Rapid and Efficient Generation of Recombinant Human Pluripotent Stem Cells by Recombinase-mediated Cassette Exchange in the AAVS1 Locus
Özet
Here we report a rapid and efficient gene editing method based on RMCE in the AAVS1 locus of human Pluripotent Stem Cells (hPSCs) that improves upon previously described systems. Using this technique, isogenic lines can be rapidly and reliably generated for proper comparative studies, facilitating transgenesis-mediated research with hPSCs.
Abstract
Even with the revolution of gene-targeting technologies led by CRISPR-Cas9, genetic modification of human pluripotent stem cells (hPSCs) is still time consuming. Comparative studies that use recombinant lines with transgenes integrated into safe harbor loci could benefit from approaches that use site-specific targeted recombinases, like Cre or FLPe, which are more rapid and less prone to off-target effects. Such methods have been described, although they do not significantly outperform gene targeting in most aspects. Using Zinc-finger nucleases, we previously created a master cell line in the AAVS1 locus of hPSCs that contains a GFP-Hygromycin-tk expressing cassette, flanked by heterotypic FRT sequences. Here, we describe the procedures to perform FLPe recombinase-mediated cassette exchange (RMCE) using this line. The master cell line is transfected with a RMCE donor vector, which contains a promoterless Puromycin resistance, and with FLPe recombinase. Application of both a positive (Puromycin) and negative (FIAU) selection program leads to the selection of RMCE without random integrations. RMCE generates fully characterized pluripotent polyclonal transgenic lines in 15 d with 100% efficiency. Despite the recently described limitations of the AAVS1 locus, the ease of the system paves the way for hPSC transgenesis in isogenic settings, is necessary for comparative studies, and enables semi-high-throughput genetic screens for gain/loss of function analysis that would otherwise be highly time consuming.
Introduction
Targeted genome recombination has facilitated rapid advances in many areas of research. In mice, the synergy between genome editing and stem cell research has allowed for an increased understanding of the complex mechanism of gene function and gene regulation. Such progress is expected in human pluripotent stem cells (hPSCs) as well, although for many years since the first isolation of human Embryonic Stem Cells (hESCs), and later human induced Pluripotent Stem Cells (hiPSCs), gene editing has constituted a technical hurdle. Recent advances in genome engineering using targeted nucleases-Zinc Finger Nucleases (ZFNs), Transcription activator-like effector nucleases (TALEN), or Clustered regularly interspaced short palindromic repeats/ CRISPR-associated protein 9 (CRISPR/Cas9) have allowed us to overcome these difficulties, making targeted recombination an efficient process1–3.
Specific loci in the mouse genome that allow stable, reliable, and ubiquitous transgene expression in the absence of adverse effects like Rosa26, Hprt1, or Col1A1, have become essential tools in the performance of genetic studies. Safe harbor loci allow comparable analysis between different lines of mice in isogenic contexts. This cannot be achieved using standard random integration methods, which are associated with well-known limitations like insertional mutagenesis, dose-dependent effects, or variegated transgene expression. Gene editing ease and flexibility in safe harbor loci is increased through the use of site-specific targeted recombinases like Cre or Flippase (FLPe), which specifically recognize target sequences (loxP or FRT, respectively) and catalyze efficient recombination between identical targets. Due to these characteristics, recombinase-mediated gene editing using loxP or FRT sequences in preintegrated safe harbor loci is a common tool used in mouse transgenesis. In addition to cassette insertion or excision mediated between identical target sequences, the use of incompatible loxP or FRT sites allows for recombinase-mediated cassette exchange (RMCE)4.
In human, attempts to identify safe harbor loci have been carried out. The mouse orthologous HPRT, despite its association with the loss-of-function Lesch-Nyhan Syndrome, and ROSA26 have been targeted in hPSCs. HPRT was reported to rapidly silence transgene expression in ESC and, like ROSA26, its ability to sustain transgene expression in terminally differentiated cells was not investigated5–7. Because a homozygous null mutation of the CCR5 gene appears to be well tolerated in humans, its value as safe harbor was assessed. CCR5 was targeted and reported to sustain stable transgene expression in different human cell lines, including ESCs8,9. However, the latter was not proven at the clonal level during long-term culture, and ubiquitous transgene expression was not demonstrated in differentiated progeny of the three germ layers. AAVS1, the natural integration site of the Adeno Associated Virus type 2 (AAV), was tested in hESCs as well, because of reported resistance to transgene silencing10. Later, many groups used the AAVS1 locus and described stable transgene expression in undifferentiated hPSCs, as well as in their differentiated progeny of all three germ layers, both in vitro and in vivo2,8,11,12. The recent results nonetheless nuance these findings, as the AAVS1 locus was found to exert variable transgene inhibition in vitro in undifferentiated hESCs and in hepatocyte progeny13.
Additional screening studies using random integration approaches and methods to determine single copy integration aimed to find genomic integration sites resistant to transgene silencing during hPSCs expansion and differentiation14,15. Overall, until now, no genomic site has been fully validated as a safe harbor in hPSCs and their progeny; identification of an appropriate site for ubiquitous stable transgene expression, not only in hPSCs but also in their differentiated progenies in vitro and in vivo, remains to be solved. Among all the studied loci and despite its limitations, AAVS1 remains the best-characterized and most-used in stem cell research.
RMCE has been successfully carried out in hPSCs in some of these loci6,7,14,16, using mostly Cre recombinase, even if there are indications that FLPe is more efficient than Cre17. In all these cases, one positive drug resistance cassette was used for the selection of recombinant colonies. Although successful, these procedures do not constitute a technical advance over standard gene-editing procedures using ZFNs, TALENs or CRISPR/Cas9, as a single antibiotic selection procedure does not rule out random integrations and requires colony screening to identify correctly targeted clones.
In this procedure, we describe methods to perform RMCE in hPSCs in the AAVS1 locus using a combination of positive (within the incoming cassette) and negative (within the preintegrated cassette) selections that allow for the generation of polyclonal transgenic lines in ±15 days with 100% efficiency and free of random integration events. Therefore, this method represents progress beyond currently described RMCE technologies in hPSCs.
Protocol
1. Preparation of the FRT-containing hPSC Master Cell Line for Transfection
- Culture hESC/iPSC master cell lines under standard procedures on or off inactivated Mouse Embryonic Fibroblasts (iMEF) using hESC or feeder-free iPSC medium, respectively (Table 1). Prepare 2 (on feeders) or 1 (feeder-free) wells of hPSCs in a 6-well plate for each experimental condition.
- Observe the cultures, and when cells reach 60 – 70% confluency, plate drug-resistant iMEF (iDR4). Briefly, coat the necessary wells of a 12-well plate with 0.5 mL of 0.1% gelatin solution and incubate for 5 min at RT. Plate 125,000 iDR4s per well and incubate O/N with iMEF medium (Table 1).
2. hPSC Transfection by Nucleofection
- The following day, pre-incubate hESC/iPSC cultures with fresh media, including 10 µM Rho-associated protein kinase (ROCK) inhibitor (Y-27632), for 1 h at 37 °C. Next, take hESC transfection solution from the fridge to pre-warm to room temperature.
- Prepare 1 mL of nucleofection plating medium (Table 1) per nucleofection condition. Take iMEF medium off the wells, wash with 1 mL of room temperature PBS, and add 500 µL of nucleofection plating medium. Transfer the other 500 µL to sterile 1.5 mL tubes and store them at 37 °C until plating. NOTE: A typical RMCE experiment contains three different conditions with molecular ratios of RMCE donor:pFLPe vectors of 2:0, 2:1, and 0:1.
- Detach hESC/iPSC into a single cell suspension.
- Take off the media, wash it with 2 mL of RT PBS, and add 1 mL trypsin 0.05% or accutase on feeder or feeder-free hPSC cultures, respectively. Incubate for 7 (trypsin) or and 2 – 5 (accutase) min at 37 °C. For the Accutase treatment — cells must remain loose, but attached.
- Carefully remove the plate from the incubator and without allowing the cells to detach, remove the dissociation agent by aspiration. Add 1 mL of hESC media and dissociate the loose cells by gently flushing the media on the plate surface, avoiding foaming. Make sure the cells are in a single cell suspension.
- Collect the cells in a clean and sterile 15 or 50 mL tube. Wash the wells with 1 mL of hESC media and collect the cells in the same 15 or 50 mL tube. Take a 50 µL aliquot for counting (during the next step) using a cell counting device. Note down the volume of collection and calculate total cell number. Spin down the cells at 300 x g for 5 min at RT.
- Resuspend the cell pellet to a suspension of 106 cells/mL with PBS using a 5 mL serological pipet and gentle pipetting. Avoid foaming. Transfer 2 mL per experimental condition (approximately 2 x 106 cells) to clean, sterile 15 mL tubes.
- While centrifuging, prepare the RMCE donor-pFLPe plasmids mixes in a maximum volume of 10 µL.
- Transfer 2.5 µg of pFLPe (6.5 Kb) to a clean, sterile 1.5 mL tube. Add a 2:1 molecular ratio of the RMCE donor vector13 to the pFLPe vector. For example, approximately 10 µg of a 12 Kb RMCE donor vector.
- Proceed with nucleofection, avoiding foaming in all steps.
NOTE: Some hPSC transfection protocols apply an additional iMEF depletion step before step 2.4. However, in practice, iMEF is a minor cell fraction in the single suspension that does not significantly alter transfection efficiency, nor will it create contaminating recombinant cells since they are non-proliferative. In addition, it's essential to proceed rapidly during the transfection process to increase viability. For those reasons, iMEF depletion is not applied.- Remove the iDR4 plate and the 1.5 mL tubes with the plating medium from the incubator. One tube at a time, pipet off the PBS carefully without disturbing the cell pellet. Remove as much as possible. Resuspend the pellet with 100 µL of transfection solution by gently pipetting. Transfer the cell suspension to the 1.5 mL tube containing the plasmid mixes and mix gently.
- Transfer the cell-DNA mix to the transfector cuvette, paying attention not to introduce bubbles. Introduce the cuvette in the nucleofector device and apply program A13 (cells cultured on feeders) or F16 (feeder-free).
NOTE: Other transfection methods or nucleofector devices may require optimization of these programs or transfection conditions. - Retake the cuvette and, using the disposable transfer pipettes available in the nucleofection kit, collect its content by applying 0.5 mL of the plating medium and aspirating all the volume. Conduct this step quickly, but gently and in one movement.
- Plate drop-wise in the 12-well plate containing the iDR4 and 500 µL of plating medium. Repeat steps 2.6.1 – 2.6.4 for the next experimental condition.
- Place the culture dish onto a shelf in the incubator and move it slowly back and forth and side to side to evenly distribute the cell suspension. Incubate it for 24 h before the next media change to allow the cells to recover in the presence of 10 µM ROCK inhibitor (Y-27632).
3. Positive and Negative Selection of Cells Undergoing RMCE
- Change culture media daily (1 mL/well), and 2 – 3 d post-transfection, start selection with 100 ng/mL of puromycin.
NOTE: Optimal concentrations of positive and negative selection reagents need to be determined experimentally for each newly generated hPSC master cell line. Perform a kill curve13, using the master cell line to determine the low dose (concentration at which minimal visual toxicity is apparent after 7 d of selection, but the culture is not overgrown), optimal dose (the lowest concentration at which all cells are dead after 7 d of selection), and high dose (concentration that caused evident toxicity, killing all cells after 2 – 3 d) for both puromycin and Fialuridine (FIAU).- Aspirate the hESC medium. Wash with 1 mL of PBS. Add hESC medium with 100 ng/mL of puromycin.
- Observe cell growth so that death and growth are balanced. Change media with puromycin daily, following step 3.1.1. When growth rate overtakes cell death, increase puromycin concentration in blocks of 25 – 50 ng/mL up to a maximum of 250 ng/mL. Continue puromycin selection for 5 – 7 d.
- 3 – 4 d after starting puromycin selection, around day 6 post-transfection, start selection with 0.5 µM FIAU. Change the media daily and maintain FIAU for no more than 7 continuous days.
4. Expansion and Characterization of RMCE Lines
- After completion of selection, around D14 – 15, resistant RMCE colonies are present and constitute the new RMCE line. Split in bulk in a 1:2 ratio following standard procedures (passage 1, P1).
- Plate 2 wells of a 12-well plate (on or off feeders, depending of the regular culture conditions of the original master cell line). Use one well for further expansion, storage, or experimental set-up (no additional description of these procedures is done in this protocol), and the other one for characterization.
- When cells from P1 are ready to split, dissociate the well for characterization into a single cell suspension as described in 2.4 and collect the cells in a 15 mL tube. NOTE: Use cells from the hPSC WT (no genetic modification) and master cell line as negative and positive controls, respectively, for the characterization.
- Spin down at 300 x g for 5 min at RT. Resuspend in 1 mL of PBS and split the sample in two for flow cytometry and DNA analysis13.
- Flow cytometry analysis13 (300 µL):
- Add fetal calf serum to the cells in PBS up to a final concentration of 5%. Transfer the cells to a clean FACS tube with a cell strainer and keep them on ice.
NOTE: Cell viability is high in these conditions, so it is possible, but not necessary, to use a fluorescent stain for non-viable cells (e.g., 7-AAD or PI). - Proceed immediately to analysis in a flow cytometer cell analyzer and record 20,000 – 30,000 events. For analysis, set the gates above GFP or tdT background florescence using the WT negative control sample.
- Add fetal calf serum to the cells in PBS up to a final concentration of 5%. Transfer the cells to a clean FACS tube with a cell strainer and keep them on ice.
- DNA analysis13 (700 µL):
- Transfer the cells to a clean 1.5 mL tube. Spin down at 300 x g for 5 min at RT and aspirate the supernatant. Proceed to DNA extraction using a commercial kit and follow the manufacturer's instructions. Store the dry pellet at -20 °C for later analysis.
- Measure DNA concentration.
- Prepare standard PCR reactions according to the conditions in Figure 3 and Table 213. Run PCR samples on a 2% agarose gel containing 1x DNA gel stain at 150 V for 15 – 25 min. Analyze the run in a gel analyzer.
Representative Results
Using AAVS1-specific ZNFs, fully characterized hESC/iPSC master cell lines containing heterotypic FRT target sequences were generated and described13. The FRT-containing master cell lines maintained pluripotency and genome integrity after the ZFN treatment and stably expressed GFP in vitro and in vivo. RMCE is performed by cotransfection of the master cell lines with FLPe recombinase and RMCE donor vectors (Figure 1A). RMCE vectors contain identical FRT sequences to the ones in the AAVS1 master cell line flanking the transgene. Figure 1B monitors RMCE using the constitutively expressing tdT pZ:F3-P CAGGS tdTPH-F. After transfection, hPSC form small groups of cells evenly distributed over the iDR4 MEF layer, and transfected hPSC express both GFP as well as transient tdT (Figure 1B, D 2). Cells are allowed to recover for 2 – 3 d post-transfection before starting selection. Positive selection is first started using a low dose of puromycin in order to favor RMCE donor insertion. Puromycin is applied gently at the start, because initial massive cell death could lead to single cells undergoing recombination, making them unable to survive the process. In the following days, puromycin concentration needs to be raised stepwise up to the optimal dose in order to allow the recombinant cells to grow into small colonies while the nonrecombined cells gradually die.
3 – 4 d after initiating positive selection (around D 6 post-transfection), negative selection is begun in order to select only for FRT-mediated recombination events. RMCE causes the loss of the thymidine kinase (Tk) suicide gene and thus the sensitivity to Fialuridine (FIAU). Negative selection applied at this point, in which small clusters of 2 – 4 GFP–/tdT+ (RMCE) cells are able to withstand optimal concentrations of FIAU (Figure 1B, D 6), prevents random integration, because in such events, the Tk gene in the AAVS1 locus remains unaffected. Occurrence of simultaneous FRT-mediated and random integration is highly unlikely, as demonstrated by the absence of random integration events later during the characterization (Figure 2B).
The mixed RMCE GFP–/tdT+ colonies continue to grow, while becoming more homogeneous, so that by D 9 – 10 post-transfection, some not fully selected mixed RMCE colonies can be found (Figure 1B). GFP–/tdT+ RMCE colonies are present only when the both RMCE donor and FLPe (2:1) are used, while RMCE does not occur in the absence of FLPe (2:0). Full selection with FIAU until day 13 – 15 post-transfection gives rise to a homogeneous GFP–/tdT+ culture. RMCE yields an average of 12.8 ± 6.8 (n = 6) PuroR/FIAUR colonies in 15 days13. Flow cytometry characterization of the newly generated RMCE line (the PuroR/FIAUR RMCE colonies combined in a nonclonal cell population) confirms that 100% of the cells present the RMCE GFP–/tdT+ phenotype (Figure 2A). When RMCE donors without constitutive expression of a fluorescent reporter are used, the resulting PuroR/FIAUR RMCE hPSC line is homogeneously GFP–.
PCR characterization of the nonclonal RMCE line demonstrates the full cassette exchange (no traces of the cassette of the master cell line can be detected) and that the selection program used generates random integration-free lines (both of the RMCE donor and the FLPe-expressing vector) (Figure 2B). These results were further proven by southern blot and, in addition, we demonstrated that the RMCE lines remain pluripotent13. This makes it no longer necessary to carry out either the genome-wide evaluation of random integration or the pluripotency test during routine RMCE experiments. In summary, using this protocol, hPSC transgenic cell lines in the AAVS1 locus free of random integration can be readily generated in 15 d with 100% efficiency and without the need of extensive characterization.
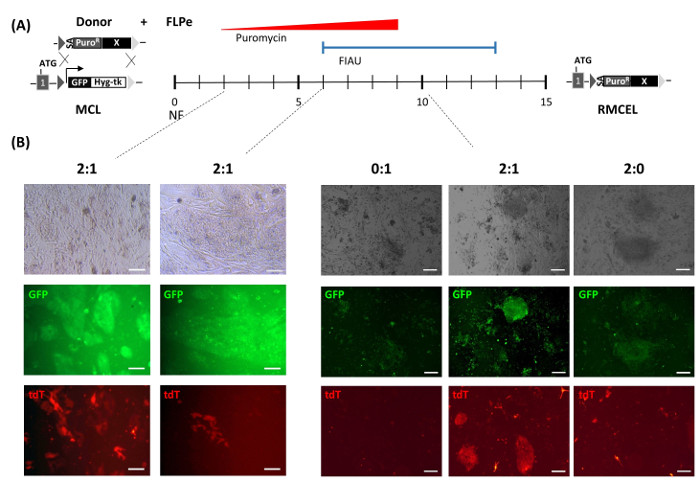
Figure 1: Schematic Overview of RMCE. (A) Timeline of the RMCE selection program. Left: the master cell line (MCL), harboring a FRT-flanked cassette (triangles) that express GFP and Hygromycin-tk (linked by 2A self-cleaving peptides), is transfected by nucleofection (NF) with the FLPe-expressing vector and the RMCE donor vector, which contains a promoterless Puromycin resistance gene (splicing acceptor – SA- PuroR) and a variable experimental cassette (X). Right: new RMCE line (RMCEL) obtained after completion of selection with Puromycin (red triangle) and FIAU (blue line). (B) RMCE using a constitutive tdT-expressing donor and different ratios of donor DNA and FLPe DNA (0:1, 2:1, 2:0) monitored by fluorescence microscopy (GFP – green fluorescence, tdT – red fluorescence) at different time points. D 2 and 6: scale bars represent 100 µm. D 10: scale bars represent 200 µm. Please click here to view a larger version of this figure.
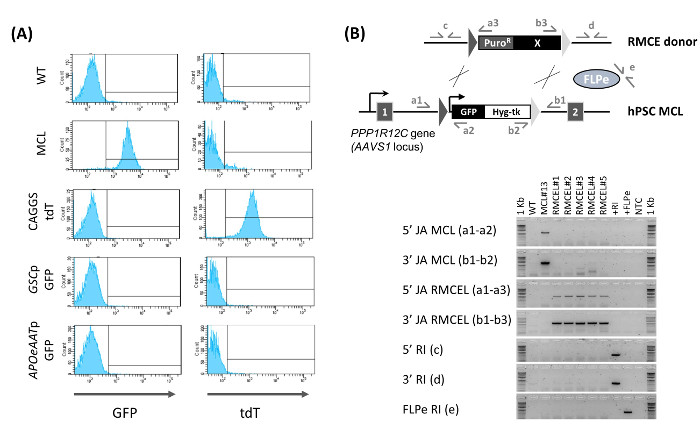
Figure 2: Characterization of RMCE Lines. (A) Determination of RMCE by flow cytometry. GFP and tdT expression is shown for the WT line, master cell line (MCL), and newly generated RMCE line at D 15 post-nucleofection using a constitutive tdT-expression vector (CAGGS tdT), or donors with GFP driven by a GOOSECOID promoter (GSCp GFP, definitive endoderm marker) or an AAT promoter (APOeAATp GFP, hepatocyte marker). (B) Confirmation of RMCE (5'/3' JA RMCEL), absence of the master cell line's (MCL) cassette (5'/3' JA MCL), and lack of random integration (5'/3'/FLPe RI) by PCR using the depicted primer pairs. DNA from WT, MCL, RMCE lines (1 – 5), donor RMCE vector (+RI), FLPe expressing vector, and no template control (NTC) were used. Figure modified from (Ordovas et al., 2015)13. Please click here to view a larger version of this figure.
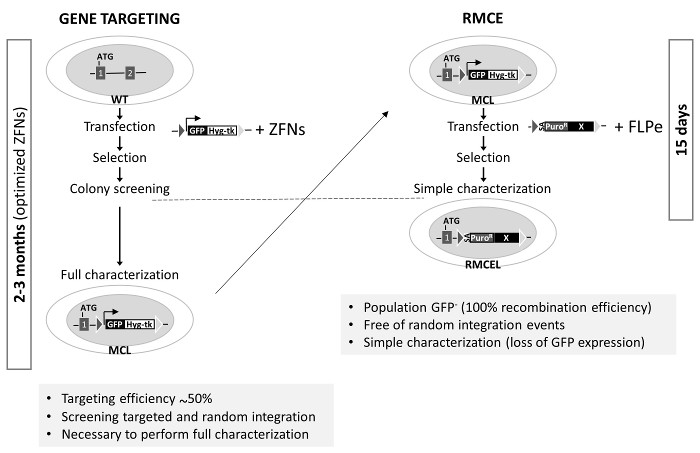
Figure 3. Generation of the RMCE-suitable Master Cell Line and Comparison of Gene Targeting with RMCE in AAVS1. Left: Gene targeting uses unmodified WT cells that are cotransfected with a donor (cassette described in Figure 1A for the generation of the master cell line, MCL) and specific optimized ZFNs. The process to complete the full characterization takes up to 3 months. Right: RMCE is performed by cotransfection of the donor with FLPe vectors, and a fully characterized line is generated in 15 d. Gene targeting efficiency in AAVS1 is from previous studies1,2. Please click here to view a larger version of this figure.
| Media | Components | Final concentration |
| hESC medium | DMEM-F12, HEPES | 80% |
| Serum replacement medium | 20% | |
| L-Glutamine | 146 mg/mL | |
| MEM Non-Essential Amino Acids Solution (100x) | 1x | |
| 2-Mercaptoethanol | 0.1 mM | |
| Penicillin/Streptomycin | 0.1% | |
| Human basic FGF | 4 ng/mL | |
| iMEF culture medium | DMEM High Glucose | |
| Fetal Bovine serum (FBS) | 15% | |
| Penicillin-Streptomycin | 0.1% | |
| L-Glutamine 200 mM | 4 mM | |
| MEM Non-Essential Amino Acids Solution (100x) | 2x | |
| 2-Mercaptoethanol | 0.1 mM | |
| Plating medium | hESC medium | 100% |
| ROCK inhibitor (Y-27632) | 10 µM |
Table 1. Media composition.
| Assay | Forward | Reverse | Amplicon | PCR Cycle | ||
| 5’JA MCL | CACTTTGAGCTCTACTGGCTTC | CGTTACTATGGGAACATACGTCA | 1.1 Kb | 95 ºC, 5’ – [95 ºC, 30’’ – 68 ºC (-0.5 ºC/cycle), 1’ 30’’]X15 – [95 ºC, 30’’ – 58 ºC, 30’’ – 72 ºC, 1’ 30’’]X25 – 72 ºC, 5’ | ||
| 3’JA MCL | TAACTGAAACACGGAAGGAG | AAGGCAGCCTGGTAGACA | 1.4 Kb | 95 ºC, 5’ – [95 ºC, 30’’ – 68 ºC (-0.5 ºC/cycle), 1’ 30’’]X15 – [95 ºC, 30’’ – 58 ºC, 30’’ – 72 ºC, 1’ 30’’]X25 – 72 ºC, 5’ | ||
| 5’JA RMCEL | CACTTTGAGCTCTACTGGCTTC | CATGTTAGAAGACTTCCTCTGC | 1.1 Kb | 95 ºC, 5’ – [95 ºC, 30’’ – 68 ºC (-0.5 ºC/cycle), 1’ 30’’]X15 – [95 ºC, 30’’ – 58 ºC, 30’’ – 72 ºC, 1’ 30’’]X25 – 72 ºC, 5’ | ||
| 3’JA RMCEL | TTCACTGCATTCTAGTTGTGG | AAGGCAGCCTGGTAGACA | 1.5 Kb | 95 ºC, 5’ – [95 ºC, 30’’ – 68 ºC (-0.5 ºC/cycle), 1’ 30’’]X15 – [95 ºC, 30’’ – 58 ºC, 30’’ – 72 ºC, 1’ 30’’]X25 – 72 ºC, 5’ | ||
| 5’RI DONOR | GTACTTTGGGGTTGTCCAG | TTGTAAAACGACGGCCAG | 0.5 Kb | 95 ºC, 5’ – [95 ºC, 30’’ – 60 ºC, 30’’ – 72 ºC, 30’’]X25 – 72 ºC, 5’ | ||
| 3’RI DONOR | CCTGAGTTCTAACTTTGGCTC | ACACAGGAAACAGCTATGAC | 0.5 Kb | 95 ºC, 5’ – [95 ºC, 30’’ – 60 ºC, 30’’ – 72 ºC, 30’’]X25 – 72 ºC, 5’ | ||
| RI FLPe | CCTAGCTACTTTCATCAATTGTG | GTATGCTTCCTTCAGCACTAC | 0.65 Kb | 95 ºC, 5’ – [95 ºC, 30’’ – 60 ºC, 30’’ – 72 ºC, 30’’]X25 – 72 ºC, 5’ | ||
Table 2. Primer Sets used for PCR Genotyping. Modified from Ordovas et al., 201513.
Discussion
Gene editing methods in safe harbor loci remain an essential tool to develop transgenesis in hPSCs. Although the safe harbor character of AAVS1 has recently been questioned for some applications13,18, this locus currently remains the best characterized in human-derived cell lines. Awareness of its limitations in hPSCs can help to obtain reliable data. Therefore, AAVS1 is still expected to be a useful site, for example, for gain- and loss-of-function studies or inducible/constitutive expression of factors that require an isogenic context within and between determined genetic backgrounds.
The AAVS1 locus has been targeted by many groups using ZFNs, TALEN or CRISPR/Cas91,2,19. These nucleases significantly increase the efficiency of homologous recombination in a determined locus. However, the process to screen and fully characterize correctly targeted clones, free of random integration and to maintain pluripotency and genome integrity can take up to 3 months (using optimized gene editing tools) (Figure 3). The last two can be caused by possible mutations generated by off-target nuclease activity. The RMCE system used here, however, offers a rapid, efficient, and elegant method to achieve this aim in only two weeks, once recombinase-specific target sequences are preintegrated into the AAVS1 locus. Use of positive/negative selection and an appropriate selection program are the main factors contributing to the simplification of gene editing in the AAVS1 locus by RMCE.
Genotypic characterization of the new transgenic lines is significantly reduced (no clonal screening is necessary), and characterization associated to off-target nuclease activity is rendered dispensable due to the specificity of FLPe for FRTs. The characterization can also be reduced in routine RMCE experiments by demonstrating the complete loss of GFP expression from the master cell line (Figure 2A), because full cassette exchange and lack of random integration have already been sufficiently demonstrated by PCR (Figure 2B) and Southern blot13. The use of positive/negative selection also constitutes the main difference with previous reports. Several groups that previously described RMCE in hPSCs, either in the AAVS1 or other loci, employed only a single positive selection step7,14,16. This does not constitute a major technical advantage over gene editing performed with nucleases, because colony screening has to be equally performed in order to demonstrate correct integration and absence of random integration at the clonal level.
Use of this RMCE system in multiple hESC/iPSC lines requires preintegration of the described FRT-containing cassette in the AAVS1 locus of each independent line. However, once generated, its rapidity and simplicity makes it possible to develop semi-high throughput genetic screens in defined isogenic settings for applications mentioned above that otherwise would be technically very time consuming. In addition, all the RMCE vectors are constructed within the pZ AAVS1 gene-targeting vector used to target the locus with ZFNs, but TALENs or CRISPR/Cas9 were also reported. The RMCE vector's duality is highly useful to generate multiple lines for a determined transgene in hPSCs with or without FRT. For example, one hit demonstrated successful in a determined genetic screen carried out by RMCE would ideally need to be tested in multiple hPSC lines to confirm the results. In the absence of multiple RMCE-suitable master cell lines, direct gene targeting of the hit using nucleases could be performed. The utility of the dual character of the RMCE vectors has been proven by our group20. In this study, gene correction was carried out in patient-derived Frontotemporal Dementia (FTD) iPSCs that bear a mutation causing PROGRANULIN (PGRN) expression deficiency. Gene complementation by ZFNs-mediated insertion in the AAVS1 locus of a cassette that restored PGRN levels, corrected the defective phenotype in corticogenesis associated with the presence of the mutation. Additionally, a comparable line was generated by RMCE in WT hESCs to be used as control for exogenous PGRN expression.
In the absence of recombinant colonies, check transfection efficiency of the RMCE donor vector. Transfection efficiencies superior to 30% yield the results indicated above. Lower transfection efficiencies decrease recombination efficiency. Finally, the RMCE system has been generated in the AAVS1 locus, but it is applicable to any other locus.
Açıklamalar
The authors have nothing to disclose.
Acknowledgements
L.O. was funded by IWT/OZM/090838, IACS BPAMER3/08/04, and the Government of Aragon FMI048/08; RB by IWT fellowship SB-121396; M.P. by FWO 1288714N; and R.S. by the Dutch Diabetes Foundation. N.H., J.V., K.C., and Q.C. were supported by IWT fellowships SB-121396, SB-101230, SB-91228, and SB-093228, respectively. Funding to C.M.V. was from FWO G.0667.07, G.0975.11, and KU Leuven (EIW-B4855-EF/05/11, ETH-C1900-PF, EME-C2161-GOA/11/012), IWT-HEPSTEM, BELSPO-IUAP-DEVREPAIR, FP7-HEMIBIO (266777).
Materials
| DR4 Mouse Embryonic Fibroblasts (MEF) | AMS bio | GSC-6204G | hPSC culture on feeers |
| CF-1 MEF, mitomycin-C treated | AMS Bio | GSC-6001M | |
| EmbryoMax 0.1% Gelatin Solution | Millipore | ES-006-B | iMEF plating |
| DMEM/F-12, HEPES | Gibco | 31330-038 | |
| KnockOut Serum Replacement | Gibco | 10828-028 | |
| L-Glutamine | Sigma Aldrich | G8540 | For hESC medium |
| L-Glutamine 200 mM | Gibco | 25030-081 | For iMEF medium |
| 2-Mercaptoethanol 50 mM | Gibco | 31350-010 | |
| MEM Non-Essential Amino Acids Solution (100X) | Gibco | 11140-035 | |
| Penicillin-Streptomycin (10,000 U/mL) | Gibco | 15140122 | |
| Human basic FGF | Peprotech | 100-18C | |
| Y-27632 in Solution | VWR | 688001-500 | |
| DMEM High Glucose | Gibco | 41965-039 | |
| Fetal Bovine Serum | Sigma Aldrich | F7524 | |
| Trypsin-EDTA (0.05%), phenol red | Gibco | 25300-054 | hPSC culture on feeders |
| Matrigel hESC qualified | VWR | 734-1440 | |
| mTeSR1 complete medium kit | Stem cell technologies | 05850 | For Feeder free culture |
| StemPro Accutase Cell Dissociation Reagent | Gibco | A1110501 | For feeder free culture |
| Y-27632 in solution | VWR | 56726 | |
| Human Stem Cell Nucleofector Kit 2 | Lonza | VVPH-5022 | |
| Puromycine Dihydrochloride | Gibco | A11138-03 | |
| FIAU (Fialuridine) | ABX | 2910 | |
| Hygromycin B | Sigma Aldrich | H3274 | |
| pFLPe | Modified to from pCAGGS-FLPe (MES4488, Open Biosystems) to remove puromycin | ||
| RMCE donors | – | – | Modified from pZ donor AAVS1 puromycin vector (PZD0020-1KT, Sigma Aldrich)13 |
| 5mL Round Bottom Polystyrene Test Tube, with Cell Strainer | Falcon | 352235 | |
| PureLink Genomic DNA kit | Invitrogen | K182001 | |
| GoTaq Flexi DNA Polymerase | Promega | M8298 | |
| Agarose | Sigma | A9539 | |
| SYBR Safe DNA Gel Stain | Invitrogen | S33102 | |
| Nucleofector 2b Device | Lonza (Amaxa) | AAB-1001 | |
| NucleoCounter NC-100 | Chemometec | NC-100 | |
| BD FACS Canto | BD Biosciences | 337175 | |
| NucleoCassette | Chemometec | 941-0002 |
Referanslar
- Hockemeyer, D., et al. Genetic engineering of human pluripotent cells using TALE nucleases. Nat. Biotechnol. 29 (8), 731-734 (2011).
- Hockemeyer, D., et al. Efficient targeting of expressed and silent genes in human ESCs and iPSCs using zinc-finger nucleases. Nat. Biotechnol. 27 (9), 851-857 (2009).
- Mali, P., et al. RNA-guided human genome engineering via Cas9. Science. 339 (6121), 823-826 (2013).
- Baer, A., Bode, J. Coping with kinetic and thermodynamic barriers: RMCE, an efficient strategy for the targeted integration of transgenes. Curr. Opin. Biotechnol. 12 (5), 473-480 (2001).
- Sakurai, K., et al. Efficient integration of transgenes into a defined locus in human embryonic stem cells. Nucleic Acids Res. 38 (7), e96 (2010).
- Irion, S., et al. Identification and targeting of the ROSA26 locus in human embryonic stem cells. Nat. Biotechnol. 25 (12), 1477-1482 (2007).
- Di Domenico, A. I., Christodoulou, I., Pells, S. C., McWhir, J., Thomson, A. J. Sequential genetic modification of the hprt locus in human ESCs combining gene targeting and recombinase-mediated cassette exchange. Cloning Stem Cells. 10 (2), 217-230 (2008).
- Lombardo, A., et al. Site-specific integration and tailoring of cassette design for sustainable gene transfer. Nat. Methods. 8 (10), 861-869 (2011).
- Lombardo, A., et al. Gene editing in human stem cells using zinc finger nucleases and integrase-defective lentiviral vector delivery. Nat. Biotechnol. 25 (11), 1298-1306 (2007).
- Simth, J. R., et al. Robust , Persistent Transgene Expression in Human Embryonic Stem Cells Is Achieved with AAVS1-Targeted Integration. Stem Cells. 26, 496-504 (2008).
- DeKelver, R. C., et al. Functional genomics, proteomics, and regulatory DNA analysis in isogenic settings using zinc finger nuclease-driven transgenesis into a safe harbor locus in the human genome. Genome Res. 20 (8), 1133-1142 (2010).
- Qian, K., et al. A simple and efficient system for regulating gene expression in human pluripotent stem cells and derivatives. Stem Cells. 32 (5), 1230-1238 (2014).
- Ordovás, L., et al. Efficient Recombinase-Mediated Cassette Exchange in hPSCs to Study the Hepatocyte Lineage Reveals AAVS1 Locus-Mediated Transgene Inhibition. Stem Cell Reports. 5 (5), 918-931 (2015).
- Du, Z. -. W., Hu, B. -. Y., Ayala, M., Sauer, B., Zhang, S. -. C. Cre recombination-mediated cassette exchange for building versatile transgenic human Embryonic Stem Cells lines. Stem Cells. 27, 1032-1041 (2009).
- Papapetrou, E. P., et al. Genomic safe harbors permit high β-globin transgene expression in thalassemia induced pluripotent stem cells. Nat. Biotechnol. 29 (1), 73-78 (2011).
- Ramachandra, C. J., et al. Efficient recombinase-mediated cassette exchange at the AAVS1 locus in human embryonic stem cells using baculoviral vectors. Nucleic Acids Res. 39 (16), e107 (2011).
- Takata, Y., Kondo, S., Goda, N., Kanegae, Y., Saito, I. Comparison of efficiency between FLPe and Cre for recombinase-mediated cassette exchange in vitro and in adenovirus vector production. Genes to Cells. 16 (7), 765-777 (2011).
- Mizutani, T., Li, R., Haga, H., Kawabata, K. Transgene integration into the human AAVS1 locus enhances myosin II-dependent contractile force by reducing expression of myosin binding subunit 85. Biochem. Biophys. Res. Commun. 465 (2), 270-274 (2015).
- Mali, P., et al. RNA-guided human genome engineering via Cas9. Science. 339 (6121), 823-826 (2013).
- Raitano, S., et al. Restoration of progranulin expression rescues cortical neuron generation in an induced pluripotent stem cell model of frontotemporal dementia. Stem Cell Reports. 4 (1), 16-24 (2015).

