Automated Robotic Liquid Handling Assembly of Modular DNA Devices
Özet
Here, an automated workflow to perform modular DNA "device" assembly using a modular cloning DNA assembly method on liquid-handling robots is presented. The protocol uses an intuitive software tool for generating liquid handler picklists for combinatorial DNA device library generation, which we demonstrate using two liquid handling platforms.
Abstract
Recent advances in modular DNA assembly techniques have enabled synthetic biologists to test significantly more of the available "design space" represented by "devices" created as combinations of individual genetic components. However, manual assembly of such large numbers of devices is time-intensive, error-prone, and costly. The increasing sophistication and scale of synthetic biology research necessitates an efficient, reproducible way to accommodate large-scale, complex, and high throughput device construction.
Here, a DNA assembly protocol using the Type-IIS restriction endonuclease based Modular Cloning (MoClo) technique is automated on two liquid-handling robotic platforms. Automated liquid-handling robots require careful, often times tedious optimization of pipetting parameters for liquids of different viscosities (e.g. enzymes, DNA, water, buffers), as well as explicit programming to ensure correct aspiration and dispensing of DNA parts and reagents. This makes manual script writing for complex assemblies just as problematic as manual DNA assembly, and necessitates a software tool that can automate script generation. To this end, we have developed a web-based software tool, http://mocloassembly.com, for generating combinatorial DNA device libraries from basic DNA parts uploaded as Genbank files. We provide access to the tool, and an export file from our liquid handler software which includes optimized liquid classes, labware parameters, and deck layout. All DNA parts used are available through Addgene, and their digital maps can be accessed via the Boston University BDC ICE Registry. Together, these elements provide a foundation for other organizations to automate modular cloning experiments and similar protocols.
The automated DNA assembly workflow presented here enables the repeatable, automated, high-throughput production of DNA devices, and reduces the risk of human error arising from repetitive manual pipetting. Sequencing data show the automated DNA assembly reactions generated from this workflow are ~95% correct and require as little as 4% as much hands-on time, compared to manual reaction preparation.
Introduction
The earliest synthetic biological genetic devices such as the Collins toggle switch1 and the Elowitz repressilator2 demonstrated that biological systems could be forward engineered to have specific, deterministic functions. Since then, synthetic biologists have strived to engineer living systems to carry out progressively more complex functionality in the service of biomaterials3, biotherapeutics4,5,6, biofuels7,8, and biosensing applications9,10,11. Achieving these applications through combining modular DNA "parts" into "devices" with specific functionality has been one of the major goals of synthetic biology. For this process to scale, there must be a technique that allows the creation of complex devices from large libraries of parts in a time-efficient, cost-efficient, and most importantly, reproducible manner.
Such an expansive assembly process is warranted because currently the field lacks a complete understanding of the rules guiding successful biological system design and composition. This is exacerbated by insufficiently characterized DNA parts12, lack of compatibility and composability of parts13, and unexpected, undesirable interactions between genetic components within synthetic devices14,15. In the absence of reliable predictive modelling, functional synthetic genetic devices are arrived at by trial and error, which demands tens, or even hundreds, of input-output signal strength variants of an intended device are screened and the "best" is selected for composition with downstream elements16. While modern standardized DNA assembly methods such as Golden Gate17, and Modular Cloning18,19,20 make this process easier, an experimental expert is still required to perform each protocol. As synthetic devices grow in size and complexity, the total available design space will become too large to construct and test manually21, and the process will be too artisanal for any significant progress which is replicable to be made in the field.
Until the advent of synthetic biology and biological part repositories such as the iGEM Parts Registry (http://partsregistry.org), the JBEI Inventory of Composable Elements22, and SynBioHub23, genetic parts were not stored in any standardized assembly format. Only a small handful of parts needed to be cloned per project, and thus, the volume of cloning done was small, and the realization of an assembled device was achievable and trivial compared to the actual research objectives. Molecular cloning was often ad hoc and performed using restriction digests based on restriction site and endonuclease availability rather than following any standardized process. The lack of standardization made it impractical to automate any cloning protocol as it was unlikely that the next cloning reaction would follow an identical protocol. Furthermore, automating DNA assembly required significant monetary investment in equipment (liquid handling robots and their associated software and labware infrastructure) as well as the time investment for the generation of instructions to develop accurate parameters for handling the various classes of liquids being processed and the precise series of instructions to run these protocols. Small scale cloning efforts did not justify these expenses. The combination of larger, more complex genetic device designs coupled with standardized assembly protocols24,25 creates an environment where the automation of these processes is very practical. Low cost robotics like the Opentrons OT-One26 are also emerging which allows even modestly funded labs to access this technology. In addition, "cloud" laboratories27 including Transcriptic and Emerald Cloud Lab as well as academic "biofoundries" such as the Edinburgh Genome Foundry, the UIUC iBioFab, and the MIT-Broad Foundry, harness robotics to assemble diverse sets of designs for a variety of customers quickly and repeatedly while maintaining a common repository of basic DNA primitives and assembly technologies for future orders.
One of the biggest challenges in automating the process of DNA assembly is the generation of the pipetting commands for the liquid handler. While the software interfaces for these devices are typically easy to use, complex pipetting instructions like those necessary for combinatorial DNA assembly require the scientist to explicitly specify each aspirate and dispense command manually. This creates a major bottleneck in the workflow, and leaves the script generation process vulnerable to the same pipetting errors as if the assembly were carried out manually. This necessitates a software tool that can automate all parts of this process, from designing the device library, to generating the pipetting instructions, and providing the researcher with the plate/reagent setups required to assemble them. In this work, we leverage our software tool to automate the design of a small combinatorial DNA device library, as well as the mixing of reagents (buffer, water, enzymes) and DNA parts (Figure 1b) into 96 one-pot modular DNA assembly reactions. Use of the tool requires no prior programming experience, is scalable and high throughput, and combinatorial by default. We show that cloning reactions prepared on two different automated liquid handling platforms yield correct sequence-verified clones with comparable frequency to reactions prepared manually (95%), and with significantly less hands-on time.
Protocol
1. Specify Parts to be Used in DNA Device Library and Generate User/Liquid Handler Instructions [15 min]
- Using any web browser, navigate to mocloassembly.com and upload Genbank files for all DNA parts that will be included in the combinatorial DNA device design.
- Once all files have been uploaded, select desired DNA parts and drag them onto the blank canvas, placing part types in the intended final order of DNA parts.
NOTE: Collections of parts, as well as individual parts, can be selected and placed on the canvas. Also, order DNA parts such that the 5' and 3' overhangs for each part match. - Click 'Assemble' on the bottom right of the page.
NOTE: The tool will only generate valid, buildable assemblies based on the four base pair overhangs that flank each part upon digestion with the BsaI enzyme. If no buildable DNA devices exist based on the parts uploaded by the scientist, the tool will indicate that no assemblies were found. - Navigate to the 'Plans' tab and download files generated by the tool. These files will include:
- Human-readable plate maps for the scientist to prepare DNA samples, as well as reagents necessary for the reactions
- A 'picklist' for the liquid handler
- Fully annotated Genbank files for all DNA devices to be assembled
NOTE: Liquid handling arm positioning when accessing any new labware must be tested. Consult the manufacturer's manual for detailed instructions on how to adjust the pipetting arm positioning in the X, Y, and Z axes as necessary.
2. Prepare Plasmid DNA and Reagents for Assembly [3 days]
- [Day 1] Using a sterile inoculation loop and working near an open flame or in a laminar flow hood, streak out bacterial glycerol stocks onto LB-agar plates supplemented with the appropriate antibiotic. Do this for all necessary DNA parts, making sure to sterilize the loop between every sample. Incubate plates at 37 °C overnight.
NOTE: Frozen bacterial glycerol stocks should be kept on ice as much as possible. Repeated freeze-thaw cycles lower the viability of the stock and should be avoided. - [Day 2] Using a sterile pipette tip, toothpick, or inoculation loop, and working near an open flame or in a laminar flow hood, inoculate 3 mL of LB broth (supplemented with appropriate antibiotic) with a single colony from the LB-agar plates prepared in 2.1. Incubate cultures overnight at 37 °C while shaking at 300 RPM.
- [Day 3] Purify plasmid DNA from the bacterial cultures using any commercially available mini-prep plasmid purification kit.
- Using the MoClo_Setup.xlsx file provided, dilute each sample of plasmid DNA to a concentration of 20 fmol/µL in water or TE buffer.
- Following the plate map of the PDF file generated by the assembly tool, place the indicated volume of each diluted DNA part into the appropriate well on a full-skirted 96-well PCR plate. Hold this SetupPlate on ice until needed, or seal with a foil adhesive seal and store at -20 °C.
- On ice, prepare the reaction mastermix with the following components: for each 20 µL of reaction add 2 µL of 10x T4 DNA ligase buffer, 0.5 µL of T4 DNA Ligase (HC), and 1 µL of BsaI enzyme. A calculator sheet is included in the MoClo_Setup.xlsx file to assist with this.
- Distribute the enzyme mastermix into the appropriate wells of a new full-skirted 96-well PCR plate, following the plate map for the ReagentPlate in the generated PDF. Keep this ReagentPlate on ice or on a 96-well cold-block.
NOTE: The ReagentPlate should only be prepared when ready to run the assembly on the liquid handler.
- Distribute the enzyme mastermix into the appropriate wells of a new full-skirted 96-well PCR plate, following the plate map for the ReagentPlate in the generated PDF. Keep this ReagentPlate on ice or on a 96-well cold-block.
3. Execute Assembly Script on the Liquid Handler [Variable]
- Place the SetupPlate(s), the ReagentPlate(s) (on a 96-well cold-block), and the necessary number of empty full-skirted 96-well PCR plates on the deck of the liquid handler. The empty plate(s) will be the OutputPlate(s) where the reactions are assembled.
- Prepare the liquid handler control software by creating instances of each sample and reagent plate prepared, making sure to name them exactly as they appear on the plate maps generated by mocloassembly.com, including a trough of clean, deionized water labeled ‘Reservoir’.
- Using the ‘Worklist’ command in the control software, load the .gwl file generated by our software tool, followed by another ‘Worklist’ command which will execute the .gwl file loaded in the first command.
- Execute the script using the controller software's 'Run' command.
NOTE: Always allow the robotic liquid handler to complete the execution of a script before trying to access the deck space. - Remove all plates from the liquid handler deck. Remaining DNA may be saved by sealing the SetupPlate(s) with aluminum sealing film and storing at -20 °C. Seal the OutputPlate(s) with adhesive film, place in a thermocycler or heat-block and run with the following cycle parameters:
37 °C for 2 h, 50 °C for 5 min, 80 °C for 10 min, hold at 4 °C
NOTE: Once the reaction thermocycling is complete, OutputPlate(s) can be stored at -20 °C until they are ready to be transformed.
4. Transform the Reactions [1 day]
- Thaw the requisite number of competent E. coli cell aliquots needed (10 µL/reaction) on ice.
NOTE: To maintain sterility, the following steps should be performed near an open flame, or within a laminar flow hood.- While cells are thawing, prepare LB-agar plates (containing appropriate antibiotic) by pipetting 50 µL of 0.1 M IPTG and 50 µL of 20 mg/mL X-GAL onto the surface. Make a master mix if plating a large number of reactions. Coat the plates evenly using a sterile glass rod or glass beads and allow the plates to rest at 37 °C for at least 15 min before plating bacteria.
NOTE: Alternatively, add IPTG and X-Gal to liquid agar before plates are poured, at 2 mM IPTG and 40 µg/mL X-Gal final concentration. Store these plates out of direct light as X-Gal is light-sensitive.
- While cells are thawing, prepare LB-agar plates (containing appropriate antibiotic) by pipetting 50 µL of 0.1 M IPTG and 50 µL of 20 mg/mL X-GAL onto the surface. Make a master mix if plating a large number of reactions. Coat the plates evenly using a sterile glass rod or glass beads and allow the plates to rest at 37 °C for at least 15 min before plating bacteria.
- On ice, aliquot 10 µL of competent cells for each reaction into a new 96-well PCR plate. This will be the Transformation Plate.
- Add 1-3 µL of each reaction from the OutputPlate(s) to the corresponding well in the Transformation Plate and incubate on ice for 5 min.
- Seal the Transformation Plate with adhesive film and heat shock in a thermocycler at 42 °C for 30 s. Then, immediately place the plate on ice for 2 min.
- To the same wells of the Transformation Plate, add 150 µL of SOC media, seal with an aluminum adhesive seal, and incubate at 37 °C while shaking at 900 RPM for 1 hr.
- Plate the full contents of each well of the Transformation Plate on the LB-agar plates prepared in 4.1.1 using a sterile glass rod or glass beads to evenly coat the surface of the plate. Incubate plates at 37 °C overnight.
5. Clone Verification [2 days]
- Prepare one or several 96-well deep-well culture blocks with 1.5 mL of LB broth (containing appropriate antibiotic).
- Similar to step 2.2, inoculate the deep-well culture block(s) with single white colonies from each LB-agar plate of transformed reactions.
- Seal the culture block(s) with a gas-permeable seal and incubate overnight at 37 °C while shaking at 900 RPM.
NOTE: The modular cloning technique utilizes blue-white screening, so positive CFUs will appear white on the LB-agar plate, while empty destination vectors will appear blue.
- Isolate plasmid DNA from bacterial cultures (as in 2.3) and submit for Sanger sequencing to verify clones.
Representative Results
Here we demonstrate the automated modular assembly of 96 DNA devices from various basic DNA parts (Figure 1b) using two automated robotic liquid handling platforms. Each transcriptional unit is a linear arrangement of promoter, ribosomal binding site, gene, and transcriptional terminator, cloned into a specific destination vector. TUs are a key component in many hierarchical genetic circuit designs28,29,30 and are therefore a natural proof of concept for this approach. Sequence-verified clones can be achieved in ~5 days from start to finish, and an overview of the presented workflow is presented in Figure 1a.
Using the described tool and protocol, we captured several metrics useful in determining the optimal assembly platform given a defined number of cloning reactions to be generated by the scientist. Figure 2a illustrates a comparison between reaction assembly times across all three modalities. Execution time is the total time to complete all pipetting steps needed to assemble 96 reactions, which includes dispensing of all DNA parts and reagents. Hands-on time refers to the total time a human was manually involved in the preparation of the cloning reactions, or setup of the software running the liquid handler. Figure 2b compares cost between the different methods. Costs presented are for a single reaction prepared by each method and include the price of enzymes and disposable pipette tips, given the lowest typical reaction volume (20 µL for liquid handler, 10 µL for manual, 250 nL for acoustic dispenser). Single clones from all 96 reactions for both manual and liquid handler-prepared sets were sequenced, with a subset of 12 sequenced for the acoustic dispenser reactions (Figure 2c). Correct sequences were obtained for 95% of the 96 manual and liquid handler sets. 83% of the acoustic dispenser sample subset were correct, however the final two reactions failed to yield any white colonies, likely due to insufficient mixing of droplets after the dispensing of DNA and reagents was complete. Figure 2d illustrates cloning reaction efficiencies, as measured by the ratio of white colonies to total number of colonies, which are comparable between manually (94%) and liquid handler-assembled (84%) reactions. Interestingly, when scaling down the final reaction volume with the acoustic dispenser, we noticed a marked drop in reaction efficiency when reactions were prepared at volumes smaller than 1 µL (Supplemental Figure 1 & Supplemental Table 2). This is likely due to evaporation during reaction thermocycling, which can cause an increase in salt concentrations in the reaction.
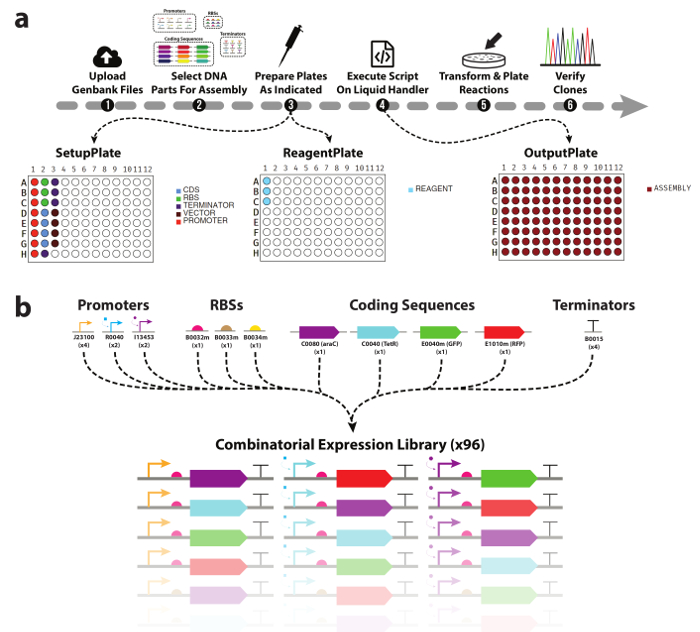
Figure 1. Automated DNA device library assembly workflow.
(a) An overview of the experimental workflow presented in this work. (1) Researchers first upload Genbank files for all DNA parts & destination vectors they wish to use. (2) Next, DNA parts to be included in the assembly are selected. (3) The tool will then generate a picklist for an automated liquid handler, as well as plate maps to assist with manual population with DNA parts and enzyme mastermix. (4) Using the DNA & reagents plates, as well as the generated picklist, researchers execute the assembly of the cloning reactions on the liquid handler. (5) Once complete, the reactions are transformed and plated for downstream analysis. (b) List of DNA parts used in this work. A total of three promoters, three ribosomal binding sites, four coding sequences, and one transcriptional terminator were used generate the combinatorial library of 96 TUs. Please click here to view a larger version of this figure.
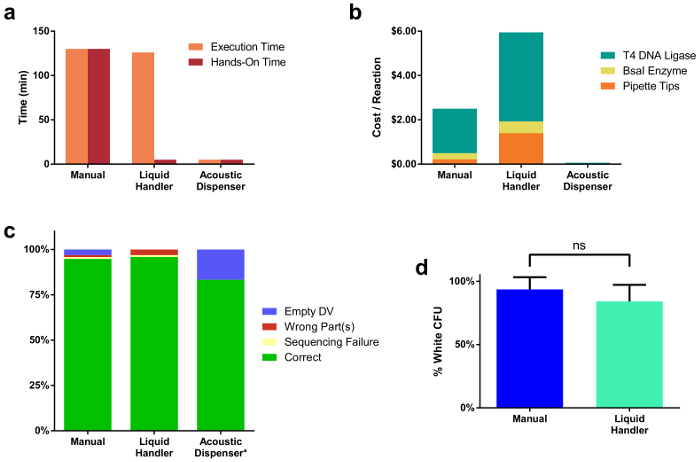
Figure 2. Comparisons across three different reaction assembly modalities.
(a) Reaction setup time for 96 reactions assembled manually, via a liquid handler, and by an acoustic dispenser. Manual assembly took 2 h 10 min, all of which was hands-on time. The liquid handler took a similar amount of time (2 h 6 min) to execute the pipetting commands, however only a fraction of that time (5 min) was hands-on. The acoustic dispenser took significantly less time to execute liquid transfers (5 min) and took minimal hands-on time (5 min). (b) Price per single cloning reaction for each assembly method. This price includes the cost of enzymes used, as well as pipette tips. (c) Sequencing results from single colonies of all 96 assembled TUs. The percentage of correct clones was comparable across all assembly methods. Note that only a subset (12) of the full 96 assemblies were transformed from the acoustic dispenser prepared samples. Two reactions failed to yield any white colonies, likely due to insufficient mixing of DNA & enzyme mastermix droplets in the OutputPlate. (d) Comparison of cloning reaction efficiencies between manual and liquid handler-prepared reactions. Please click here to view a larger version of this figure.
Supplemental Figure 1. Reaction efficiency decreases with lowering reaction volume.
Reaction efficiencies drop markedly when scaling down reaction volumes below 1 µL. This trend is seen with two different final DNA concentrations; however, scientists may opt to use smaller volumes to save on reagent costs if lower efficiencies can be tolerated. Please click here to download this file.
Supplemental Table 1.
Table of DNA and ENZYME liquid class parameters for various pipetting volumes. Please click here to download this file.
Supplemental Table 2.
Reaction efficiency calculations. Raw CFU numbers are given for 12 of the 96 transformed reactions prepared manually and via the liquid handler. CFU numbers are also provided for testing of smaller final reaction volumes on the acoustic dispenser. Please click here to download this file.
Discussion
In conclusion, the automation of the complete DNA device creation process, from in-silico design to liquid handling, is a viable goal with currently existing technology. Software and modern robotics allow the creation of workflows that are cost-efficient, time-efficient, and scalable, while also producing more consistently reproducible results than manual methods. While automation may not always be the most cost-effective choice for executing a protocol, it does improve experimental reproducibility and frees up valuable researcher time. However, depending on the hardware utilized, the use of automation can sometimes drive cost and execution time well below what can be achieved through conventional manual methods. Furthermore, automation captures protocols explicitly in a formalized manner preventing ad-hoc, artisanal, and anecdotal best practice based experimentation. Here the automated assembly of modular DNA devices is demonstrated, and the protocol, electronic files, and physical DNA resources needed for the reader to perform these, and similar, experiments on their own are provided.We hope the availability of our tool, and the publication of this protocol will serve as a resource and move the field toward a more transparent and communal future in the area of DNA assembly processes and liquid handling robotics.
The usefulness of automation hardware is largely dependent on the configuration & capabilities of that hardware. For instance, our liquid handler uses system fluid displacement to actuate aspirate and dispense commands. The pistons that drive the system fluid are relatively large 1 mL syringes which, while useful for a range of volumes, imposes a 2 µL lower limit for accurate dispensing of reagents. As a consequence, we scaled up the total volume of cloning reactions set up on the liquid handler to 20 µL, since every dispense command needed to be ≥2 µL. This effectively doubled the cost per reaction for liquid handler-prepped reactions, however the amount of hands-on time required to execute those reactions was significantly reduced. In an effort to address this issue, we repeated the reaction setup for all 96 reactions on an acoustic liquid dispenser. This device uses sound energy to dispense fluid directly from one plate to another, and can achieve dispense volumes far below (2.5 nL) what is possible with standard air displacement based manual pipettes. Using this device, we were able to scale down the total volume of our reactions to 250 nL, a 40-fold reduction compared to manually prepared reactions of 10 µL. Because of the small volumes dispensed, and the lack of tip changes between pipetting steps, the acoustic dispenser was able to generate the same 96 reactions in a fraction of the time (<5 min). The smaller reaction volumes also save on wasted reagents, since we typically only transform 1-3 µL of the reaction. That being said, the use of automation hardware, in conjunction with intuitive software, can make the generation of large numbers of DNA assembly reactions accessible to a much wider academic audience.
Here, we demonstrate the utility of our software tool, however there are a number of features which would help broaden its usefulness. First, each time the tool is used to generate a combinatorial assembly, new DNA part plates must be generated, requiring the scientist to manually populate these plates for every assembly run. It would be helpful, instead, if the scientist could specify the location of parts in a DNA plate to be used in the assembly. This would allow the use of high-throughput plasmid DNA purification kits since researchers could inoculate cultures from a kit like the CIDAR MoClo Library, and purify all samples together while maintaining the well location of each part specified in the kit. Second, the tool currently only supports the use of 96-well plates. For larger projects where several hundred DNA devices need to be built, the number of DNA, reagent, and destination plates may exceed the deck capacity of the liquid handler. This problem could be, at least partially, alleviated by support for higher density plate formats (384 or 1536-well). Lastly, the tool currently only supports a single type of liquid handler and DNA assembly strategy. While it is relatively easy to convert the tool-produced liquid handler instructions to an acoustic dispenser format using spreadsheet software, we hope to expand native support to many different liquid handlers which would greatly broaden its applicability, as would compatibility with other common DNA assembly techniques like Gibson assembly31.
A crucial part of this automated assembly ecosystem is a set of software tools that translates high-level assembly plans into automation friendly protocols that are explicitly scheduled to run on liquid handling robots. Though a number of software tools exist that allow researchers to design assemblies in-silico including Benchling, MoClo Planner, and Raven32, few have the ability to translate those designs into executable instructions to run on a liquid handler. To that end, work such as PR-PR Automation and Puppeteer33,34,35 have begun to make these tools available. In addition, commercial entities working in this area are looking at ways to introduce "cloud labs" which provide experimental services to large groups of end-users via automation. The protocol outlined in this paper could serve as a piece of any of these efforts provided they are presented as a service.
While automated assembly of DNA devices is of immediate and obvious value to synthetic biology, our protocol is useful for the larger community of molecular biologists as well. Automating DNA assembly allows large numbers of known, but similar, genetic devices to be created in parallel and can enable the rapid synthesis of expression libraries for screening and testing purposes in drug development studies. We hope that our software tool will make larger combinatorial-based DNA assembly efforts more accessible, and serve as a useful resource to both the synthetic biology, as well as larger academic community.
Açıklamalar
The authors have nothing to disclose.
Acknowledgements
We thank Swapnil Bhatia, Alejandro Pelaez, and Johnson Lam for work on the Puppeteer project, as well as Swati Carr, Rachael Smith, and Thomas Costa for help with this manuscript. This work was funded by NSF CAREER Award #1253856. It is also funded by the NSF Expeditions in Computing Award #1522074.
Materials
| Hardware / Software | |||
| Freedom EVO 150 Liquid Handling Robot | Tecan | Custom liquid handler fitted with an 8-channel pipetting arm http://lifesciences.tecan.com/products/liquid_handling_and_robotics/freedom_evo |
|
| Freedom EVOware Standard (Version 2.4 Service Pack 2) | Tecan | Software used to control the Freedom EVO 150 liquid handler http://lifesciences.tecan.com/products/software/freedom_evoware |
|
| Echo 550 | Labcyte | Acoustic Liquid Dispenser http://www.labcyte.com/products/liquidhandling/echo-550-liquid-handler |
|
| Sorvall Legend RT | Sorvall | Large benchtop swing-bucket sentrifuge | |
| MasterCycler Pro | Eppendorf | 950040015 | Thermocycler with 96-well heat block https://online-shop.eppendorf.us/US-en/PCR-44553/Cyclers-44554/Mastercycler-pro-PF-5193.html |
| ECHOTHERM Chilling/Heating Dry Bath | Torrey Pines Scientific | Heating/Chilling block for EVO 150 deck https://www.torreypinesscientific.com/products/chilling-and-heating-dry-baths/echotherm-ric20-series-remote-controlled-chillingheating-dr |
|
| Tabletop Microcentrifuge 5418 | Eppendorf | 5418000017 | Stardard 18-well microcentrifuge https://online-shop.eppendorf.com/OC-en/Centrifugation-44533/Centrifuges-44534/Centrifuges-5418–5418R-PF-9257.html |
| Name | Company | Catalog Number | Yorumlar |
| Kaynaklar | |||
| mocloassembly.com | Lattice Automation | Web-tool for combinatorial DNA assembly mocloassembly.com |
|
| CIDAR MoClo Parts Kit | AddGene | 1000000059 | Kit of bacterial glycerol stocks for all DNA parts used in this study https://www.addgene.org/cloning/moclo/densmore/ |
| CIDAR ICE Registry | CIDAR Lab | Registry of plasmid DNA maps https://ice.cidarlab.org/folders/8 https://synbiohub.programmingbiology.org/public/bubdc_ice/bubdc_ice_folder_8/current |
|
| Name | Company | Catalog Number | Yorumlar |
| Labware | |||
| 50 μL Conductive Tips | Tecan | 30 057 818 | Sterile 50 μL conductive tips for Tecan liquid handler http://lifesciences.tecan.com/products/consumables/disposable_tips/liquid_handling_disposable_tips |
| 2 mL Deep 96-well Culture Plates | USA Scinetific | 5678-0285 | Bacterial culture plates used for culturing of large numbers of samples http://www.thermoscientific.com/en/product/nunc-1-3-2-0ml-deepwell-plates-shared-wall-technology.html |
| 1.5 mL Microcentrifuge Tubes | USA Scinetific | 1615-5599 | Disposable microcentrifuge tubes http://www.usascientific.com/Seal-Rite-1.5-ml-tube-colors.aspx |
| Breathe Easier sealing membrane | Sigma-Aldrich | Z763624-100EA | Breathable sealing membrane for bacterial culture plates http://www.sigmaaldrich.com/catalog/product/sigma/z763624?lang=en®ion=US |
| Full-Skirted, Low-Profile, 96-Well PCR Plates | GeneMate | T-3183-2 | PCR plates used for all steps https://www.bioexpress.com/store/catalog/product.jsp?catalog_number=T-3183-R |
| Alluminum Sealing Foil for PCR Plates | GeneMate | T-2451-1 | Alluminum seals for PCR plate storage https://www.bioexpress.com/store/catalog/product.jsp?catalog_number=T-2451-1 |
| Polyolefin Sealing Film for PCR Plates | GeneMate | T-2450-1 | Plastic seals for PCR plates during cycling https://www.bioexpress.com/store/catalog/product.jsp?catalog_number=T-2450-1 |
| PCR Cooler | Eppendorf | 22510525 | 96-well cold block https://online-shop.eppendorf.us/US-en/Temperature-Control-and-Mixing-44518/Accessories-44520/PCR-Cooler-PF-55940.html |
| Name | Company | Catalog Number | Yorumlar |
| Reagents | |||
| GenCatch Plasmid DNA Mini-Prep Kit | Epoch Life Sciences | 2160250 | Plasmid DNA purification kit http://www.epochlifescience.com/Product/PurificationKit/dna_mini.aspx |
| T4 DNA Ligase (HC) | Promega | M1794 | High concentration T4 DNA Ligase https://www.promega.com/products/cloning-and-dna-markers/molecular-biology-enzymes-and-reagents/t4-dna-ligase/?catNum=M1794 |
| BbsI Restriction Enzyme | New England Biolabs | R0539L | BbsI enzyme at 10,000 units/ml https://www.neb.com/products/r0539-bbsi |
| BsaI Restriction Enzyme | New England Biolabs | R0535L | BsaI enzyme at 10,000 units/ml https://www.neb.com/products/r3535-bsai-hf |
| T4 DNA Ligase Buffer Pack | Promega | C1263 | 10x T4 DNA ligase buffer https://www.promega.com/products/cloning-and-dna-markers/cloning-tools-and-competent-cells/t4-dna-ligase/ |
| Isopropyl-ß-D-thiogalactopyranoside (IPTG) | Zymo Research | I1001-25 | 0.5M IPTG Solution http://www.zymoresearch.com/buffers-solutions/chemicals/isopropyl-ss-d-thiogalactopyranoside-iptg |
| 5-bromo-4-chloro-3-indolyl ß-D-galactopyranoside (X-GAL) | Zymo Research | X1001-25 | 20 mg/ml X-GAL solution http://www.zymoresearch.com/buffers-solutions/chemicals/5-bromo-4-chloro-3-indolyl-ss-d-galactopyranoside-x-gal |
| Kanamycin Sulfate | Zymo Research | A1003-25 | 35 mg/ml Kanamycin solution http://www.zymoresearch.com/buffers-solutions/antibiotics/kanamycin-sulfate |
| Carbenicillin (Disodium Salt) | Fisher | BP26481 | 1 g Carbenicillin (Ampicillin analog) https://www.fishersci.com/shop/products/carbenicillin-disodium-salt-fisher-bioreagents-3/p-25005#?keyword=carbenicillin |
| SOC Broth Media | Teknova | S0225 | Powder media used to make SOC broth http://www.teknova.com/SOC-BROTH-MEDIA-p/s0225.htm |
| LB Broth (Lennox) Media | Sigma-Aldrich | L3022-1KG | Powder media used to make LB broth http://www.sigmaaldrich.com/catalog/product/sigma/l3022?lang=en®ion=US |
| LB Broth with agar (Lennox) Media | Sigma-Aldrich | L2897-1KG | LB with agar mix used for making solid media plates http://www.sigmaaldrich.com/catalog/product/sigma/l2897?lang=en®ion=US |
| Alpha-Select Gold Efficiency Competent Cells | Bioline | BIO-85027 | High efficiency chemically competent E. coli cells http://www.bioline.com/us/alpha-select-gold-efficiency.html |
| Name | Company | Catalog Number | Yorumlar |
| Primers | |||
| Primer VF | 5'-TGCCACCTGACGTCTAAGAA-3' Primers used for Sanger sequencing and colony PCRs |
||
| Primer VR | 5'-ATTACCGCCTTTGAGTGAGC-3' Primers used for Sanger sequencing and colony PCRs |
Referanslar
- Gardner, T. S., Cantor, C. R., Collins, J. J. Construction of a genetic toggle switch in Escherichia coli. Nature. 403 (6767), 339-342 (2000).
- Elowitz, M. B., Leibler, S. A synthetic oscillatory network of transcriptional regulators. Nature. 403 (6767), 335-338 (2000).
- Lutolf, M. P., Hubbell, J. A. Synthetic biomaterials as instructive extracellular microenvironments for morphogenesis in tissue engineering. Nat Biotechnol. 23 (1), 47-55 (2005).
- Anderson, J. C., Clarke, E. J., Arkin, A. P., Voigt, C. A. Environmentally controlled invasion of cancer cells by engineered bacteria. J Mol Biol. 355 (4), 619-627 (2006).
- Xie, Z., Wroblewska, L., Prochazka, L., Weiss, R., Benenson, Y. Multi-input RNAi-based logic circuit for identification of specific cancer cells. Science. 333 (6047), 1307-1311 (2011).
- Ro, D. K., et al. Production of the antimalarial drug precursor artemisinic acid in engineered yeast. Nature. 440 (7086), 940-943 (2006).
- Georgianna, D. R., Mayfield, S. P. Exploiting diversity and synthetic biology for the production of algal biofuels. Nature. 488 (7411), 329-335 (2012).
- Savage, D. F., Way, J., Silver, P. A. Defossiling fuel: how synthetic biology can transform biofuel production. ACS Chem Biol. 3 (1), 13-16 (2008).
- Fussenegger, M., et al. Streptogramin-based gene regulation systems for mammalian cells. Nat Biotechnol. 18 (11), 1203-1208 (2000).
- Boorsma, M., et al. A temperature-regulated replicon-based DNA expression system. Nat Biotechnol. 18 (4), 429-432 (2000).
- Malphettes, L., et al. A novel mammalian expression system derived from components coordinating nicotine degradation in arthrobacter nicotinovorans pAO1. Nucleic Acids Res. 33 (12), e107 (2005).
- Brophy, J. A., Voigt, C. A. Principles of genetic circuit design. Nat Methods. 11 (5), 508-520 (2014).
- Lou, C., Stanton, B., Chen, Y. J., Munsky, B., Voigt, C. A. Ribozyme-based insulator parts buffer synthetic circuits from genetic context. Nat Biotechnol. , (2012).
- Cardinale, S., Arkin, A. P. Contextualizing context for synthetic biology–identifying causes of failure of synthetic biological systems. Biotechnol J. 7 (7), 856-866 (2012).
- Carr, S. B., Beal, J., Densmore, D. M. Reducing DNA context dependence in bacterial promoters. PLoS One. 12 (4), e0176013 (2017).
- Yeung, E., Ng, A., Kim, J., Sun, Z. Z., Murray, R. M. . Decision and Control (CDC), 2014 IEEE 53rd Annual Conference on. , 5405-5412 (2014).
- Engler, C., Gruetzner, R., Kandzia, R., Marillonnet, S. Golden gate shuffling: a one-pot DNA shuffling method based on type IIs restriction enzymes. PLoS One. 4 (5), e5553 (2009).
- Weber, E., Engler, C., Gruetzner, R., Werner, S., Marillonnet, S. A modular cloning system for standardized assembly of multigene constructs. PLoS One. 6 (2), e16765 (2011).
- Iverson, S. V., Haddock, T. L., Beal, J., Densmore, D. M. CIDAR MoClo: Improved MoClo Assembly Standard and New E. coli Part Library Enable Rapid Combinatorial Design for Synthetic and Traditional Biology. ACS Synth Biol. 5 (1), 99-103 (2016).
- Casini, A., Storch, M., Baldwin, G. S., Ellis, T. Bricks and blueprints: methods and standards for DNA assembly. Nat Rev Mol Cell Biol. 16 (9), 568-576 (2015).
- Bhatia, S. P., Smanski, M., Voigt, C. A., Densmore, D. M. Genetic design via combinatorial constraint specification. ACS Synth Biol. , (2017).
- Ham, T. S., et al. Design, implementation and practice of JBEI-ICE: an open source biological part registry platform and tools. Nucleic Acids Res. 40 (18), e141 (2012).
- Madsen, C., et al. The SBOL Stack: A Platform for Storing, Publishing, and Sharing Synthetic Biology Designs. ACS Synth Biol. 5 (6), 487-497 (2016).
- Knight, T. . Draft standard for BioBrick biological parts. , (2007).
- Gibson, D. G., et al. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods. 6 (5), 343-345 (2009).
- Ma, A. C., et al. FusX: A Rapid One-Step Transcription Activator-Like Effector Assembly System for Genome Science. Hum Gene Ther. 27 (6), 451-463 (2016).
- Check Hayden, E. The automated lab. Nature. 516 (7529), 131-132 (2014).
- Nielsen, A. A., et al. Genetic circuit design automation. Science. 352 (6281), aac7341 (2016).
- Woodruff, L. B. A., et al. Registry in a tube: multiplexed pools of retrievable parts for genetic design space exploration. Nucleic Acids Res. 45 (3), 1553-1565 (2017).
- Hasty, J., McMillen, D., Collins, J. J. Engineered gene circuits. Nature. 420 (6912), 224-230 (2002).
- Gibson, D. G. Enzymatic assembly of overlapping DNA fragments. Methods Enzymol. 498, 349-361 (2011).
- Appleton, E., Tao, J., Haddock, T., Densmore, D. Interactive assembly algorithms for molecular cloning. Nat Methods. 11 (6), 657-662 (2014).
- Vasilev, V., Liu, C., Haddock, T., Bhatia, S., Adler, A., Yaman, F., Beal, J., Babb, J., Weiss, R., Densmore, D. A Software Stack for Specification and Robotic Execution of Protocols for Synthetic Biological Engineering. SynBERC Fall Retreat, Harvard University. , (2011).
- Beal, J., et al. An end-to-end workflow for engineering of biological networks from high-level specifications. ACS Synth Biol. 1 (8), 317-331 (2012).
- Bhatia, S., Densmore, D. Pigeon: a design visualizer for synthetic biology. ACS Synth Biol. 2 (6), 348-350 (2013).

