) is a widely used method to analyze protein complexes31,32. Unlike sodium dodecylsulfate polyacrylamide gel electrophoresis (SDS-PAGE), native PAGE separates proteins on the basis of their shape, size, and charge. It also retains native protein structure without denaturation31,33,34,35. The protocol presented takes advantage of these features of native PAGE and detects both monomeric and dimeric forms of IRF5. This method is particularly important for detecting early activation events because there is no suitable commercially available antibody that can detect endogenous phosphorylated IRF5. Previously, several published studies used native PAGE to assess IRF5 dimerization. However, the majority of these studies depended on the overexpression of exogenous epitope-tagged IRF5 to analyze activation status2,13,24,36,37. This work presents a step-by-step protocol for analyzing endogenous IRF5 dimerization via a modified native PAGE technique in a human plasmacytoid dendritic cell (pDC) line, where IRF5 activity has been shown to be crucial for its function1,38,39,40. This same technique has been applied to other cell lines23.
Protocol
NOTE: The protocol described here uses CAL-1 pDC cell line treated with resiquimod (R848), an agonist for TLR7/8. This protocol has been applied to other human and murine cell types, including RAW 264.7 (murine macrophage line), THP-1 (human monocytic cell line), BJAB (human B cell line), Ramos (human B cell line), and MUTZ-3 (human dendritic cell line)23.
1. Stimulation of CAL-1 Cells
- Maintain CAL-1 cell cultures in a T75 flask at 37 °C and 5% CO2 under sterile conditions with 20−25 mL of RPMI 1640 medium containing 5% fetal bovine serum (FBS), 25 mM HEPES, and 1x mercaptoethanol (i.e., complete RPMI 1640 medium).
- Transfer the cells into a 50 mL conical tube.
NOTE: CAL-1 cells are non-adherent. For adherent cell types, standard trypsinization can be performed to harvest cells. - Centrifuge the cells at 200 x g for 5 min at room temperature (RT). Remove supernatant and resuspend the cell pellet in 8 mL of the complete RPMI 1640 medium to obtain a homogeneous single cell suspension.
- Count the cells using a hemocytometer. Seed the cells at a density of 1 x 106 cells per well in a 6 well plate with 4 mL of preheated complete RPMI 1640 medium in each well. Incubate for 20−24 h at 37 °C and 5% CO2 to allow the confluency to reach 90%−95% (corresponding to approximately 1.5 x 106 cells).
- Stimulate the cells by adding 4 µL of 1 mg/mL R848 per well of the 6 well plate (final concentration of 1 µg/mL). Also set up an unstimulated control well with cells without the R848 treatment.
- Ensure the R848 is evenly dispersed by gently rocking the plate side to side. Then, incubate the cells for 2−16 h in the incubator at 37 °C and 5% CO2.
2. Extraction of Cellular Proteins
- Transfer the cell suspensions from the 6 well plate into 5 mL centrifuge tubes.
- Centrifuge at 200 x g for 5 min at RT. Remove the supernatant and resuspend the cell pellet in 1 mL of phosphate-buffered saline (PBS) to obtain a homogeneous single cell suspension.
- Transfer the cell suspension into a 1.5 mL centrifuge tube.
- Spin down briefly at 12,000 x g for 0.5−1 min at 4 °C and carefully remove the supernatant.
- Prepare the lysis buffer containing 6.25 mL of 1 M Tris-HCl pH 7.4 (final concentration of 25 mM), 7.5 mL of 5 M NaCl (final concentration of 150 mM), 0.5 mL of 0.5 M EDTA (final concentration of 1 mM), 2.5 mL of NP-40 (final concentration of 1%) and 7.5 mL of glycerol (final concentration of 5%) in 250 mL of deionized water (ddH2O). Add 100x protease inhibitor single-use cocktail to a final concentration of 1x to the lysis buffer just before use. Keep the prepared lysis buffer on ice.
NOTE: The lysis buffer without the protease can be stored at 4 °C. - Resuspend the cell pellet in 30 µL of ice-cold lysis buffer and mix by pipetting up and down.
- Incubate on ice for 15−20 min.
- Clarify the lysate by centrifuging at 12,000 x g for 15−20 min at 4 °C. Transfer the supernatant into a new prechilled 1.5 mL centrifuge tube. Keep the extracts on ice at all times.
NOTE: Cell lysates can be stored at -20 °C or -80 °C. Do not boil the samples. - Measure protein concentration using Bradford reagent.
3. Analysis of IRF5 Dimerization by Native PAGE
- Prepare the upper (-) and lower (+) chamber electrophoresis buffers. The upper chamber buffer consists of 0.3% sodium deoxycholate (NaDOC) in 1x native PAGE running buffer, and the lower chamber consists of only 1x native PAGE running buffer.
NOTE: Prepare a fresh upper chamber buffer for every new run. - Rinse a 3%−12% native PAGE gel thoroughly with water without distorting the wells. Set the gel into the mini gel tank and remove the comb. Prerun the gel in a 4 °C cold room or on ice at 150 V for 30 min.
NOTE: Prerunning removes excessive ammonia and persulfate ions that can interfere with the running of the gel. - During the prerun, prepare the samples for loading by mixing the cellular proteins kept on ice with 4x native sample buffer.
- After the prerun, load 10−15 µg of protein with a final volume of 10−15 µL per sample.
NOTE: Overloading of protein can cause smearing. - Run gel at 85 V for 30 min, then 150 V for 2 h.
NOTE: Considering differences in equipment and cell lines used by different labs, minor modifications to the concentration of protein samples, voltage and running time may be appropriate to optimize this protocol. Lowering pre-running and running voltage while increasing running time may help to improve dimer resolution and result consistency. - Soak the gel in SDS running buffer (25 mM Tris pH 8.3, 250 mM Glycine, 0.1% SDS) for 30 min at RT.
NOTE: No agitation is required. Occasionally, the intensity of the band may not be proportional to the amount of protein loaded due to inefficient transfer in the presence of deoxycholate (DOC), which mainly affects the monomeric form of IRF5. Soaking the gel in SDS running buffer prior to the transfer solves this problem. The gel is fragile. Handle with extreme care from the bottom (i.e., higher percentage) end of the gel.
4. Immunoblot Analysis of IRF5
- Activate the polyvinylidene difluoride (PDVF) membrane by soaking it in methanol for approximately 5 min.
- Make a cut on one corner of the membrane to indicate its orientation. Assemble the transfer sandwich according to the sequential order detailed in the manufacturer’s protocol with extra care to ensure that no air bubbles are trapped within.
- Place the transfer cassette into the tank and transfer at 20 V for 1 h on ice.
NOTE: Perform all incubations and washes in subsequent steps with a rocking shaker. - Remove the membrane from the cassette with plastic forceps after the transfer is completed. Block the membrane in blocking buffer (TBS) for 45 min at RT.
NOTE: TBS buffer with 5% BSA can also be used as a blocking buffer. - Incubate the membrane with the primary antibody listed in Table 1. Wash the membrane with 1x TBST washing buffer (20 mM Tris, pH 7.0, 150 mM NaCl and 0.1% Tween 20) for 3 min while rocking. Repeat the wash 2x.
| Dillution | Dillution buffer | Incubation | Yorumlar | |
| Primary antibody(Anti-IRF5) | 1/1,000 | TBS blocking buffer | Overnight at 4 °C or 2 h at RT | Diluted antibodies can be reused several times if stored at 4 °C in the presence of 0.02% sodium azide. |
| Secondary antibody(Anti-rabbit) | 1/10,000 | TBS blocking buffer | 45 min at RT | Diluted antibodies can be reused several times if stored at 4 °C in the presence of 0.02% sodium azide. |
| NOTE: Dilution need to be optimized as it varies between manufacturers. | ||||
Table 1: Specifications of the antibodies used in the immunoblotting procedure.
- Incubate the membrane with the secondary antibody listed in Table 1. Wash the membranes for 3 min in 1x TBST washing buffer. Repeat the wash 2x.
- Scan the blot using an appropriate gel documentation system.
Representative Results
The immunoblot (IB) with an anti-IRF5 antibody was performed on CAL-1 cells unstimulated or stimulated with 1 µg/mL R848 for 2 h (Figure 1). Cell lysates were prepared, and the native PAGE was performed. In unstimulated CAL-1 cells, IRF5 was detected as a single band on the native PAGE, corresponding to its monomeric form. Upon treatment of CAL-1 cells with R848 for 2 h, the level of IRF5 monomer decreased with a concurrent increase in the accumulation of a slowly migrating band that corresponded to the dimeric form of IRF5.
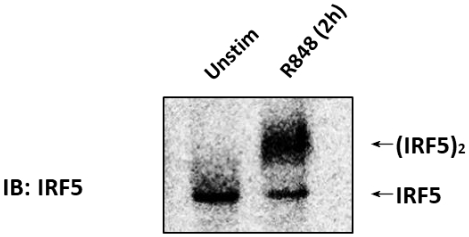
Figure 1: Endogenous IRF5 dimerized in CAL-1 cells when stimulated with TLR7/8 agonist. CAL-1 cells were untreated or treated with R848 for the indicated time. Protein samples were resolved by native PAGE and followed by IB using the anti-IRF5 antibody. Please click here to view a larger version of this figure.
The immunoblot with anti-IRF5 antibody was performed on IRF5-overexpressing 293T cells untransfected and transfected with various constructs. Cell lysates were prepared and native PAGE was performed (Figure 2). No IRF5 was detected in untransfected 293T cells, demonstrating the specificity of the anti-IRF5 antibody. A single band corresponding to monomeric IRF5 was only detected in the 293T cells overexpressing IRF5. When constructs encoding IRF5-activating proteins, including NRIG (constitutively active RIG-I), MAVS, and IKKβ were cotransfected, a slowly migrating band corresponding to the dimeric form of IRF5 appeared. However, NMDA5 (constitutively active MDA5), a related protein to RIG-I, did not induce IRF5 dimerization when cotransfected.
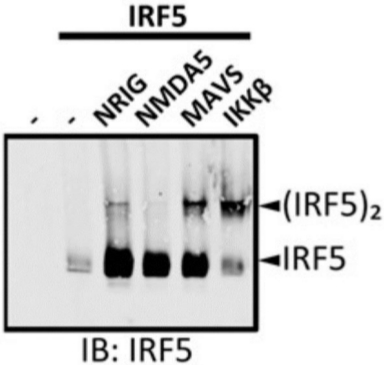
Figure 2: Cotransfection of IRF5-activating factors induced IRF5 dimerization in 293T cells. The 293T cells were untransfected (lane 1) or transfected with IRF5 (lane 2) along with various IRF5 regulators (lanes 3−6). Protein samples were resolved by native PAGE and followed by IB using the anti-IRF5 antibody. NRIG = N-terminal of RIG-I; NMDA5 = N-terminal of MDA5. (Originally published in The Journal of Immunology. KT Chow, C Wilkins, M Narita, R Green, M Knoll, YM Loo and M Gale Jr. 2018. Differential and Overlapping Immune Programs Regulated by IRF3 and IRF5 in Plasmacytoid Dendritic Cells. J. Immunol. 201 (10) 3036-3050. Copyright © 2018 The American Association of Immunologists, Inc.23). Please click here to view a larger version of this figure.
Discussion
The protocol described here is a modified native PAGE that distinguishes both monomeric and dimeric forms of endogenous IRF5. There have been few studies reporting the detection of endogenous IRF5 activation using the specialized imaging flow cytometry technique23,27,28,30. This protocol uses a common technique and commonplace reagents and tools to assess the endogenous IRF5 activation state during the early events of activation. The protocol entails simple modifications to a standard native PAGE protocol to enable distinction between the monomeric and dimeric forms of IRF5. It can be easily adapted to studies using other cell lines23. This modified native PAGE protocol can resolve endogenous IRF5 in its two forms clearly without non-specific protein interferences (Figure 2). Endogenous IRF5 from unstimulated cells was detected as a clear single band in this gel system, whereas treatment with R848 for 2 h resulted in the appearance of a band corresponding to IRF5 dimers (Figure 1).
A native PAGE dimerization assay for IRF3, a similar transcription factor to IRF5 in the same family, has been developed and widely used in the past two decades32. Despite extensive testing and troubleshooting, we were unable to apply the same protocol that employs the Laemmli Tris-glycine system to resolve IRF5 monomer and dimer. The protocol described in this article uses Bis-Tris gradient gels, which have a very different chemistry to the Tris-glycine single percentage gels used in the IRF3 protocol. The different pH and chemical composition of the gel electrophoretic systems may be crucial in distinguishing the various forms of IRF3 and IRF5. Indeed, IRF3 and IRF5, while similar, have different properties (e.g., isoelectric points and modification sites) likely resulting in different behavior while being separated on different gel systems.
A 1x native PAGE running buffer containing DOC was used for the gel run. The buffer needs to be prepared fresh or kept in a clean and protein-free environment to avoid the appearance of white precipitates clouding the solution in the upper chamber as a result of DOC precipitating non-specific proteins. It is highly recommended that the extracted endogenous IRF5 samples be subjected to the native PAGE as soon as possible, preferably within a week with minimal freeze-thaw cycles. Otherwise, there may be significant protein degradation. The degradation is observed with storage at both -80 °C and -20 °C. In addition, the pH of the SDS running buffer and the TBST washing buffer should be adjusted at RT.
The ideal final volume of sample loaded in each well was 10−15 µL, but slight adjustments might be required depending on different cell types. After the initial run at 85 V for 30 min, it is recommended to continue running the gel at 150 V for approximately 2 to 3 h to attain distinct separation and resolution of the IRF5 monomer and dimer. After the run is finished, it is of utmost importance to handle the gel meticulously from its bottom end due to its differential levels of density, ranging from 3% at the top and increasing towards 12% at the bottom. In this case, it is preferable to remove the gel from the plate by immersing it in the used running buffer, which acts as an impact cushion to minimize friction and allows the gel to float away from the plate to avoid breakage.
A few minor drawbacks of this protocol include the limited selection of gels available to achieve the desired results. Homemade gels and a few other brands of commercially available gels have been tested without success. In our hands, use of a commercial running buffer and gel system contributed to the robustness and reproducibility of this protocol, although extensive testing of homemade buffers has not been performed. Attention to detail is essential, and experience is key to success in obtaining clear results. Lastly, the resolution of IRF5 required a long time (i.e., 2−3 h) to get an ideal separation of the monomer and dimer. Further enhancement and modifications in the future can improve the efficiency and minimize the drawbacks of this protocol.
In conclusion, this protocol is a robust assay for the detection of endogenous IRF5 monomer and dimer. It is suitable for applications in various human and murine cell types expressing endogenous IRF5. It will be a valuable tool to study the IRF5 regulatory pathways and signaling components in various cell types.
Açıklamalar
The authors have nothing to disclose.
Acknowledgements
The work was supported by funding from the Croucher Foundation and City University startup funds. We thank all members of the Chow laboratory for help with the experiment and critical reading of the manuscript.
Materials
| 2-Mercaptoethanol | Life Technologies, HK | 21985023 | |
| 300 W/250 V power supply 230 V AC | Life Technologies, HK | PS0301 | |
| Anti-IRF5 antibody | Bethyl Laboratories, USA | A303-385 | |
| BIOSAN Rocker Shaker (cold room safe) | EcoLife, HK | MR-12 | |
| EDTA Buffer, pH 8, 0.5 M 4 X 100 mL | Life Technologies | 15575020 | |
| Glycerol 500 mL | Life Technologies | 15514011 | |
| Glycine | Life Technologies, HK | 15527013 | |
| Goat anti-Mouse IgG DyLight 800 Conjugated Antibody | LAB-A-PORTER/Rockland, HK | 610-145-002-0.5 | |
| Goat anti-Rabbit IgG DyLight 800 Conjugated Antibody | LAB-A-PORTER/Rockland, HK | 611-145-002-0.5 | |
| Halt protease inhibitor cocktail (100x) | Thermo Fisher Scientific, HK | 78430 | |
| HEPES | Life Technologies, HK | 15630080 | |
| LI-COR Odyssey Blocking Buffer (TBS) | Gene Company, HK | 927-50000 | |
| Mini Tank blot module combo; Transfer module, accessories | Life Technologies, HK | NW2000 | |
| NativePAGE 3-12% gels, 10 well kit | Life Technologies, HK | BN1001BOX | |
| NativePAGE Running Buffer 20x | Life Technologies, HK | BN2001 | |
| NativePAGE Sample Buffer 4x | Life Technologies, HK | BN2003 | |
| NP-40 Alternative, Nonylphenyl Polyethylene Glycol | Tin Hang/Calbiochem, HK | #492016-100ML | |
| PBS 7.4 | Life Technologies, HK | 10010023 | |
| Polyvinylidene difluoride (PVDF) membrane | Bio-gene/Merck Millipore, HK | IPFL00010 | |
| Protein assay kit II (BSA) | Bio-Rad, HK | 5000002 | |
| R848 | Invivogen, HK | tlrl-r848 | |
| RPMI 1640 | Life Technologies, HK | 61870127 | |
| Sodium Chloride | ThermoFisher | BP358-1 | |
| Sodium deoxycholate ≥97% (titration) | Tin Hang/Sigma, HK | D6750-100G | |
| Tris | Life Technologies, HK | 15504020 | |
| TWEEN 20 | Tin Hang/Sigma, HK | #P9416-100ML |
Referanslar
- Takaoka, A., et al. Integral role of IRF-5 in the gene induction programme activated by Toll-like receptors. Nature. 434 (7030), 243-249 (2005).
- Ren, J., Chen, X., Chen, Z. J. IKKbeta is an IRF5 kinase that instigates inflammation. Proceedings of the National Academy of Sciences of the United States of America. 111 (49), 17438-17443 (2014).
- Negishi, H., Taniguchi, T., Yanai, H. The Interferon (IFN) Class of Cytokines and the IFN Regulatory Factor (IRF) Transcription Factor Family. Cold Spring Harbor Perspective Biology. 10 (11), (2018).
- Clark, D. N., et al. Four Promoters of IRF5 Respond Distinctly to Stimuli and are Affected by Autoimmune-Risk Polymorphisms. Frontiers in Immunology. 4, 360 (2013).
- Bo, M., et al. Rheumatoid arthritis patient antibodies highly recognize IL-2 in the immune response pathway involving IRF5 and EBV antigens. Scientific Reports. 8 (1), 1789 (2018).
- Duffau, P., et al. Promotion of Inflammatory Arthritis by Interferon Regulatory Factor 5 in a Mouse Model. Arthritis and Rheumatolpgy. 67 (12), 3146-3157 (2015).
- Feng, D., et al. Irf5-deficient mice are protected from pristane-induced lupus via increased Th2 cytokines and altered IgG class switching. European Journal of Immunology. 42 (6), 1477-1487 (2012).
- Richez, C., et al. IFN regulatory factor 5 is required for disease development in the FcgammaRIIB-/-Yaa and FcgammaRIIB-/- mouse models of systemic lupus erythematosus. The Journal of Immunology. 184 (2), 796-806 (2010).
- Tada, Y., et al. Interferon regulatory factor 5 is critical for the development of lupus in MRL/lpr mice. Arthritis and Rheumatology. 63 (3), 738-748 (2011).
- Weiss, M., et al. IRF5 controls both acute and chronic inflammation. Proceedings of the National Academy of Sciences of the United States of America. 112 (35), 11001-11006 (2015).
- Schoenemeyer, A., et al. The interferon regulatory factor, IRF5, is a central mediator of toll-like receptor 7 signaling. Journal of Biological Chemistry. 280 (17), 17005-17012 (2005).
- Balkhi, M. Y., Fitzgerald, K. A., Pitha, P. M. Functional regulation of MyD88-activated interferon regulatory factor 5 by K63-linked polyubiquitination. Molecular and Cellular Biology. 28 (24), 7296-7308 (2008).
- Lopez-Pelaez, M., et al. Protein kinase IKKβ-catalyzed phosphorylation of IRF5 at Ser462 induces its dimerization and nuclear translocation in myeloid cells. Proceedings of the National Academy of Sciences of the United States of America. 111 (49), 17432-17437 (2014).
- McGettrick, A. F., O’Neill, L. A. Localisation and trafficking of Toll-like receptors: an important mode of regulation. Current Opinion Immunology. 22 (1), 20-27 (2010).
- Baccala, R., Hoebe, K., Kono, D. H., Beutler, B., Theofilopoulos, A. N. TLR-dependent and TLR-independent pathways of type I interferon induction in systemic autoimmunity. Nature Medicine. 13 (5), 543-551 (2007).
- Gilliet, M., Cao, W., Liu, Y. J. Plasmacytoid dendritic cells: sensing nucleic acids in viral infection and autoimmune diseases. Nature Reviews Immunology. 8 (8), 594-606 (2008).
- Kawai, T., Akira, S. Toll-like Receptors and Their Crosstalk with Other Innate Receptors in Infection and Immunity. Immunity. 34 (5), 637-650 (2011).
- Liu, Z., Davidson, A. Taming lupus-a new understanding of pathogenesis is leading to clinical advances. Nature Medicine. 18 (6), 871-882 (2012).
- del Fresno, C., et al. Interferon-beta production via Dectin-1-Syk-IRF5 signaling in dendritic cells is crucial for immunity to C. albicans. Immunity. 38 (6), 1176-1186 (2013).
- Wang, X., et al. Expression Levels of Interferon Regulatory Factor 5 (IRF5) and Related Inflammatory Cytokines Associated with Severity, Prognosis, and Causative Pathogen in Patients with Community-Acquired Pneumonia. Medical Science Monitor. 24, 3620-3630 (2018).
- Zhao, Y., et al. Microbial recognition by GEF-H1 controls IKKepsilon mediated activation of IRF5. Nature Communications. 10 (1), 1349 (2019).
- Almuttaqi, H., Udalova, I. A. Advances and challenges in targeting IRF5, a key regulator of inflammation. FEBS Journal. 286 (9), 1624-1637 (2019).
- Chow, K. T., et al. Differential and Overlapping Immune Programs Regulated by IRF3 and IRF5 in Plasmacytoid Dendritic Cells. The Journal of Immunology. 201 (10), 3036-3050 (2018).
- Cheng, T. F., et al. Differential Activation of IFN Regulatory Factor (IRF)-3 and IRF-5 Transcription Factors during Viral Infection. The Journal of Immunology. 176 (12), 7462-7470 (2006).
- Chang Foreman, H. C., Van Scoy, S., Cheng, T. F., Reich, N. C. Activation of interferon regulatory factor 5 by site specific phosphorylation. PLoS One. 7 (3), 33098 (2012).
- Lin, R., Yang, L., Arguello, M., Penafuerte, C., Hiscott, J. A CRM1-dependent nuclear export pathway is involved in the regulation of IRF-5 subcellular localization. Journal of Biological Chemistry. 280 (4), 3088-3095 (2005).
- Stone, R. C., et al. Interferon regulatory factor 5 activation in monocytes of systemic lupus erythematosus patients is triggered by circulating autoantigens independent of type I interferons. Arthritis and Rheumatology. 64 (3), 788-798 (2012).
- De, S., et al. B Cell-Intrinsic Role for IRF5 in TLR9/BCR-Induced Human B Cell Activation, Proliferation, and Plasmablast Differentiation. Frontiers in Immunology. 8, 1938 (2017).
- Fabie, A., et al. IRF-5 Promotes Cell Death in CD4 T Cells during Chronic Infection. Cell Reports. 24 (5), 1163-1175 (2018).
- Cushing, L., et al. IRAK4 kinase activity controls Toll-like receptor-induced inflammation through the transcription factor IRF5 in primary human monocytes. Journal of Biological Chemistry. 292 (45), 18689-18698 (2017).
- Li, C., Arakawa, T. Application of native polyacrylamide gel electrophoresis for protein analysis: Bovine serum albumin as a model protein. International Journal of Biological Macromolecules. 125, 566-571 (2019).
- Iwamura, T., et al. Induction of IRF-3/-7 kinase and NF-kappaB in response to double-stranded RNA and virus infection: common and unique pathways. Genes to Cells. 6 (4), 375-388 (2001).
- Subhadarshanee, B., Mohanty, A., Jagdev, M. K., Vasudevan, D., Behera, R. K. Surface charge dependent separation of modified and hybrid ferritin in native PAGE: Impact of lysine 104. Biochimica et Biophysica Acta – Proteins and Proteomics. 1865 (10), 1267-1273 (2017).
- Reynolds, J. A., Tanford, C. Binding of Dodecyl Sulfate to Proteins at High Binding Ratios – Possible Implications for State of Proteins in Biological Membranes. Proceedings of the National Academy of Sciences of the United States of America. 66 (3), 1002 (1970).
- Manning, M., Colon, W. Structural basis of protein kinetic stability: resistance to sodium dodecyl sulfate suggests a central role for rigidity and a bias toward beta-sheet structure. Biyokimya. 43 (35), 11248-11254 (2004).
- Balkhi, M. Y., Fitzgerald, K. A., Pitha, P. M. IKKalpha negatively regulates IRF-5 function in a MyD88-TRAF6 pathway. Cellular Signalling. 22 (1), 117-127 (2010).
- Paun, A., et al. Functional characterization of murine interferon regulatory factor 5 (IRF-5) and its role in the innate antiviral response. Journal of Biological Chemistry. 283 (21), 14295-14308 (2008).
- Yasuda, K., et al. Murine dendritic cell type I IFN production induced by human IgG-RNA immune complexes is IFN regulatory factor (IRF)5 and IRF7 dependent and is required for IL-6 production. The Journal of Immunology. 178 (11), 6876-6885 (2007).
- Steinhagen, F., et al. IRF-5 and NF-kappaB p50 co-regulate IFN-beta and IL-6 expression in TLR9-stimulated human plasmacytoid dendritic cells. European Journal of Immunology. 43 (7), 1896-1906 (2013).
- Gratz, N., et al. Type I interferon production induced by Streptococcus pyogenes-derived nucleic acids is required for host protection. PLoS Pathogens. 7 (5), 1001345 (2011).

