Nanoparticle-mediated siRNA Gene-silencing in Adult Zebrafish Heart
Özet
It remains a major challenge to develop conditional gene-knockout or effective gene-knockdown in adult zebrafish organs. Here we report a protocol for performing nanoparticle-mediated siRNA gene-silencing in adult zebrafish heart, thus providing a new loss-of-function method for studying adult organs in zebrafish and other model organisms.
Abstract
Mammals have a very limited capacity to regenerate the heart after myocardial infarction. On the other hand, the adult zebrafish regenerates its heart after apex resection or cryoinjury, making it an important model organism for heart regeneration study. However, the lack of loss-of-function methods for adult organs has restricted insights into the mechanisms underlying heart regeneration. RNA interference via different delivery systems is a powerful tool for silencing genes in mammalian cells and model organisms. We have previously reported that siRNA-encapsulated nanoparticles successfully enter cells and result in a remarkable gene-specific knockdown in the regenerating adult zebrafish heart. Here, we present a simple, rapid, and efficient protocol for the dendrimer-mediated siRNA delivery and gene-silencing in the regenerating adult zebrafish heart. This method provides an alternative approach for determining gene functions in adult organs in zebrafish and can be extended to other model organisms as well.
Introduction
Myocardial infarction has become a major health threat, leading to a tremendous economic burden around the world1. The adult mammalian heart fails to regenerate and replenish the lost cardiomyocytes on a macroscopic scale after the injury, leading to the formation of scar tissues and subsequent heart failure. Unlike mammals, the zebrafish is capable of heart regeneration, primarily through the robust myocardial proliferation after different types of heart injury, making it an ideal model organism for investigating the molecular mechanisms of heart regeneration2,3,4,5,6,7,8. Deciphering the endogenous mechanisms underlying zebrafish heart regeneration is an exciting area of research in the search for novel therapeutic strategies to improve human heart regeneration9.
Genetic manipulation methods are available in zebrafish. These consist of morpholinos (MO) that are also widely used in frogs, chick, and mammals besides in zebrafish10,11,12,13. MO has efficient knockdown of target gene expression in the adult zebrafish fin, brain, and retina14,15,16,17,18,19. Locked-nucleic acid (LNA) is another artificial oligonucleotide used to knock down endogenous gene expression not only in zebrafish embryos but also in adult animal organs20,21,22,23,24. However, the lack of effective loss-of-function methods for adults' hearts remains an obstacle in studying the molecular mechanisms of organ regeneration. At present, small-molecule inhibitors or transgenic expression of dominant-negative mutants are primarily used to block the function of a certain gene or pathway to study its function in the adult zebrafish heart regeneration25,26,27. However, not all genes or signaling pathways are applicable for these methods.
Small-interfering RNAs (siRNAs) are widely used for the loss-of-function analysis in mammalian cells and embryos of model organisms, as well as adult organs for preclinical studies in animal models28,29,30,31,32. siRNAs have been effectively used to silence genes in tumors33,34,35 and in cardiomyocytes36,37,38,39,40 via different delivery systems. Recently, we developed efficient siRNA-encapsulated nanoparticle gene-silencing in the regenerating adult heart using several different nanoparticles41,42,43, providing a novel tool for functional studies of genes in adult zebrafish organs. Based on our previous studies41,42,43, here we present a simple, practical, yet powerful protocol for siRNA gene-silencing in the regenerating adult zebrafish heart using f-PAMAM-PEG-R9 dendrimers. Aldh1a2 (aldehyde dehydrogenase 1 family, member A2) gene was upregulated after zebrafish apex resection and ablation of Aldh1a2 blocked the cardiac regeneration44. Here we take aldh1a2 gene as an example to test the gene knockdown efficiency mediated by nanoparticle-encapsulated siRNA injection. This protocol contains a procedure for zebrafish heart resection, chemical synthesis of nanoparticles, and a delivery method on siRNA-encapsulated nanoparticles into adult zebrafish heart.
Protocol
All animal procedures used a zebrafish protocol approved by the Institutional Animal Care and Use Committee at Peking University, which is fully accredited by Association for Assessment and Accreditation of Laboratory Animal Care.
1. Preparation of Tricaine Solution
- To prepare tricaine stock solution, add 400 mg ethyl 3-aminobenzoate methanesulfonate powder to 97.9 mL distilled water, and then add 2.1 mL of 1 M Tris (pH 9.5) to adjust the pH to ~7. Store the stock solution at 4 °C.
- To prepare tricaine working solution, add 4.2 mL tricaine stock solution to 100 mL aquarium system water.
2. Adult Zebrafish Ventricular Resection
- Prepare instruments and materials for the surgery which include a piece of sponge, a Petri dish filled with tricaine working solution, a plastic pipette, a stereomicroscope, two sharp forceps, iridectomy scissors, and elbow tweezers (Figure 1).
- Prepare a small groove of the size of 4 cm x 0.5 cm and 0.5 cm in depth on a sponge to hold the zebrafish during the ventricular resection surgery.
- Fill a 3 L zebrafish tank with half a tank of zebrafish system water that is made by using 60 mg/L sea salts in dH2O for the recovery of the injured fish.
- Anesthetize the fish in a 10 cm Petri dish with tricaine solution until the gill movements decrease to one per second and keep the fish for at least 30 s to ensure that it is fully anesthetized.
- Transfer the anesthetized fish with the elbow tweezers to the groove in a moist sponge ventral side up. Visually locate the margin of the beating heart, and then locate the heart under a stereomicroscope.
- Use the iridectomy scissors to make a small incision of about 1 mm that penetrates the skin and muscles, and carefully tear the silvery epithelial layer of the pericardial sac with the forceps tips to access the heart. Do not insert the forceps too deep into the cavity to avoid the heart injury.
NOTE: At this point, keep in mind to avoid extensive bleeding. Exclude the fish from the experiment if extensive bleeding occurs, since the excessive bleeding is mainly caused by the accidental injury to the heart and so will result in misleading results. - Expose the ventricle by gently holding its apex with sharp forceps using the non-dominant hand or by applying gentle abdominal pressure. Then, with the other hand, remove ~20% of the ventricle at the apex using the iridectomy scissors.
NOTE: No extensive bleeding should occur at this point. - Use a wet napkin to cover the incision, as the wounds bleed profusely for 15–45 s before clotting, and then put the fish back into the tank with system water.
NOTE: It is not necessary to suture the incision. - Make sure that the fish regains gill movements within 1 min and then resumes swimming. If the fish does not show gill movements after 50 s in water, facilitate it by squirting water into the gills using a plastic pipette until the fish resumes active ventilation.
- Maintain the fish in the recycle system after the ventricular resection.
NOTE: No special cares are necessary compared with the normal fish. The death rate of adult zebrafish ventricular resection is about 5–10%. Almost all death happens within 24 h after the surgery.
3. Preparation of siRNA-encapsulated Nanoparticles
- Synthesize the f-PAM4-PEG-R9 dendrimer.
NOTE: The synthesis of f-PAM4-PEG-R9 dendrimer materials was performed as described previously37 with some modifications. The length of α, ω-dipyridyl disulfido polyethylene glycol (Py-PEG-Py) were reduced to a molecular weight of 1,000 to maximize the encapsulation of siRNAs (Figure 2B). - Dry 10 mg of the cystamine core of G4.0 polyamidoamine (termed PAMAM) using vacuum dryer at 45 °C for 1 h and then dissolve the PAMAM in 2 mL Dulbecco's phosphate-buffered saline (DPBS, a balanced salt solution used for a variety of cell culture applications).
- Dissolve 1.7 mg tris(2-carboxyethyl) phosphine (TCEP) in 1 mL DPBS, mix with the PAMAM solution prepared in 3.2 and leave at room temperature with gentle stirring for 8 h.
NOTE: TCEP is ten-fold in molar ratio excess to the disulfide bonds of the PAMAM. - Remove excessive TCEP by ultrafiltration. Add DPBS to the above PAMAM-TCEP mixture to reach the volume of 10 mL and the solution is transferred into a 3 kDa MWCO (molecular weight cut off) ultrafiltration tube. After the centrifugation at 3,000 g for 15 min, discard the solution in the collection tube. Add DPBS to the filter device up to 10 mL and gently pipette the sample multiple times and centrifuge again. Repeat this step for 4 times. After the last filtration, aspirate the sample from the filter device.
NOTE: The product of this step is reduced PAMAM (termed f-PAM4). - Dissolve 14 mg Py-PEG-Py in 1 mL DPBS, mix with the f-PAM4 solution and leave it with gentle stirring at room temperature for 3 h. Then remove the excessive Py-PEG-Py by the ultrafiltration step as described in 3.4. After the last filtration, aspirate the sample from the filter device.
NOTE: The product is named as f-PAM4-PEG-Py. The amount of Py-PEG-Py is 10-fold in the molar ratio to the thiol groups in the f-PAM4. - Dissolve 6.4 mg of Cysteine terminated R9 Peptide (Ac-CRRRRRRRRR-NH2) in 1 mL DPBS, mix with f-PAM4-PEG-Py and gently stir for 8 h at room temperature. Purify the final product, called f-PAM4-PEG-R9, by the ultrafiltration step as described in 3.4 except that DPBS is changed to deionized water. Lyophilize the product by using a freeze vacuum dryer.
NOTE: The products in 3.5 and 3.6 before lyophilization are stored at -20 ℃ for at least 1 month. After the lyophilization of f-PAM4-PEG-R9, the samples are stored at -20 ℃ for at least 2 months. - To determine the degree of PEG and peptide modification, dissolve 5 mg of the f-PAM4-PEG-Py in the D2O (Deuterium oxide) and transfer it into the NMR (Nuclear Magnetic Resonance) sample tube. Perform 1H-NMR spectral analysis at 400 MHz and analyze the data using NMR data analysis software37.
NOTE: Specifically, the representative PAMAM peaks (-NCH2CH2CO-), PEG peaks (-OCH2CH2-), and arginine residue peaks (-HCCH2CH2CH2NH-) in the peptides are expected to be in the ranges 2.4–2.6, 3.7–3.8, and 1.6–1.8 ppm, respectively. Determine the degree of modification of the PEG and R9 relative to the PAMAM by calculating the integrals of the PEG and R9 peaks to the PAMAM peaks. The modification degree of PEG is 100% and the R9 modification is at least 50%. The 1H-NMR spectral analyses are performed in the core facility of the institution. - Dissolve the f-PAM4-PEG-R9 dendrimer powder in PBS to 4 mg/mL and make aliquots of the solution for one-time use.
NOTE: The dendrimer solution can be stored at -20 °C for at least one month, without repeated freezing and thawing. - Dissolve the cyanine dyes-labeled siRNAs (termed Cy5-siRNA), Aldh1a2 siRNA or scrambled negative control siRNA in distilled water to 10 µM.
NOTE: Use the cyanine dyes-labeled siRNAs to evaluate whether the nanoparticle encapsulated siRNA can enter the fish heart (Figure 2A-B). This protocol takes aldh1a2 as an example gene to determine the efficiency of delivery of f-PAM4-PEG-R9 dendrimer-encapsulated siRNA. The sequences of the siRNAs are: Cy5-siRNA antisense strand: 5′-ACGUGACACGUUCGGAGAAdTdT-3′, siAldh1a2 antisense strand: 5'-UUCAGGACAACCGUGUUCCdTdT-3' and scrambled negative control siRNA antisense strand: 5'-ACGUGACACGUUCGGAGAAdTdT-3'. - To prepare the siRNA-encapsulated nanoparticle solution, mix siRNA with an equal volume of dendrimer solution, and incubate the sample at room temperature for 20 min. Freshly prepare the siRNA-nanoparticle solution before use.
NOTE: The volume of siRNA-encapsulated nanoparticle solution depends on the number of fish going to be injected. For example, prepare at least 120 µL dendrimer solution (mix 60 µL siRNA solution and 60 µL dendrimer solution) for the injection of 10 fish (~10 µL per fish), counting in the loss of solution.
4. Injection of siRNA-encapsulated Nanoparticles into Adult Zebrafish Heart
- As described above, prepare a piece of sponge with a groove, a 10 cm Petri dish filled with tricaine solution, a plastic pipette, a stereomicroscope, elbow tweezers, and an insulin syringe (Figure 1), as well as a fish tank with system water for housing the fish after injection.
- Allow the injured zebrafish prepared in steps 2.1–2.10 to recover for 1 day after ventricular resection, and then anesthetize the 1 dpa (days post-amputation) fish in tricaine solution as described in 2.4.
- Transfer the anesthetized fish, ventral side up, to the groove in the moist sponge using the elbow tweezers. Locate the beating heart under the stereomicroscope.
- Load 10 µL of the siRNA-encapsulated nanoparticle solution into the insulin syringe. Avoid bubbles as much as possible.
- Immobilize the fish by gently using the elbow tweezers to hold the thoracic region with the non-dominant hand. Do not clamp the fish too tightly.
NOTE: Keep in mind that no extensive bleeding should occur during the injection. - Hold the filled insulin syringe at ~30° and inject ~10 µL of solution into the thoracic cavity using the other hand (Supplemental Figure 1).
NOTE: The solution sometimes flows out during the injection, but this does not affect the siRNA effect. The siRNA-nanoparticle solution can be injected once a day for several days to one month, depending on a specific experimental plan. Inject a fish with one type of siRNA each time. If more than two types of siRNA need to be injected into one fish, inject one type of siRNA each time, and then inject another type of siRNA 1 h later. In this protocol, only one type of siRNA per fish was injected. - Transfer the fish to the tank with the system water.
NOTE: The fish are expected to restart ventilation within 1 min, and then resume swimming. The injection of siRNA-encapsulated nanoparticles or siRNA normally does not cause death in adult zebrafish.
5. Heart Collection, Fixation, and Evaluation of the Efficiency of siRNA Delivery
- Prepare 1 mL of 4% paraformaldehyde solution in a microcentrifuge tube for each group.
- Anesthetize the fish in tricaine solution at selected days after injections. Place the fish supinely into the groove in a moist sponge. Make a large incision in the thoracic region and open widely with the tweezers. Grip the outflow tract and pull out the whole heart carefully with the tweezers.
- Rinse the heart with PBS and rapidly remove extra liquid with a napkin. Put up to 10 hearts into a 1.5 mL microcentrifuge tube with 1 mL paraformaldehyde. Gently invert the tube several times, and keep overnight at 4 °C.
- Measure the fluorescence intensity of Cy5-siRNAs-injected hearts using the in vivo imaging system (Supplemental Figure 2). Use 3–4 zebrafish hearts for each group in this study.
- Mount the fixed hearts with either paraffin or OCT compound (Embedding Medium for Frozen Tissue Specimens to ensure Optimal Cutting Temperature).
6. Evaluation of Nanoparticle-mediated siRNA Gene-silencing
- Prepare an insulated container and long tweezers. Pour in liquid nitrogen to a third of the volume of the container.
- Anesthetize the fish in tricaine solution at 2 dpa. Dissect the hearts as described in 5.2, and then remove the outflow tract and atrium.
NOTE: The fish can recover from the surgery in one day and then injected at 1 dpa with either scrambled siRNA-encapsulated nanoparticles (siNC) as a negative control or aldh1a2 siRNA-encapsulated nanoparticles (siAldh1a2). - Transfer the ventricles into a 1.5 mL microcentrifuge tube using the tweezers, cover the tube lid, and immediately place the tube into the insulated container with liquid nitrogen. Let the tubes float on the liquid nitrogen for a few min. Use the long tweezers to remove tubes from the container.
- Repeat steps 6.2 and 6.3 until all ventricles are collected. Use 6–10 hearts for each group.
- Extract the total RNA of hearts with RNA isolation reagent.Assess the expression levels of the respective genes by quantitative RT-PCR using primers Gapdh F: GATACACGGAGCACCAGGTT; Gapdh R: GCCATCAGGTCACATACACG; Aldh1a2 F: TGAGCGAGGAGCAGCAGAGA; Aldh1a2 R: TCCACGAAGAAGCCTTTAGTAGCA.
Representative Results
To determine the efficiency of the dendrimer-mediated siRNA delivery, we resected the apex of the ventricle of the zebrafish heart, then injected about 10 µL of dendrimer only (mock group), Cy5-siRNA only (naked group), or f-PAMAM-PEG-R9 dendrimer-encapsulated Cy5-siRNA (Cy5-siRNA group) intrapleurally, respectively (Figure 2A-B). The fluorescence signal was detectable in the hearts injected with dendrimer-encapsulated Cy5-siRNA at 3, 24, and 48 hpi (hours post-injection), while it was hardly detectable in hearts from the mock and naked groups at 48 hpi (Figure 2C-D), suggesting that the f-PAMAM-PEG-R9 dendrimer effectively facilitates the delivery of siRNAs into the adult zebrafish heart and is stable for at least two days.
To investigate the effect of dendrimer-encapsulated siRNA on gene silencing, we chose aldh1a2 (retinoic acid-synthesizing enzyme) as a target gene, which has been reported to be required for heart regeneration after ventricular resection44. As shown in Figure 3A, the fish were allowed to recover for one day after ventricular resection and then microinjected with dendrimer-encapsulated siRNA. We found that the aldh1a2 mRNA expression level decreased in the hearts treated with f-PAMAM-PEG-R9 dendrimer-encapsulated siAldh1a2 compared with that of dendrimer-encapsulated scrambled siRNA (siNC) at 2 dpa (Figure 3B), demonstrating that f-PAMAM-PEG-R9 dendrimer-mediated siRNAs achieve gene-specific silencing in adult zebrafish heart.
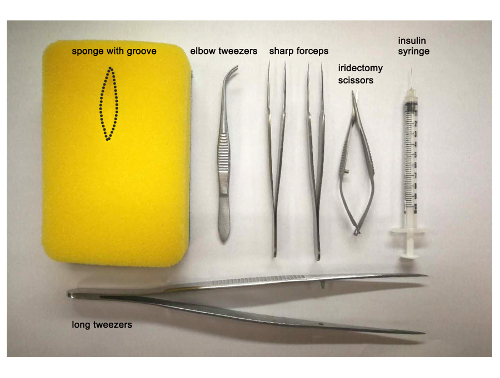
Figure 1: Heart surgery and thoracic injection instruments. Photograph of the instruments used for ventricular resection in the adult zebrafish heart: sponge with a groove, elbow tweezers, sharp forceps, iridectomy scissors, long tweezers, and the insulin syringe used for dendrimer-encapsulated siRNA delivery. Please click here to view a larger version of this figure.
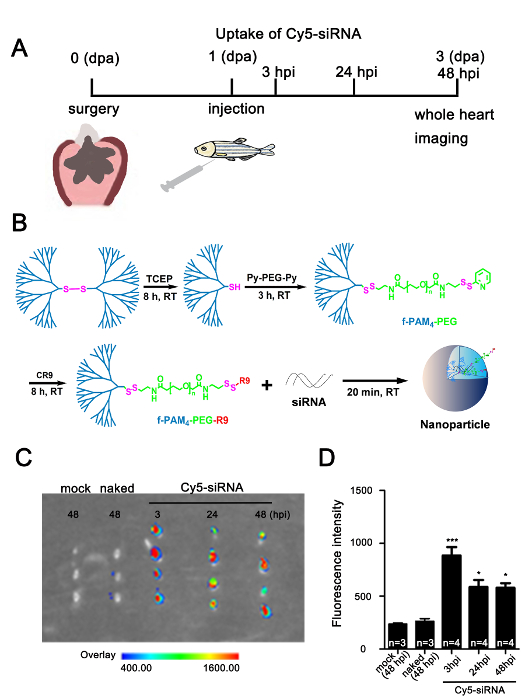
Figure 2: Effective f-PAMAM-PEG-R9 dendrimer-assisted delivery of siRNA into the injured adult zebrafish heart. (A) Scheme for investigating the uptake efficiency of nanoparticle-encapsulated Cy5-siRNA in adult zebrafish heart after ventricular resection (dpa: days post-amputation; hpi: hours post-injection). (B) Synthesis of f-PAMAM-PEG-R9 dendrimer and preparation of dendrimer-encapsulated Cy5-siRNA. (C) Cy5 fluorescence imaging of hearts injected with dendrimers only (mock), Cy5-siRNA only without dendrimer encapsulation (naked), and dendrimer-encapsulated fluorescence labeled Cy5-siRNA (Cy5-siRNA) detected by the in vivo imaging system, showing that fluorescence signals were nearly undetectable in the mock and naked groups at 48 hpi, while strong signals retained in the dendrimer-encapsulated Cy5-siRNA groups from 3 to 48 hpi. The arbitrary scale of fluorescence signals shows from weak (blue) to strong (red). (D) Quantification of Cy5-siRNA fluorescence signals in hearts assessed by the in vivo imaging system as in panel C (*P <0.05, ***P <0.001; data are mean ± s.e.m.; one-way analysis of variance followed by Bonferroni's multiple comparison tests; n = 3-4 hearts). Please click here to view a larger version of this figure.
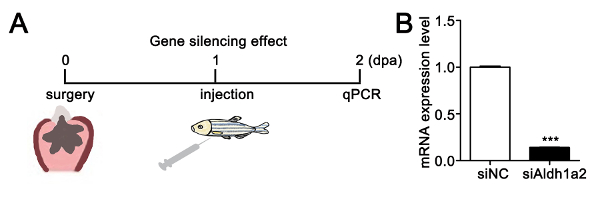
Figure 3: Efficient siRNA silencing of aldh1a2 in the injured adult zebrafish heart. (A) Scheme for investigating the gene-silencing effect of aldh1a2 siRNA. (B) qPCR reveals that aldh1a2 mRNA decreased in nanoparticle-encapsulated siAldh1a2 hearts compared with nanoparticle-encapsulated scrambled siRNA hearts (siNC). The aldh1a2 mRNA expression level was normalized by GAPDH (***P <0.001; data are mean ± s.e.m. with paired Student's t-test). Please click here to view a larger version of this figure.
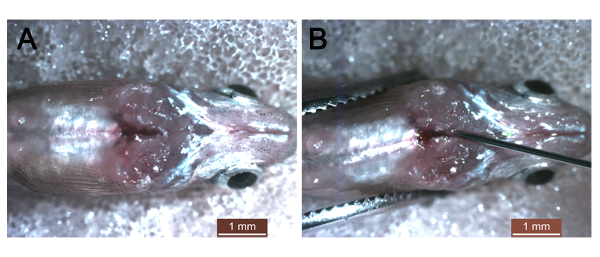
Supplementary Figure 1: Images of thoracic cavity injection. (A) The thoracic cavity in adult fish at 1 day post amputation. (B) Injection of nanoparticle-encapsulated siRNA into the thoracic cavity. Scale bars: 1 mm. Please click here to view a larger version of this figure.
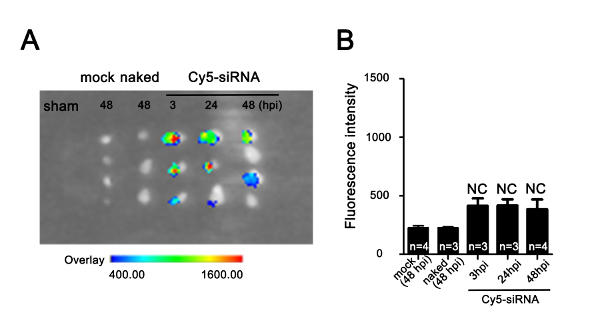
Supplementary Figure 2: Uptake efficiency of nanoparticle-encapsulated Cy5-siRNA in uninjured adult zebrafish heart. (A) Cy5 fluorescence imaging of hearts injected with dendrimers only (mock), naked Cy5-siRNA without dendrimer encapsulation (naked), and dendrimer-encapsulated Cy5-siRNA under the in vivo imaging system. The arbitrary scale of fluorescence signals is from weak (blue) to strong (red). (B) Quantification of Cy5-siRNA fluorescence signals in hearts measured by the in vivo imaging system as in panel A (NC: no significant different; data are mean ± s.e.m.; one-way analysis of variance followed by the Bonferroni's multiple comparison test; n = 3-4 hearts). Please click here to view a larger version of this figure.
Discussion
The zebrafish is fully capable of regenerating a variety of organs including the adult heart5. While transgenic and genetic methods are well-developed for studying gene functions in the embryos of zebrafish, investigators are still faced with the daunting task of generating conditional mutant alleles in zebrafish45,46. Thus, transgenic dominant-negative mutants or small-molecule inhibitors are frequently used to address gene functions in adult zebrafish organs25,27,42. Over the past a few years, we have developed an alternative method for the loss-of-function analysis of genes in the adult zebrafish heart using nanoparticle-mediated siRNA delivery and gene silencing41,42,43. Here, a detailed protocol for this method using dendrimer-mediated siRNA gene silencing in zebrafish hearts in particular and possibly in other zebrafish organs, in general, is presented.
The siRNA molecule cannot enter into the cells by itself due to its negative charge and would be easily degraded by the nuclease. With mono-disperse molecules, tunable structures and properties, dendrimers have been considered as promising siRNA carriers 47. The PAMAM dendrimers have rich cationic peripheries and buffering amines inside which could mediate siRNA encapsulation in physiological conditions and induce a "proton sponge" effect to release siRNA from endosomes in the cytoplasm respectively48,49,50. In this article, the PAMAM was modified to enhance the stability and delivery efficiency. The f-PAMAM-PEG-R9 system provides a hemispherical PAMAM dendrimer head with the positive charge for siRNA encapsulation to prevent the degradation by the nuclease. The PEG arm and the R9 transmembrane peptide was used to improve the hydrophilicity of the nanoparticles for stabilization and promote the uptake by cells respectively.
The quality and type of nanoparticles are critical for effective siRNA delivery and gene-silencing in the zebrafish heart. We have successfully investigated three structurally-diverse nanoparticles for this purpose: PEG-PLA, f-PAMAM-PEG-R9, and PEI-HYD-RGD nanoparticles41,42,43. Although they are not commercially produced, a regular organic chemistry lab can easily synthesize and purify nanoparticles as described previously35,37,43. Here, we chose the f-PAMAM-PEG-R9 dendrimer as an example to show the simple, efficient, dendrimer-mediated siRNA delivery and gene-silencing in the adult zebrafish heart after ventricular resection, because it is relatively easy to prepare siRNA-loaded dendrimer-complexes, such as by incubating the mixture at room temperature for ~20 min. On the other hand, the pH has to be adjusted for siRNA-loaded nanocomplexes if PEI-HYD-RGD is used43. Similarly, emulsifying and purification procedures are essential to obtain uniform nanoparticle-encapsulated siRNA solution if PEG-PLA nanoparticles are used35,41.
Another critical step is to minimize or have no bleeding during the injection of dendrimer-encapsulated siRNA as described in 4.5 since extensive bleeding interrupts heart regeneration and other biological systems. Thus, we suggest excluding fish from experimental groups if bleeding occurs during injection. In general, this simple injection procedure is easily mastered after a few trials.
The major limitations of this method are in the siRNA technology itself, such as potential off-target effects and incomplete deletion of genes of interest. Otherwise, highly reproducible biological replication of gene-silencing of several genes has been demonstrated in our work and that of others41. Importantly, this protocol is rapid, simple, and efficient, and the injection treatment is well-tolerated by adult zebrafish. Experimental analysis of siRNA gene-silencing efficiency may include the assessment of mRNA expression levels by in situ hybridization or quantitative RT-PCR, or of protein expression levels using Western blots, immunohistochemical staining, or other functional analyses. Together, we fully endorse nanoparticle-facilitated siRNA delivery as an alternative tool for loss-of-function studies in the adult zebrafish heart in particular and this approach may also be extended to other organs in adult zebrafish and other model organisms.
Açıklamalar
The authors have nothing to disclose.
Acknowledgements
The authors thank Dr. IC Bruce for critical comments and reading the manuscript. This work was supported by grants from the National Natural Science Foundation of China (31430059, 31701272, 31730061, 81470399, and 31521062), AstraZeneca Asia, and Emerging Market Innovative Medicine and Early Development.
Materials
| tricaine | Sigma | E10521 | Store at 4°C |
| stereomicroscope | Leica | S8AP0 | |
| sharp forcep | WPI | 14098 | |
| iridectomy scissors | WPI | 501778 | |
| elbow tweezers | Suzhou Liuliu | SE05Cr | |
| α,ω-dipyridyl disulfido polyethylene glycol(Py-PEG-Py) | Biomatrik (Jiaxing) Inc. | 5239 | |
| core of G4.0 polyamidoamine (PAMAM) | Andrews ChemServices | AuCS-297 | |
| vacuum drying equipment | Yiheng | DZF-6020 | |
| Dulbecco's phosphate-buffered saline (DPBS) | Gibco | 14190144 | |
| tris(2-carboxyethyl)phosphine(TCEP) | Alfar Aesar | 51805-45-9 | Causes severe skin burns and eye damage. Causes serious eye damage. |
| ultrafiltration tube | Millipore | UFC900308 | |
| freeze dryer | Martin Christ | Alpha 2-4 Ldplus | |
| NMR spectrometer | Bruker | AV400 | |
| Deuterium oxide(D2O) | J&K | 174611 | |
| NMR sample tube | J&K | WG-1000-7-50 | |
| 3 kDa MWCO ultrafiltration tube | Merck | UFC900308 | |
| sea salts | Instant Ocean® | SS15-10 |
Referanslar
- Writing Group Members. Executive Summary: Heart Disease and Stroke Statistics–2016 Update: A Report From the American Heart Association. Circulation. 133 (4), 447-454 (2016).
- Chablais, F., Veit, J., Rainer, G., Jazwinska, A. The zebrafish heart regenerates after cryoinjury-induced myocardial infarction. BMC Dev Biol. 11, 21 (2011).
- Gonzalez-Rosa, J. M., Martin, V., Peralta, M., Torres, M., Mercader, N. Extensive scar formation and regression during heart regeneration after cryoinjury in zebrafish. Development. 138 (9), 1663-1674 (2011).
- Parente, V., et al. Hypoxia/reoxygenation cardiac injury and regeneration in zebrafish adult heart. PLoS One. 8 (1), 53748 (2013).
- Poss, K. D., Wilson, L. G., Keating, M. T. Heart regeneration in zebrafish. Science. 298 (5601), 2188-2190 (2002).
- Raya, A., et al. Activation of Notch signaling pathway precedes heart regeneration in zebrafish. Proc Natl Acad Sci U S A. 100, 11889-11895 (2003).
- Schnabel, K., Wu, C. C., Kurth, T., Weidinger, G. Regeneration of cryoinjury induced necrotic heart lesions in zebrafish is associated with epicardial activation and cardiomyocyte proliferation. PLoS One. 6 (4), 18503 (2011).
- Wang, J., et al. The regenerative capacity of zebrafish reverses cardiac failure caused by genetic cardiomyocyte depletion. Development. 138 (16), 3421-3430 (2011).
- Gonzalez-Rosa, J. M., Burns, C. E., Burns, C. G. Zebrafish heart regeneration: 15 years of discoveries. Regeneration (Oxf). 4 (3), 105-123 (2017).
- Heasman, J., Kofron, M., Wylie, C. Beta-catenin signaling activity dissected in the early Xenopus embryo: a novel antisense approach. Dev Biol. 222 (1), 124-134 (2000).
- Nasevicius, A., Ekker, S. C. Effective targeted gene ‘knockdown’ in zebrafish. Nat Genet. 26 (2), 216-220 (2000).
- Coonrod, S. A., Bolling, L. C., Wright, P. W., Visconti, P. E., Herr, J. C. A morpholino phenocopy of the mouse mos mutation. Genesis. 30 (3), 198-200 (2001).
- London, C. A., et al. A novel antisense inhibitor of MMP-9 attenuates angiogenesis, human prostate cancer cell invasion and tumorigenicity. Cancer Gene Ther. 10 (11), 823-832 (2003).
- Kizil, C., Otto, G. W., Geisler, R., Nusslein-Volhard, C., Antos, C. L. Simplet controls cell proliferation and gene transcription during zebrafish caudal fin regeneration. Dev Biol. 325 (2), 329-340 (2009).
- Thummel, R., et al. Inhibition of zebrafish fin regeneration using in vivo. electroporation of morpholinos against fgfr1 and msxb. Dev Dyn. 235 (2), 336-346 (2006).
- Kizil, C., Brand, M. Cerebroventricular microinjection (CVMI) into adult zebrafish brain is an efficient misexpression method for forebrain ventricular cells. PLoS One. 6 (11), 27395 (2011).
- Kizil, C., Iltzsche, A., Kaslin, J., Brand, M. Micromanipulation of gene expression in the adult zebrafish brain using cerebroventricular microinjection of morpholino oligonucleotides. J Vis Exp. (75), e50415 (2013).
- Craig, S. E., et al. The zebrafish galectin Drgal1-l2 is expressed by proliferating Muller glia and photoreceptor progenitors and regulates the regeneration of rod photoreceptors. Invest Ophthalmol Vis Sci. 51 (6), 3244-3252 (2010).
- Thummel, R., Bailey, T. J., Hyde, D. R. In vivo electroporation of morpholinos into the adult zebrafish retina. J Vis Exp. (58), e3603 (2011).
- Rayburn, E. R., Zhang, R. Antisense, RNAi and gene silencing strategies for therapy: mission possible or impossible. Drug Discov Today. 13 (11-12), 513-521 (2008).
- Seth, P. P., et al. Short antisense oligonucleotides with novel 2′-4′ conformationaly restricted nucleoside analogues show improved potency without increased toxicity in animals. J Med Chem. 52 (1), 10-13 (2009).
- Prakash, T. P., et al. Antisense oligonucleotides containing conformationally constrained 2′,4′-(N-methoxy)aminomethylene and 2′,4′-aminooxymethylene and 2′-O,4′-C-aminomethylene bridged nucleoside analogues show improved potency in animal models. J Med Chem. 53 (4), 1636-1650 (2010).
- Yamamoto, T., Nakatani, M., Narukawa, K., Obika, S. Antisense drug discovery and development. Future Med Chem. 3 (3), 339-365 (2011).
- Itoh, M., Nakaura, M., Imanishi, T., Obika, S. Target gene knockdown by 2′,4′-BNA/LNA antisense oligonucleotides in zebrafish. Nucleic Acid Ther. 24 (3), 186-191 (2014).
- Han, P., et al. Hydrogen peroxide primes heart regeneration with a derepression mechanism. Cell Res. 24 (9), 1091-1107 (2014).
- Jopling, C., et al. Zebrafish heart regeneration occurs by cardiomyocyte dedifferentiation and proliferation. Nature. 464 (7288), 606-609 (2010).
- Lepilina, A., et al. A dynamic epicardial injury response supports progenitor cell activity during zebrafish heart regeneration. Cell. 127 (3), 607-619 (2006).
- McManus, M. T., Sharp, P. A. Gene silencing in mammals by small interfering RNAs. Nat Rev Genet. 3 (10), 737-747 (2002).
- de Fougerolles, A., Vornlocher, H. P., Maraganore, J., Lieberman, J. Interfering with disease: a progress report on siRNA-based therapeutics. Nat Rev Drug Discov. 6 (6), 443-453 (2007).
- Kim, D. H., Rossi, J. J. Strategies for silencing human disease using RNA interference. Nat Rev Genet. 8 (3), 173-184 (2007).
- McCaffrey, A. P., et al. Inhibition of hepatitis B virus in mice by RNA interference. Nat Biotechnol. 21 (6), 639-644 (2003).
- Raoul, C., et al. Lentiviral-mediated silencing of SOD1 through RNA interference retards disease onset and progression in a mouse model of ALS. Nat Med. 11 (4), 423-428 (2005).
- Hu-Lieskovan, S., Heidel, J. D., Bartlett, D. W., Davis, M. E., Triche, T. J. Sequence-specific knockdown of EWS-FLI1 by targeted, nonviral delivery of small interfering RNA inhibits tumor growth in a murine model of metastatic Ewing’s sarcoma. Cancer Res. 65 (19), 8984-8992 (2005).
- Schiffelers, R. M., et al. Cancer siRNA therapy by tumor selective delivery with ligand-targeted sterically stabilized nanoparticle. Nucleic Acids Res. 32 (19), 149 (2004).
- Yang, X. Z., et al. Systemic delivery of siRNA with cationic lipid assisted PEG-PLA nanoparticles for cancer therapy. J Control Release. 156 (2), 203-211 (2011).
- Ko, Y. T., Hartner, W. C., Kale, A., Torchilin, V. P. Gene delivery into ischemic myocardium by double-targeted lipoplexes with anti-myosin antibody and TAT peptide. Gene Ther. 16 (1), 52-59 (2009).
- Liu, J., et al. Functionalized dendrimer-based delivery of angiotensin type 1 receptor siRNA for preserving cardiac function following infarction. Biomaterials. 34 (14), 3729-3736 (2013).
- Nam, H. Y., Kim, J., Kim, S. W., Bull, D. A. Cell targeting peptide conjugation to siRNA polyplexes for effective gene silencing in cardiomyocytes. Mol Pharm. 9 (5), 1302-1309 (2012).
- Nam, H. Y., McGinn, A., Kim, P. H., Kim, S. W., Bull, D. A. Primary cardiomyocyte-targeted bioreducible polymer for efficient gene delivery to the myocardium. Biomaterials. 31 (31), 8081-8087 (2010).
- Won, Y. W., McGinn, A. N., Lee, M., Bull, D. A., Kim, S. W. Targeted gene delivery to ischemic myocardium by homing peptide-guided polymeric carrier. Mol Pharm. 10 (1), 378-385 (2013).
- Diao, J., et al. PEG-PLA nanoparticles facilitate siRNA knockdown in adult zebrafish heart. Dev Biol. 406 (2), 196-202 (2015).
- Xiao, C., et al. Chromatin-remodelling factor Brg1 regulates myocardial proliferation and regeneration in zebrafish. Nat Commun. 7, 13787 (2016).
- Wang, F., et al. A Neutralized Noncharged Polyethylenimine-Based System for Efficient Delivery of siRNA into Heart without Toxicity. ACS Appl Mater Interfaces. 8 (49), 33529-33538 (2016).
- Kikuchi, K., et al. Retinoic acid production by endocardium and epicardium is an injury response essential for zebrafish heart regeneration. Dev Cell. 20 (3), 397-404 (2011).
- Hoshijima, K., Jurynec, M. J., Grunwald, D. J. Precise Editing of the Zebrafish Genome Made Simple and Efficient. Dev Cell. 36 (6), 654-667 (2016).
- Zu, Y., et al. TALEN-mediated precise genome modification by homologous recombination in zebrafish. Nat Methods. 10 (4), 329-331 (2013).
- Kesharwani, P., Gajbhiye, V., Jain, N. K. A review of nanocarriers for the delivery of small interfering RNA. Biomaterials. 33 (29), 7138-7150 (2012).
- Luong, D., et al. PEGylated PAMAM dendrimers: Enhancing efficacy and mitigating toxicity for effective anticancer drug and gene delivery. Acta Biomater. 43, 14-29 (2016).
- Luo, K., He, B., Wu, Y., Shen, Y., Gu, Z. Functional and biodegradable dendritic macromolecules with controlled architectures as nontoxic and efficient nanoscale gene vectors. Biotechnol Adv. 32 (4), 818-830 (2014).
- Shcharbin, D., Shakhbazau, A., Bryszewska, M. Poly(amidoamine) dendrimer complexes as a platform for gene delivery. Expert Opin Drug Deliv. 10 (12), 1687-1698 (2013).

