Preparation and Characterization of Nanoliposomes for the Entrapment of Bioactive Hydrophilic Globular Proteins
Özet
This study describes classical hydration using the thin lipid film method for nanoliposome preparation followed by nanoparticle characterization. A 47 kDa-hydrophilic and globular protein, tarin, is successfully encapsulated as a strategy to improve stability, avoid fast clearance, and promote controlled release. The method can be adapted to hydrophobic molecules encapsulation.
Abstract
Liposome nanocapsules have been applied for many purposes in the pharmaceutical, cosmetic, and food industries. Attributes of liposomes include their biocompatibility, biodegradability, non-immunogenicity, non-toxicity, and ability to entrap both hydrophilic and hydrophobic compounds. The classical hydration of thin lipid films in an organic solvent is applied herein as a technique to encapsulate tarin, a plant lectin, in nanoliposomes. Nanoliposome size, stability, entrapment efficiency, and morphological characterization are described in detail. The nanoliposomes are prepared using 1,2-dioleoyl-sn-glycerol-3-phosphoethanolamine (DOPE), 1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N-[amino(polyethylene glycol)-2000] (ammonium salt; DSPE-MPEG 2000), and cholesterylhemisuccinate (CHEMS) as the main constituents. Lipids are first dissolved in chloroform to obtain a thin lipid film that is subsequently rehydrated in ammonium sulfate solution containing the protein to be entrapped and incubated overnight. Then, sonication and extrusion techniques are applied to generate nanosized unilamellar vesicles. The size and polydispersity index of the nanovesicles are determined by dynamic light scattering, while nanovesicle morphology is assessed by scanning electron microscopy. Entrapment efficiency is determined by the ratio of the amount of unencapsulated protein to original amount of initially loaded protein. Homogeneous liposomes are obtained with an average size of 155 nm and polydispersity index value of 0.168. A high entrapment efficiency of 83% is achieved.
Introduction
The number of studies investigating efficient drug delivery systems has risen in recent years. However, limitations such as rapid clearance, poor biodistribution, and solubility at physiological pH and insufficient cellular uptake still need to be surpassed. The use of nanosystems has emerged as recent progress in cancer therapeutics, applied to increase the intracellular concentration of drugs inside cancerous cells while minimizing toxicity in healthy cells. Moreover, nanoparticles obtained from a different range of materials (i.e., polymers, dendrimers, liposomes, viruses, carbon nanotubes, and metals such as iron oxide and gold) are currently being applied to enhance anticancer effects and reduce systemic toxicity1. Liposome nanocapsules in particular have been applied for many purposes in the pharmaceutical, cosmetic, and food industries. In recent years, various nutraceutical products such as vitamins, enzymes, and herbal extracts have been formulated using liposome technology2.
Liposomes are spherical vesicles consisting of one or more concentric lipid bilayers spontaneously formed by the dispersion of phospholipids in aqueous media3,4. The polar heads of the phospholipids are located on the outer and inner surfaces of the membranes, in contact with the aqueous environment. In contrast, fatty acid chains form the hydrophobic core of the membranes and are protected from water5. Some attributes of liposomes that make them attractive drug delivery systems include their biocompatibility, biodegradability, non-immunogenicity, non-toxicity, and ability to entrap both hydrophilic and hydrophobic compounds6.
Liposomes can be prepared using various processes steps such as agitation, sonication, extrusion, lyophilization, freezing, and thawing. Classical methods include reverse phase evaporation, solvent injection, and detergent dialysis. The most applied method is thin lipid film hydration, also known as Bangham's method, which is used to obtain vesicular-lipid forms7,8,9,10,11. Lamellarity (the number of phospholipid bilayers) and particle size are classical parameters used to characterize liposomes as either 1) unilamellar vesicles (ULVs), formed by a unique phospholipid bilayer and varying in size as follows: i) small unilamellar vesicles (SUVs, ~0.02-0.20 µm), ii) large unilamellar vesicles (LUVs, ~0.2-1.0 µm), and iii) giant unilamellar vesicles (GUVs, >1 µm); or 2) multilamellar vesicles (MLVs, >0.1 µm)3,12. Vesicle size is an important parameter when considering for therapeutic use, such as in cancer treatment, in which sizes of <200 nm are ideal to allow nanovesicles to cross the endothelial barrier and reach tumoral tissues4.
Herein, the encapsulation procedure following the classical hydration of a thin lipid film technique7 was described using tarin, a plant lectin characterized as a hydrophilic globular protein13,14,15. Nanosized vesicles are produced by including sonication and extrusion steps in the main technique, resulting in stable liposomal nanovesicles with high entrapment efficiency16.
Protocol
1. Preparation of tarin liposomal nanocapsules16
NOTE: All preparations should be prepared in triplicate in order to obtain a larger volume (7 mL) and enable the sample to be centrifuged in an ultracentrifuge (see details below).
- Weigh the liposome components using an analytical balance, as shown in Table 1.
- Dissolve the lipid components in chloroform using a 250 mL volumetric flask that fits in a rotary evaporator to avoid loss of material.
- Stir the mixture at 150 rpm for 15 min.
- Remove the chloroform using a rotary evaporator under the following conditions:
- Adjust the volumetric flask mouth to the standard position (25°) for optimal efficiency, while in contact with the water from the heating bath.
NOTE: The equipment arm should be inclined at 25° to maintain contact between the volumetric flask and water bath, while not affecting evaporation efficiency or damaging the sample. The standard position can vary according to the equipment brand. - Set the condenser temperature to a minimum of 3 °C.
- Set the heating bath temperature to 40 ± 1 °C.
- Set the rotation to 120 rpm.
- Adjust the vacuum to 207 mbar and boiling point to 20 °C.
- After ~25 min, remove the flask and discard the evaporated solvent, remaining in the condenser, appropriately.
NOTE: A thin and opaque film, consisting of the liposome components, is formed in this step and can be easily visualized. The evaporated solvent remaining in the condenser must be stored into disposal recipients (chlorinated) to be handled by a specialized company for appropriate discarding.
- Adjust the volumetric flask mouth to the standard position (25°) for optimal efficiency, while in contact with the water from the heating bath.
- Hydrate the lipid film to reach a 0.01 M lipid concentration in 0.3 M ammonium sulfate solution (pH = 7.4) containing tarin at 1 mg/mL to a final volume of 10 mL.
- Stir the mixture for 40 min and incubate overnight at 4 °C.
NOTE: This step can be considered a stop point. Overnight incubation is not obligatory.
- Stir the mixture for 40 min and incubate overnight at 4 °C.
- After incubation, sonicate the suspension for 1 min at 25 °C (room temperature; RT) to reduce vesicle size and avoid aggregation.
NOTE: Size reduction was performed in an ultrasonic sonicator under the following conditions: 130 W and 40 kHz. - Perform a 12-cycle extrusion through a 0.2 µm polycarbonate pore membrane.
NOTE: Before the extrusion process, test the mini-extruder assembly using water to avoid sample leakage. A 0.1 µm pore membrane is also suitable. In this case, pre-heat the mini-extruder holder above the lipid transition temperature to facilitate extrusion, while maintaining the physicochemical characteristics of both lipids and proteins.- Fit the parts of the mini extruder, as described in the manufacturer's manual.
- Place the polycarbonate membrane between two pre-wet filter supports and place it into the holder.
- Insert a 1 mL empty syringe into the device, fill the other syringe to its total volume with the liposomal suspension, and insert it on the opposite side.
- Perform a 12-cycle extrusion through a 0.2 µm polycarbonate pore membrane. Push the sample from one syringe to another, slowly. Collect the extruded suspension in a pre-cooled tube.
NOTE: The polycarbonate membrane should be replaced only when sample transference from one syringe to another becomes difficult. The liposomal suspension should become clear during the extrusion process as a result of size reduction due to the formation of SUVs. About 0.2 mL of the sample can be lost during this step.
- Separate liposomes by ultracentrifugation.
NOTE: Maintain the samples in an ice bath until the ultracentrifuge is ready to be used. Separate SUVs from the remaining components and ammonium sulfate by ultracentrifugation using an ultracentrifuge with a swing-bucket rotor (see details below).- Weigh the sample in the titanium tube that fits the rotor and balance the tubes. Check the minimum volume required according to the rotor used to avoid damaging the tubes, and adjust the liposomal suspension volume with ammonium sulphate, if necessary.
NOTE: The swing bucket should always be supported on the stand when outside of the centrifuge to avoid scratching the "zebra stripes" (i.e., the black and white stripes on the bottom), which are used by the centrifuge to determine rotation speed. - Turn on the vacuum before using the centrifuge to allow it to refrigerate.
- Fit the titanium tubes into the swing bucket.
NOTE: Lift the tubes to the position they assume when running to ensure that they are perfectly fitted. - Release the vacuum, open the centrifuge door, and place the rotor inside.
NOTE: Pay attention to the circle mark on the bottom of the rotor, which must fit in the opposite direction of the same circle mark into the centrifuge itself. - Close the centrifuge door, press Vacuum and wait until the vacuum reaches from 200 to <20 microns or from 26 Pa to <3 Pa.
- Adjust the parameters in the ultracentrifuge display to 150,000 x g (the equivalent to 29,600 rpm for the aforementioned swing bucket) for 90 min at 4 °C (acceleration: max, deceleration: max).
NOTE: Always convert the speed to x g if the specific centrifuge is set in rpm. Use the centrifuge website to convert the unit according to the rotor used. - Press Recall, check the conditions, and press Start to run.
NOTE: Wait until the centrifuge reaches the desired speed. - After 90 min, release the vacuum by pressing the Vacuum button and open the centrifuge door when the vacuum reaches 200-700 microns (equivalent to 26-93 Pa).
- Switch off the centrifuge, remove the rotor from the inside, and leave it on the bench with the buckets to dry.
- Maintain the ultracentrifuged samples on ice.
- Weigh the sample in the titanium tube that fits the rotor and balance the tubes. Check the minimum volume required according to the rotor used to avoid damaging the tubes, and adjust the liposomal suspension volume with ammonium sulphate, if necessary.
- Carefully, separate supernatant from the pellet by turning the tube upside down into a disposable 15 mL centrifuge tube to separate the supernatant and pellet.
NOTE: Store the supernatant containing the unencapsulated protein at 4 °C. It will be used to determine the encapsulation efficiency. The pellet appears as a translucent jelly. - Suspend the pellet containing the encapsulated protein in HEPES buffered saline (3 mL of 1x HBS).
NOTE: The HBS 2x (stock solution) is prepared by diluting the following amounts of reagents in distilled water: 140 mM NaCl, 1.5 mM Na2HPO4, 50 mM HEPES, then adjusting the pH to 7.4 and final volume to 100 mL. Na2HPO4 can be replaced by NaHCO3or omitted to avoid interference if liposome concentration is to be determined. The HBS 2x stock solution should be diluted in distilled water to obtain HBS 1x before use.
2. Encapsulation efficiency
NOTE: Determine the encapsulation efficiency using Peterson's protocol17in order to avoid lipid interference in protein quantification. All samples (BSA standards and liposome supernatant) should be analyzed in triplicate. Also prepare a blank tube.
- Preparation of stock reagents and working solutions17
- For stock reagents, prepare copper-tartrate-carbonate (CTC) by mixing 10 mL of 20% sodium carbonate, 200 µL of 0.1% copper sulfate, 200 µL of 0.2% potassium tartrate with 9.6 mL of distilled water. Prepare 100 mL of 10% sodium dodecyl sulfate (SDS) and 0.8 N sodium hydroxide (NaOH).
- For working solutions, prepare 10 mL of 0.15% sodium deoxycholate (DOC) and 72% trichloroacetic acid (TCA). Dissolve 10 mg of bovine serum albumin (BSA) in 10 mL of distilled water to obtain a 1 mg/mL standard solution. Prepare reagent A by adding equal parts of CTC, NaOH, SDS, and H2O. Prepare reagent B by diluting Folin-Ciocalteu phenol reagent 1:5 in distilled water.
NOTE: Reagent A requires 1 mL for each reaction tube, while reagent B requires 0.5 mL. To determine the final volumes of reagents A and B, first define the number of reaction tubes to be used, considering three distinct concentrations of BSA, blank, and samples in triplicate. Reagent A must be well-homogenized before use and can be stored at 25 °C (RT) for 2 weeks. Reagent B is also stable at 25 °C (RT) if stored in an amber bottle.
- Precipitation
NOTE: This step is performed in microcentrifuge tubes.- Dilute the liposome supernatant with water to a final volume of 1 mL containing 5-100 µg of protein.
NOTE: The blank tube should be filled with 1 mL of distilled water. - Add 0.1 mL of 0.15% DOC, homogenize by vortexing, and incubate for 10 min at RT.
- Add 0.1 mL of 72% TCA, mix well, and centrifuge at 3,000 x g and RT for 15 min.
NOTE: DOC-TCA promotes protein precipitation, forming two distinct phases. The target protein can be recovered by centrifugation, avoiding lipid interference. - Carefully discard the supernatant by verting the tube downwards and laying it on an absorbent paper. Save the pellet for the subsequent step.
NOTE: The pellet can be very difficult to see, but the tube should be turned upside down even if it is not visible.
- Dilute the liposome supernatant with water to a final volume of 1 mL containing 5-100 µg of protein.
- Spectrophotometry
- Suspend the pellet obtained from step 2.2.4 in 1 mL of distilled water. Mix thoroughly to make sure the pellet is dissolved and transfer the sample to a new test tube.
- Prepare dilutions of albumin (BSA) standards to a final volume of 1 mL.
NOTE: Protein standards must be prepared between 5-100 µg/mL. - Add 1 mL of reagent A to the tubes from step 2.3.1 and 2.3.2 without exception, mix well, and incubate for 10 min at RT.
NOTE: SDS can relieve possible lipid interferences while aiding in protein solubilization. - Add 0.5 mL of reagent B to the tubes from step 2.3.1 and 2.3.2, mix well, and incubate for 30 min at RT while protected from the light.
NOTE: The Follin-Ciocalteu phenol reagent is a mixture of phosphomolybdate and phosphotungstate used for colorimetric assays of some nitrogen-containing compounds, such as proteins. Copper complexation increases the reactivity of phenols towards this reagent, producing a blue/purple complex according to protein concentration. - Determine absorbances at 750 nm using a spectrophotometer.
- Calculate the protein concentration in the supernatant based on the standard curve as follows.
- Plot absorbance value (Abs) vs. BSA concentration (mg/mL) to obtain the angular coefficient (k) considering a linear tendency line.
- Determine the supernatant protein concentration (C) by the ratio between the absorbance value and angular coefficient (k), then multiply by the total volume as follows:
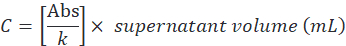
- Determine encapsulation efficiency according to the following formula:
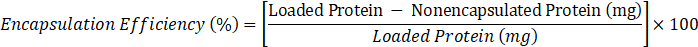
- where loaded protein = 10 mg, nonencapsulated protein = value of C obtained in step 2.3.6.2.
NOTE: In this case, a total of 10 mg tarin dissolved in ammonium sulfate solution (1 mg/mL) is used to perform the encapsulation procedure, since this concentration is sufficient to obtain satisfactory in vitro effects13,16,18.
3. Size and stability determination
NOTE: Size distribution and polidispersity index (PdI) of the liposomal preparations are evaluated by dynamic light scattering (DLS). A PdI close to 0.1 indicates a homogeneous preparation. For stability determination, store liposomes at 4 °C and check size distribution and size average regularly.
- Turn on the DLS equipment 30 min before use to warm up the laser lamp.
- Transfer the liposomal preparation obtained in step 1.10 to a disposable sizing cuvette.
- Set the equipment parameters as follows: dispersant type = water (RI = 1.33); material = lipids (RI = 1.45); and RT.
- Press Start and wait while the equipment finishes its reading.
- Remove the cuvette and turn off the equipment.
NOTE: Either transfer the sample from the cuvette back to the disposable 15 mL centrifuge tube for subsequent analyses, or discard it if there is a sufficient amount for new reads.
4. Morphological characterization
NOTE: Liposome characterization is performed according to Murtey and Ramasamy19. Samples containing nanoliposomes obtained in step 1.10 are prepared in triplicate.
- Fix the glass coverslips (13 mm diameter) in the bottom of a Petri dish with double-sided tape. Cut the tape into small pieces (appropriate size to fix the coverslips), remove the protective paper underneath, and fix it on the bottom of the Petri dish. With the aid of tweezers, remove the protective paper on top of the tape and fix the coverslips on it.
NOTE: Be careful in the following steps not to release the coverslips, and use a strong tape. - Coat the coverslips with poly-L-lysine. Place wet filter papers inside the Petri dish to maintain moisture and incubate for 1 h at RT (25 °C).
- After coating, rinse the coverslips with distilled water.
- Fill the coverslips with a drop of the sample from step 1.10 and allow them to dry for 1 h at RT.
- To fix the samples, cover them with 4% glutaraldehyde prepared in 0.1 M phosphate buffer, pH = 7.2. Place wet filter papers inside the Petri dish and seal the dish to maintain moisture levels. Incubate at 4 °C for 48 h.
- Rinse the coverslips 3x for 5 min with the same phosphate buffer.
- Dehydrate samples as follows: 35% ethanol 1x for 15 min, 50% ethanol 1x for 15 min, 75% ethanol 1x for 15 min, 95% ethanol 2x for 15 min, and absolute ethanol 3x for 20 min.
- Chemically dry the samples by immersion 2x in 1−2 mL of hexamethyldisilazane (HMDS) for 10 min.
NOTE: The HMDS should be manipulated carefully inside a fume hood. Samples should be allowed to dry overnight inside a desiccator or inside the fume hood at RT. - Mount the dried samples on a stub with a carbon conductive adhesive tape.
- Sputter the surface of the coverslip in a vacuum with an electrically conductive layer (20 nm thickness) of gold-palladium.
- Record images with a scanning electron microscope (SEM) at low vacuum mode and low voltage (20 kV).
Representative Results
Figure 1 describes the nanoliposome preparation16,20,21. Phospholipids, 1,2-dioleoyl-sn-glycerol-3-phosphoethanolamine (DOPE), 1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N-[amino(polyethylene glycol)-2000] (ammonium salt; DSPE-MPEG 2000), and cholesterylhemisuccinate (CHEMS), the main liposome constituents, were first dissolved in chloroform to obtain the lipid film. The lipid film was then rehydrated in ammonium sulfate solution containing the hydrophilic protein (tarin) to be entrapped, and the incubation was performed overnight. Then, sonication and extrusion techniques were applied to generate small unilamellar vesicles. The ultracentrifugation step separated the liposomal preparation from free lipids and unencapsulated protein, while the supernatant was used for the determination of entrapment efficiency.
Nanoliposomes produced using the aforementioned methodology exhibited a size distribution ranging from 51−396 nm and an average size of 155 nm (Table 2). The preparation was homogeneous, since the polydispersity index was 0.168. A high entrapment efficiency of 83% can be reached if the liposomes are extruded through a 0.2 µm pore size membrane (Table 2).
Morphological nanoliposome characteristics were evaluated by SEM. Figure 2A,B displays round-shaped liposomal vesicles in the range of 121 nm and analyzed at 20 kV, whereas Figure 2C,D displays inadequately prepared samples. Nanoliposomes were simply air dried without previous fixation or any other treatment described in this study. As a result, larger and damaged vesicles in the range of 332 µm and analyzed at 5 kV were observed.
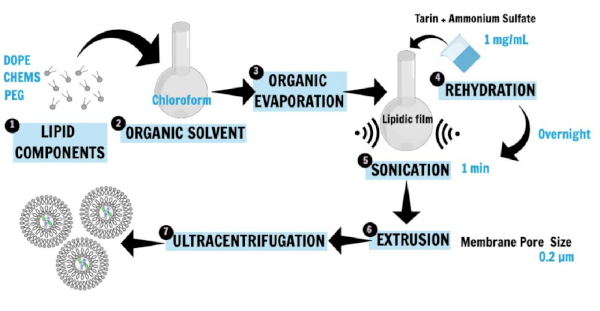
Figure 1: Schematic representation of nanoliposome preparation. DOPE, PEG, and CHEMS, the main liposome constituents, were first dissolved in chloroform to obtain the lipid film (1, 2, 3). The lipid film was then rehydrated in a saline buffer containing the hydrophilic protein (tarin) to be entrapped, and the incubation was performed overnight (4). Then, sonication and extrusion techniques were applied to generate SUVs (5, 6). The ultracentrifugation step separated the liposomal preparation from free lipids and unencapsulated protein, while the supernatant was used for the determination of entrapment efficiency (7). This figure has been modified from Correa et al.16. Please click here to view a larger version of this figure.
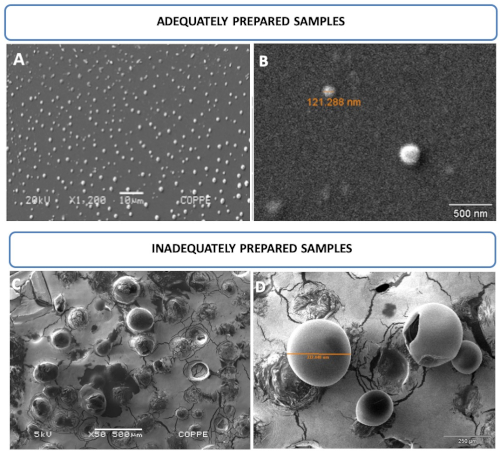
Figure 2: Nanoliposome photomicroscopy by SEM. (A,B) Images of round-shaped liposomal vesicles in the range of 121 nm and analyzed at 20 kV. (C,D) Images of inadequately prepared samples. Mistreated samples allowed for the observation of larger and/or damaged vesicles, which cannot resist vacuum and/or voltage conditions at 5 kV. This figure has been modified from Correa et al.16. Please click here to view a larger version of this figure.
| Liposome Components | Weight (g) | Concentration (mM) | Final volume |
| DOPE | 0.0420 | 5.7 | 10 mL |
| MPEG 2000-DSPE | 0.1059 | 3.8 | |
| CHEMS | 0.0024 | 0.5 |
DOPE – 1,2-dioleoyl-sn-glycerol-3-phosphoethanolamine); MPEG 2000-DSPE – 1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N- [amino (polyethylene glycol)-2000] (ammonium salt); CHEMS – cholesterylhemisuccinate.
Table 1: Preparation of tarin liposomal nanocapsules.
| Membrane pore size (µm) | Size distribution (nm) | Average size (nm) | Polydispersity index (PdI) | Peak (nm) | Entrapment Efficiency |
| 0.2 | 51 – 396 | 155 | 0. 168 | 94 ± 39 | 0.83 |
Size and polydispersity index were evaluated by dynamic light scattering, while encapsulation efficiency was determined according to Peterson17.
Table 2: Size, polidispersity index, and entrapment efficiency of the nanoliposome preparation.
Discussion
The protocol described herein was tested by Correa et al.16 to encapsulate tarin, an immunomodulatory and antitumoral lectin purified from Colocasia esculenta22. The methodology yielded successful results, allowing for the production of stable nanoliposomes of appropriate size for therapeutic applications. The formulation presents controlled release at different pH levels under physiological conditions. It also potentiates tarin pharmacological properties, such as inhibition of human glioblastoma U-87 MG and breast cancer MDA-MB-231 cell lines and stimulation of mice bone marrow cells. The liposomal preparation exhibited no toxic effects in healthy mice cells16.
The classical method, first described by Bangham et al.7, allows for the production of large multilamellar liposome vesicles, heterogeneous in size and shape. Adaptations of this method, as reported in the present study, are successfully applied by including additional steps such as sonication and extrusion through a 0.2 µm polycarbonate membrane. This allows production of a more homogeneous dispersion regarding size in the nanometer range16,23,24. Therefore, to ensure successful results, the encapsulation protocol and liposomal formulation described here should be strictly followed.
The nanoliposome composition was carefully selected in order to ensure the formation of a bilayer membrane with DOPE, MPEG 2000-DSPE, and CHEMS as the main constituents. These are natural animal membrane bilayer constituents and the latter can confer fluidity to nanoliposome architecture, ensuring broad application for bioactive compound delivery in human beings.
Nanoliposome pegylation is essential to guarantee liposome structure stability. The absence of PEG leads to size enlargement, a high polydispersity index, and low entrapment efficiency. Optimal results can be obtained with DOPE as the main liposome component. However, this is a high-cost phospholipid. The financial costs of nanoliposome production can be achieved by replacing DOPE with other similar lipids such as DOPC (1,2-dioleoyl-sn-glycero-3-phosphocholine). CHEMS is a cholesterol molecule naturally found in animal cell membranes, which should not be excluded from the formulation, since it is important to ensure lipid bilayer fluidity and malleability16.
Other aspects of the encapsulation protocol can also be adapted. The chloroform used to dissolve the liposomal components can easily be replaced by methanol with no effects on size average, homogeneity, and entrapment efficiency. However, some protein leakage can occur at storage under 4 °C16. The overnight incubation step with ammonium sulfate solution containing tarin is not mandatory; however, for convenience it can be performed with no damage to nanoliposomal biophysical characteristics, encapsulation, or stability efficiency losses, as demonstrated by Correa et al.16. The extrusion step is performed at room temperature, which can decrease flow rate between the syringes if a 0.1 µm pore size membrane is used.
To overcome this issue, use of a 0.2 µm pore size membrane or heating of the extruder holder above the lipid transition temperature should be considered. The analyst must be careful not to damage the lipids or protein that can be inactivated and lose biological activity. Alternatively, liposomal preparation can be dialyzed against HBS instead of ultracentrifugation, using a cut-off membrane according to protein molecular weight. The choice of chemical nature of the buffer in which nanoliposomes are suspended after ultracentrifugation is directly related to its subsequent application. Since perspectives of this study include in vivo and in vitro assays, suspension in HEPES buffered saline was adequate to ensure no cytotoxic effects and a pH range close to physiological conditions.
Liposomes should be finely treated, similar to living cells, to obtain higher quality SEM images. Fixation and drying procedures are important to ensure the visualization of smaller intact vesicles that support values higher than 20 kV under vacuum conditions. Figure 2A,B displays nanosized vesicles compatible with the extrusion procedure. Visualization of vesicles ranging from 51-396 nm is possible if adequate sample preparation following this procedure is performed. The steps include fixation, drying by increasing ethanol concentrations, and chemical dehydration to avoid the formation of aggregates and ruptured vesicles caused by the vacuum and electron beam. On the other hand, Figure 2C,D shows liposome vesicles dried under room temperature and not subjected to any treatments described here, which means that they were prepared inadequately. As a result of the inadequate procedure, giant vesicles are formed, even after extrusion through a 0.2 µm pore size membrane. Ruptured vesicles are also observed in both panels as a result of vacuum and electron beam damage.
Nanoliposome vesicles have been explored as an encapsulation and delivery system for hydrophobic molecules, including resveratrol (3,5,4'-trihydroxystilbene), a bioactive compound against colorectal cancer cells. The encapsulation procedure can overcome the poor solubility of lipophilic compounds in addition to providing biocompatibility, biodegradability, non-immunogenicity, and non-toxicity characteristics inherent to liposome nanocapsules25. Protocol adaptations must be taken into consideration depending on the administration route and purpose, such as the development of new liposome formulations for oral administration.
Açıklamalar
The authors have nothing to disclose.
Acknowledgements
The authors are thankful to the COPPE/UFRJ, Electronic Microscopy Laboratory, and Multiuser Materials Characterization Laboratory facilities; to Dr. Adalberto Vieyra, Dr. Jennifer Lowe, and Rafael Lindoso, professors at Universidade Federal do Rio de Janeiro, UFRJ, Brazil, for use of the ultracentrifuge; to Dr. Alexandre Guedes Torres and Daniel Perrone, professors at the Universidade Federal do Rio de Janeiro, UFRJ, Brazil, for use of the rotary evaporator; to Professor Roland Bodmeier and Dr. Andriy Dashevskiy from Freie Universität in Berlin, who helped with resources, provided new methodologies, and supervised ACNTF during a 6 month Erasmus+ fellowship in Germany; to Dr. Rossana Thiré and Aline Fernandes, professor and technician at the Universidade Federal do Rio de Janeiro, UFRJ, Brazil, for use of Zetasizer Malvern; to Bluma Guenther and Taissa Rodrigues, professor and technician at the Universidade Federal do Rio de Janeiro, UFRJ, Brazil, for use of the SEM; to Dr. Rachel Ann Hauser Davis, researcher at Fundação Oswaldo Cruz, for narration. This study was financed in part by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior, Brasil (CAPES) – Finance Code 001 (grant No. 1627392; 1811605); by Fundação Carlos Chagas Filho de Amparo à Pesquisa do Estado do Rio de Janeiro (FAPERJ) (grant No. E-26/202.815/2018; E-26/202.815/2018; E-26/203.039/2015 and E-26/202.860/2016); by Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) (grant No. 406601/2018-6), and Financiadora de Estudos e Projetos (FINEP).
Materials
| Ammonium Sulfate | Sigma-Aldrich Co | A4418 | |
| Analitycal Ballance Mettler H10Tw | Mettler Inc. | 417870 | |
| Beckman DU-640 Spectrophotometer | Beckman Coulter | 8043-30-1090 | |
| Bovine serum albumin (BSA) | Sigma-Aldrich Co | 5470 | |
| BUCHI Rotavapor R-300 Rotary Evaporator with Controller and V-300 Pump | Thermo Fischer Scientific | 05-001-022PM | |
| CHEMS (cholesterylhemisuccinate) | Sigma-Aldrich Co | C6512 | |
| Chloroform | Sigma-Aldrich Co | 48520-U | CAUTION |
| Copper (II) Sulfate (Pentahydrate) | Sigma-Aldrich Co | 209198 | |
| Coverslips (13mm diameter) | Thermo Scientific Nunc | EW-01839-00 | |
| DOPE(1,2-dioleoyl-sn-glycerol-3-phosphoethanolamine) | Lipoid GMBH | 565600.1 | |
| Ethanol Absolute | Sigma-Aldrich Co | 32205 | |
| Folin -Ciocalteu phenol reagent | Sigma-Aldrich Co | F9252 | |
| Glutaraldehyde | Sigma-Aldrich Co | G5882 | |
| HEPES | Sigma-Aldrich Co | H3375 | |
| Hexamethyldisilazane (HMDS) | Sigma-Aldrich Co | 440191 | CAUTION |
| JEOL JSM-6460 LV Sacnning Electron Microscope | JEOL LTD | ||
| Mini Extruder 7 | Avanti Polar Lipids | 610000 | |
| MPEG 2000-DSPE 1,2-distearoyl-sn-glycero-3- phosphoethanolamine-N-[amino(polyethylene glycol)-2000] (ammonium salt) | Lipoid GMBH | 588200.1 | |
| Optima L-90k Ultracentrifuge | Beckman Coulter | PN LL-IM-12AB | |
| Phosphate Buffer | Sigma-Aldrich Co | P3619 | |
| Poli-L-lysine | Sigma-Aldrich Co | P8920 | |
| Potassium L-tartrate monobasic | Sigma-Aldrich Co | 243531 | |
| Sodium Carbonate | Sigma-Aldrich Co | S7795 | |
| Sodium chloride | Sigma-Aldrich Co | S7653 | |
| Sodium Deoxycholate (DOC) | Sigma-Aldrich Co | D6750 | |
| Sodium Dodecyl Sulfate | Sigma-Aldrich Co | L3771 | |
| Sodium Hydroxide | Sigma-Aldrich Co | S8045 | |
| Sodium phosphate dibasic anhydrous | Sigma-Aldrich Co | RES20908-A7 | |
| TESCAN VEGA 3 Scanning Electron Microscope | Tescan | #657874 | |
| Trichloroacetic Acid (TCA) | Sigma-Aldrich Co | 91230 | |
| Zetasizer Nano ZSP | Malvern Panalytical LTD | ||
| Ultrasonic cleaning bath model 2510 | Branson |
Referanslar
- Wang, X., Wang, Y., Chen, Z. G., Shin, D. M. Advances of cancer therapy by nanotechnology. Cancer Research and Treatment: Official Journal of Korean Cancer Association. 41 (1), 1 (2009).
- Keller, B. C. Liposomes in nutrition. Trends in Food Science & Technology. 12 (1), 25-31 (2001).
- Frézard, F., Schettini, D. A., Rocha, O. G., Demicheli, C. Lipossomas: propriedades físico-químicas e farmacológicas, aplicações na quimioterapia à base de antimônio. Quimica Nova. 28 (3), 511-518 (2005).
- Ferreira, D. d. S., Lopes, S. C. d. A., Franco, M. S., Oliveira, M. C. pH-sensitive liposomes for drug delivery in cancer treatment. Therapeutic Delivery. 4 (9), 1099-1123 (2013).
- Papachristos, A., Pippa, N., Ioannidis, K., Sivolapenko, G., Demetzos, C. Liposomal forms of anticancer agents beyond anthracyclines: present and future perspectives. Journal of Liposome Research. 25 (2), 166-173 (2015).
- Akbarzadeh, A., et al. Liposome: classification, preparation, and applications. Nanoscale Research Letters. 8 (1), 102 (2013).
- Bangham, A., Standish, M. M., Watkins, J. C. Diffusion of univalent ions across the lamellae of swollen phospholipids. Journal of Molecular Biology. 13 (1), 238-252 (1965).
- Szoka, F., Papahadjopoulos, D. Procedure for preparation of liposomes with large internal aqueous space and high capture by reverse-phase evaporation. Proceedings of the National Academy of Sciences of the United States of America. 75 (9), 4194-4198 (1978).
- Batzri, S., Korn, E. D. Single bilayer liposomes prepared without sonication. Biochimica et Biophysica Acta (BBA) – Biomembranes. 298 (4), 1015-1019 (1973).
- Zumbuehl, O., Weder, H. G. Liposomes of controllable size in the range of 40 to 180 nm by defined dialysis of lipid/detergent mixed micelles. Biochimica et Biophysica Acta (BBA) – Biomembranes. 640 (1), 252-262 (1981).
- Szebeni, J., et al. Oxidation and denaturation of hemoglobin encapsulated in liposomes. Biochimica et Biophysica Acta (BBA) – General Subjects. 798 (1), 60-67 (1984).
- Coelho, J. F., et al. Drug delivery systems: Advanced technologies potentially applicable in personalized treatments. EPMA Journal. 1 (1), 164-209 (2010).
- Pereira, P. R., et al. Purification and characterization of the lectin from taro (Colocasia esculenta) and its effect on mouse splenocyte proliferation in vitro and in vivo. The Protein Journal. 33 (1), 92-99 (2014).
- Pereira, P. R., et al. Structural analysis and binding properties of isoforms of tarin, the GNA-related lectin from Colocasia esculenta. Biochimica et Biophysica Acta (BBA) – Proteins and Proteomics. 1854 (1), 20-30 (2015).
- Pereira, P. R., et al. High-resolution crystal structures of Colocasia esculenta tarin lectin. Glycobiology. 27 (1), 50-56 (2016).
- Correa, A., Vericimo, M. A., Dashevskiy, A., Pereira, P. R., Paschoalin, V. M. F. Liposomal Taro Lectin Nanocapsules Control Human Glioblastoma and Mammary Adenocarcinoma Cell Proliferation. Molecules. 24 (3), 471 (2019).
- Peterson, G. L., et al. A simplification of the protein assay method of Lowry et al. which is more generally applicable. Analytical Biochemistry. 83 (2), 346-356 (1977).
- Merida, L. A., et al. Tarin stimulates granulocyte growth in bone marrow cell cultures and minimizes immunosuppression by cyclo-phosphamide in mice. PLoS ONE. 13 (11), e0206240 (2018).
- Murtey, M. D., Ramasamy, P., Janecek, M., Kral, R. Sample Preparations for Scanning Electron Microscopy – Life Sciences. Modern Electron Microscopy in Physical and Life Sciences. , (2016).
- Andrade, C. A., Correia, M. T., Coelho, L. C., Nascimento, S. C., Santos-Magalhães, N. S. Antitumor activity of Cratylia mollis lectin encapsulated into liposomes. International Journal of Pharmaceutics. 278 (2), 435-445 (2004).
- dos Santos Ferreira, D., et al. Development of a bone-targeted pH-sensitive liposomal formulation containing doxorubicin: physicochemical characterization, cytotoxicity, and biodistribution evaluation in a mouse model of bone metastasis. International Journal of Nanomedicine. 11, 3737 (2016).
- Pereira, P. R., Corrêa, A. C. N. T. F., Vericimo, M. A., Paschoalin, V. M. F. Tarin, a Potential Immunomodulator and COX-Inhibitor Lectin Found in Taro (Colocasia esculenta). Comprehensive Reviews in Food Science and Food Safety. 17 (4), 878-891 (2018).
- Olson, F., Hunt, C., Szoka, F., Vail, W., Papahadjopoulos, D. Preparation of liposomes of defined size distribution by extrusion through polycarbonate membranes. Biochimica et Biophysica Acta (BBA) – Biomembranes. 557 (1), 9-23 (1979).
- Mui, B., Chow, L., Hope, M. J. Extrusion technique to generate liposomes of defined size. Methods in Enzymology. 367, 3 (2003).
- Soo, E., et al. Enhancing delivery and cytotoxicity of resveratrol through a dual nanoencapsulation approach. Journal of Colloid and Interface Science. 462, 368-374 (2016).

