Targeting Biofilm Associated Staphylococcus aureus Using Resazurin Based Drug-susceptibility Assay
Özet
Most bacterial infections produce a biofilm. By virtue of their environment, biofilm associated bacteria are often phenotypically drug resistant. Novel antibacterial molecules that kill bacteria in biofilms are thus a high priority. We establish an assay to quickly screen for antimicrobial compounds that are effective at eradicating biofilms.
Abstract
Most pathogenic bacteria are able to form biofilms during infection, but due to the difficulty of manipulating and assessing biofilms, the vast majority of laboratory work is conducted with planktonic cells. Here, we describe a peg plate biofilm assay as performed with Staphylococcus aureus. Bacterial biofilms are grown on pegs attached to a 96-well microtiter plate lid, washed through gentle submersion in buffer, and placed in a drug challenge plate. After subsequent incubation they are again washed and moved to a final recovery plate, in which the fluorescent dye resazurin serves as a viability indicator. This assay offers greatly increased ease-of-use, reliability, and reproducibility, as well as a wealth of data when conducted as a kinetic read. Moreover, this assay can be adapted to a medium-throughput drug screening approach by which an endpoint fluorescent readout is taken instead, offering a path for drug discovery efforts.
Introduction
Pathogenic microbes can form biofilms in vivo leading to non-acute chronic infections 1. Biofilm-associated infection is a serious risk factor of medical procedures that involve implantation of foreign objects (e.g., artificial bone replacements, breast implants) or the installation of tracheal tubes or urinary catheters 2. In these contexts, anti-infective therapy is almost always necessary as biofilm-based infections are rarely cleared on their own, even in immunocompetent individuals. Staphylococcus aureus is one of the most frequently observed pathogens implicated in biofilm-related complications occurring during the use of invasive medical devices 3.
Unfortunately, the very nature of biofilms as a protective barrier makes them more resistant to treatment than planktonic cells 1,4, and evaluation of predicted clinical efficacy is a critical part of both initial drug development as well as drug resistance surveillance. There is increasing acknowledgement that laboratory conditions, focused on planktonic cultures, may not faithfully represent real-world disease 5. Further compounding the problem, replication of biofilm phenotypes is difficult, and existing biofilm models are tedious and suffer from high inter- and intra-assay variability 6. Thus, many researchers, through necessity, default to planktonic cell assays of drug susceptibility, thereby potentially neglecting an important aspect of bacterial virulence and disease.
Here we describe the protocol for assaying bacteria, specifically S. aureus, in pre-grown biofilms utilizing a 96-well based biofilm system 7,8. While the protocol for biofilm formation and challenge follows essentially the recommendation of the manufacturer, we present an alternative information-rich methodology for quantifying the viability of the biofilm after challenge. Briefly, bacteria are cultured in the peg plate, where biofilms form on the protruding pegs attached to the plate's lid. After biofilm formation, the pegs are gently dipped in wells of a fresh plate filled with PBS to remove planktonic cells. The peg-lid, with biofilms attached, is transferred to a new challenge plate, containing various concentrations of antibiotics to be assayed. After a second incubation, lids are again removed, washed, and transferred to a recovery plate containing resazurin dye, where they undergo a final incubation. Resazurin conversion can be recorded kinetically or taken as an endpoint reading after a defined recovery period. This dye-based method of quantifying the viability of biofilms differs considerably from the tedious CFU (colony forming units) count-based methodology described in the original protocol 7. OD600 measurements of the drug challenge plate and resazurin conversion kinetics serve as viability readouts of planktonic and biofilm cells, respectively, offering a fast, reliable, information-rich and technically simple assay for biofilm survival.
Protocol
1. Biofilm Initiation
- Grow a culture of biofilm producing organism in a nutrient rich medium. Inoculate Staphylococcus aureus strain Newman in 10 ml of Mueller-Hinton medium from a glycerol stock. Perform all work involving handling of S. aureus with gloves and within a biosafety cabinet.
- Incubate for 16 hr at 37 °C on a rotary shaker (100-200 rpm).
- Determine OD600 of the culture using a spectrophotometer and a standard cuvette (path length: 1cm).
- Calculate the volume (V) of culture needed to prepare 20 ml of inoculum with OD600(inoculum) of 0.1 in chelexed Roswell Park Memorial Institute media (CRPMI)9 using the formula: [V = 20 ml*0.1/OD600(culture)]
Note: Reference 9 provides details on CRPMI preparation.- Add the calculated culture volume (from above) to a centrifuge tube and pellet cells by spinning for 1 min at 10,000 x g. To prepare biofilm inoculum, aspirate the spent growth medium and resuspend cell pellet in 20 ml CRPMI.
- Pour inoculum into a 25 ml reservoir.
- Carefully remove the lid from a sterile 96-well peg plate by pulling it straight up.
Note: Care must be taken not to contaminate the pegs hanging down from the lid. The lid can be placed upside down on a clean surface within a biosafety cabinet. - Using a 200 µl multi-channel pipette, add 125 µl inoculum to each well of rows A to G of the bottom plate.
- Fill 125 µl sterile CRPMI medium in each well of row H as a sterility control.
- Take the peg-lid and hold it above the plate bottom with the pegs facing downwards. Carefully align the individual pegs with their respective wells. Avoid physical contact between the plate and the pegs. Once aligned, lower the lid carefully down. Avoid misalignments as readjustments performed after the pegs have touched the bottom plate may contaminate sterility controls in row H.
- Seal the lid to the plate with paraffin film to prevent evaporation and place in a sealable plastic bag, sealing tightly. Keep the air volume in the bag as low as possible to minimize evaporation.
- Incubate without shaking at 37 °C for 48 hr in a 5% CO2 incubator.
Note: 48 hr of growth has been optimal for S. aureus, but will vary based upon the organism used.
Note: Although static incubation is a good starting point to optimize the assay, gently shaking at 150 rpm on a microplate shaker is a possible variation that may enhance biofilm formation of some S. aureus isolates.
2. Preparation of the Biofilm Challenge Plate
- On the day of the biofilm challenge take a fresh 96-well plate and add 100 µl of CRPMI, equilibrated to RT, to each well of rows B to H. To avoid inconsistent results, utilize a 96-well plate that can hold 200 µl of media when the pegs are inside, and features wells with dimensions identical to those of the biofilm initiation plate.
- Dilute up to 4 test compounds separately in 1.0 ml of CRPMI to 2x the desired final concentration, in order to account for a final dilution in step 2.7.
Note: A good starting point is 8 fold above the inhibitory concentration of planktonic cells. - Add 200 µl of compound 1 in wells A1 to A3, compound 2 in wells A4 to A6, compound 3 in wells A7 to A9 and compound 4 in wells A10 to A12.
- Serially dilute the test compounds by using a multichannel pipette and transfer 100 µl from row A to row B and mix.
- In that manner, continue to transfer 100 µl well to well. Mix after each transfer and stop in row F.
- After mixing, expel 100 µl from row F into a waste container. Do not transfer any liquid to rows G and H.
- Add 100 µl of CRPMI to each well of the plate using a multichannel pipette to bring the volume to 200 µl. See Figure 1 for final plate layout.
3. Biofilm Challenge
- Add 200 µl of phosphate buffered saline (PBS) to a fresh sterile 96-well plate that features wells with dimensions identical to those of the biofilm initiation plate.
- Remove the plastic bag and paraffin film from incubated biofilm initiation plate.
- Lift the lid straight up. Avoid contact between the pegs and the plate bottom as this may damage the biofilm. Keep bottom part for subsequent OD600 analysis (see 3.8).
- Rinse off planktonic cells by placing the peg-lid onto the plate containing PBS (from step 3.1). Submerge pegs for 5 sec, lift out of PBS, and submerge once more, being careful not to hit the pegs against the well sides.
- Place the rinsed peg-lid into the challenge plate. Avoid unnecessary contact between pegs and bottom plate as this will damage the biofilm leading to inconsistent results.
- Seal plate with paraffin film and place into a sealable plastic bag as described in step 1.10.
- Incubate the plate without shaking for 24 hr at 37 °C in a 5% CO2 incubator.
- Take bottom of biofilm initiation plate from 3.3. Resuspend the bacteria in each well using a multichannel pipette and measure the OD600 with a suitable microplate reader to confirm growth occurring in all wells but the sterility controls in row H.
4. Biofilm Determination
- Use a plate reader equipped with an incubation chamber and capable of taking kinetic fluorescence readings for detecting resazurin conversion. Set up a 24 hr kinetic fluorescence measurement using a 530 nm excitation and 590 nm emission.
- Set the chamber temperature to 37 °C and have the plate read every 20 min. Set the plate reader to measure from the bottom of the plate according to manufacturer's instructions.
- Prepare wash plate by adding 200 µl of PBS with a multichannel pipette to a sterile 96 well plate.
- Prepare biofilm recovery medium by adding 400 µl of 0.8 mg/ml resazurin stock solution to 20 ml of CRPMI medium to a final concentration of 16 µg/ml resazurin.
Note: Resazurin is a known skin irritant. Use a lab coat, splash goggles, and gloves while handling. CRPMI as the biofilm recovery media was used because it provides the lowest background signal, but it can be replaced by Mueller Hinton media if appropriate. - Add 150 µl of biofilm recovery medium with a multichannel pipette to each well of a fresh 96-well plate.
- Transfer challenge plate from incubator into a biosafety cabinet and carefully remove plastic bag and sealing film.
- Remove the peg-lid from the challenge plate and rinse off planktonic cells in PBS as described in 3.4. Keep the bottom of the challenge plate for subsequent OD600 analysis (see 4.10).
- After rinsing, place the peg-lid into the plate bottom containing the biofilm recovery medium.
- Wrap the side of the plate with paraffin film, being careful not to obstruct the bottom of the perimeter wells. Immediately after wrapping the plate, place it on the plate reader and start a kinetic read over a 24 hr period by initiating the previously made resazurin kinetic protocol from step 4.1.
- To quantify growth of planktonic cells that may or may not occur during challenge, read the OD600 of the entire challenge plate bottom using a micro plate reader. Follow instructions given in step 3.8.
Note: Carefully resuspend the content of each well as cells shedding off the biofilm tend to grow in clumps at the bottom of the well. Any bubbles within the well will strongly interfere with OD600 readings and must be avoided or removed prior to the read.
Representative Results
The resazurin assay is sensitive enough to reliably detect very few viable cells capable of replicating in drug free medium after challenge. In this methodology, we define the minimal biofilm eradicating concentration as the lowest concentration at which no resazurin conversion is seen within 24 hr. The resazurin assay relies on oxidative molecules found in metabolically active cells which convert the dye color from blue to pink. Dye conversion is thus an indicator of cell growth, and can be quantified using a fluorescent micro plate reader. In this paper, neocuproine and Cu-neocuproine (neocuproine complexed with copper) were tested for their activity toward biofilm associated S. aureus applying a layout that is based on Figure 1. The data obtained during execution of the protocol are an OD600 reading of the biofilm initiation plate, which serves as a quality control (data not shown), an OD600 reading of the plate bottom after challenge has been completed (Figure 2A), and a kinetic recording of resazurin conversion into its fluorescent metabolite resorufin of the recovery plate (Figure 2B). Further, an endpoint image of the challenge plate can be used for visual inspection of resazurin conversion (Figure 2C) if a yes/no evaluation of resazurin conversion is sufficient for the purpose of the experiment. Alternatively, if kinetic recording capabilities are unavailable or multiple plates are run in parallel, fluorescence can be determined after 24 hr by a single endpoint read. As seen in Figure 2A, OD readings from the challenge plate indicate that neocuproine alone does not inhibit growth of planktonic cells; however, Cu-neocuproine inhibits growth at 1.3 µM, as expected 10. A similar pattern is observed from the resazurin kinetic of the biofilm plate, indicating that only Cu-neocuproine is able to eliminate metabolically active biofilm-associated cells (Figure 2B).
In addition to providing biofilm eradication concentrations, the kinetic assay provides information on the metabolic state of the biofilm which is indicative of the number of viable bacteria. When comparing biofilms treated with gentamicin at increasing concentrations to the untreated controls, a lag of resazurin conversion can be seen (Figure 3). For 2.5 µg/ml gentamicin, there is a 13 hr delay until the well reaches the same conversion of resazurin as the untreated control. This lag likely comes from a reduced number of viable cells present in the treated biofilm compared to the untreated.
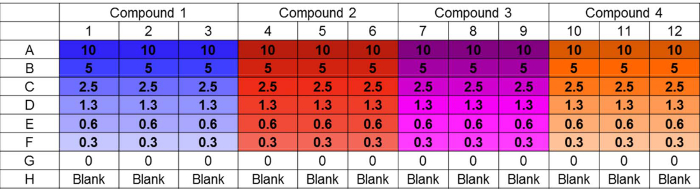
Figure 1. Challenge plate layout. Example layout of challenge plate accommodating parallel testing of four compounds in triplicate at six distinct concentrations. Untreated and sterility controls are also included. The layout design supports convenient in-plate preparation of serial dilutions using a multichannel pipet. Concentrations are in µM. Please click here to view a larger version of this figure.
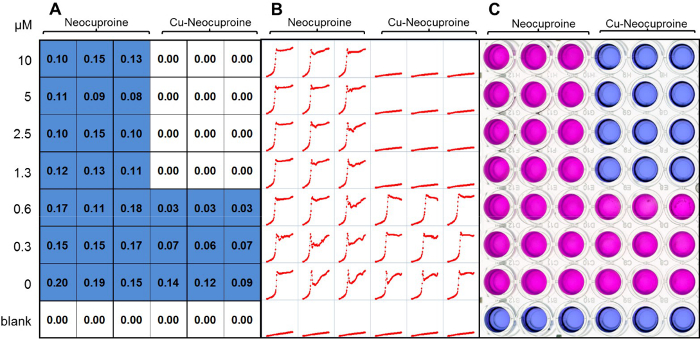
Figure 2. Determining minimum biofilm eradication concentration. Neocuproine and Cu-neocuproine were tested for biofilm eradication potential. (A) The OD600 was taken from the challenge plate after removal of the peg-lid to quantify the growth of biofilm dissociated cells. (B) Resazurin conversion was monitored kinetically in individual wells over a 24 hr period. Individual minigraphs show fluorescence (RFU) over time (24 hr). The lack of resazurin conversion indicates the absence of survivors. Resazurin conversion indicates the presence of viable cells shedding off the biofilm and growing in the recovery medium. (C) If a kinetic readout is not possible or not needed, then a simple yes/no answer for the presence of viable cells can be determined visually from the color conversion of the blue resazurin to its pink, reduced form. Please click here to view a larger version of this figure.
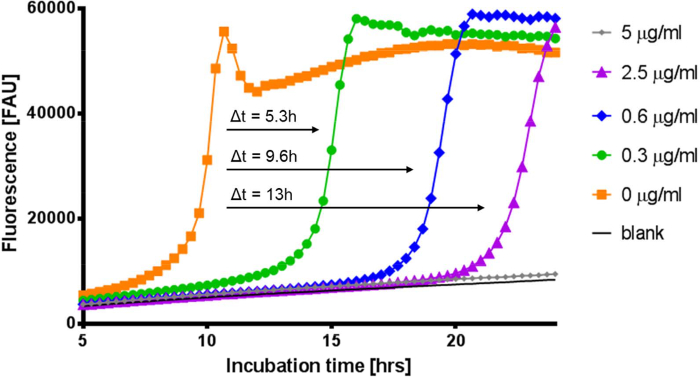
Figure 3. Kinetic resazurin assay reveals inhibitory effects on bacteria in a biofilm. Compounds may not fully eradicate biofilm embedded cells, but can still reduce their viability. This can be seen when comparing the delay of resazurin conversion of wells treated with different concentrations of gentamicin. Wells that have a delay in resazurin conversion indicate a reduced number of viable cells. Black (only media), Orange (0 µg/ml), Green (0.3 µg/ml), Blue (0.6 µg/ml), Purple (2.5 µg/ml), and Gray (5 µg/ml). Δt refers to the time differential of exponential resazurin conversion between untreated and gentamicin treated samples at the approximate half maximum fluorescence value. Please click here to view a larger version of this figure.
Discussion
Here, we have described a modified biofilm assay for determining activity of tested inhibitors on S. aureus biofilms focusing on the metabolic state of biofilm associated cells. While the described biofilm initiation and challenge procedures mostly mimicked manufacturer recommendations, the use of resazurin dye to detect and quantify biofilm-associated cells that survive a 24 hr inhibitor challenge dramatically simplifies the recommended procedure of cell recovery (via water bath sonification) and subsequent enumeration of viable cells via counting of CFUs. This manufacturer-recommended procedure involves at least 5 additional manipulation steps, each of them error prone, labor intensive, and impractical to automate. The advantage of simplifying the read-out procedure of the peg plate assay increases its attractiveness for high throughput operations and applications.
Though a lengthy protocol by number of steps, this procedure is rather conceptually simple, with few critical steps. Most crucially, the ability to repeatedly grow biofilms of the same quality is essential for obtaining reproducible results. Furthermore, one must exercise care whenever removing or replacing the peg-lid. Contact at any point with a well's wall would disrupt the biofilm, skewing results. More so, adequate washing is crucial: failure to remove planktonic cells at any point removes any ability to measure biofilm-associated bacteria. Conversely, though, washing must not be too rigorous; additional wash steps may begin removing biofilm, producing inconsistent results.
The peg plate approach has multiple uses, depending on the desired resulting data. The assay is described here as an information-rich kinetic measurement of metabolic resazurin conversion into a fluorescent dye 11. This conversion is associated with a color shift from blue to pink (Figure 2C), which, in addition to the quantitative fluorescence read, allows for an easy initial qualitative readout simply by observing the conversion. After removing the peg lid, this conversion can also be quantified by reading the absorbance at 600 nm should fluorescence capabilities not be available. Although the fluorescence eventually plateaus upon maximum dye conversion in each well (with survivors), the onset of dye conversion could be used to estimate the metabolically active survivor population relative to the untreated control (Figure 3). In our hands we have found that each ~100 min delay in the onset of resazurin conversion corresponds to approximately a 10-fold decrease in the number of metabolically active cells in the starting inoculum relative to the untreated control (as determined on planktonic cell culture serially diluted in 10-fold increments using biofilm recovery medium (data not shown)).
Beyond kinetic readouts, though, the modified peg plate method is more amenable to high-throughput drug screen projects than the original protocol. By measuring resazurin conversion as an endpoint instead of a kinetic, a user can generate a binary analysis of the samples to track simple activity of compounds (i.e., dye conversion/no conversion), which removes the need for 24 hr in a fluorescence plate reader, permitting conventional incubations. This methodology allows for determining the minimal biofilm eradicating concentration without the need of sonification and plating. The kinetic read also tracks the metabolic state of the biofilm depending on the treatment condition and inhibitor concentration. The reduced number of manipulation steps, increased reproducibility, and quantitative readout method (as opposed to plating each well individually 12) allows users to screen potentially thousands of compounds per day in a single concentration screen. In fact, peg-style plates have been previously used in high-throughput drug screens for biofilm dispersants, though resazurin (as used here) is a much more economical choice than the proprietary luciferase reagent utilized 13.
While a general improvement over the existing manufacturer protocol, the assay featured here does have certain limitations one must keep in mind. Inherently, we are only measuring metabolic capability of biofilm-associated cells. This assay does not measure biofilm mass or otherwise weaken the biofilm architecture or integrity. Those effects would not necessarily be detected. Metabolically inactive, but still viable, persister cells, which have been shown to exist in biofilms 14,15 will also be undetected. Further, compounds that induce quiescence without actually sterilizing a biofilm might erroneously appear as false positives, depending on length of the induced state. Viability indicators are ultimately surrogates for more iron-clad measurement methods such as flow cytometry or CFU enumeration, and the end user must determine which method is appropriate. Finally, as the resazurin conversion relies upon endogenous NADH, depletion of intracellular stores could be detrimental for bacteria. While we have not observed deleterious effects against S. aureus, other bacterial species might react differently. Users must empirically test and validate this protocol for reliability and reproducibility given varying organisms and lab environments.
Açıklamalar
The authors have nothing to disclose.
Acknowledgements
We thank Saran Kupul for technical assistance. This work was supported in part by NIH grant R01-AI104952 to FW. Further support was provided by the University of Alabama at Birmingham (UAB) Center for AIDS Research (CFAR), an NIH funded program (P30 AI027767) that was made possible by the following NIH Institutes: NIAID, NIMH, NIDA, NICHD, NHLDI, NIA.
Materials
| Mueller-Hinton Medium | Oxoid | CM0405 | Follow recommendations of manufacturer |
| RPMI-medium | Corning | 17-105-CV | |
| CRPMI | Ref 9 | RPMI-1640 medium chelexed for 1h with Chelex 100 resin and then supplemented with 10% unchelexed RPMI-1640 | |
| Chelex 100 Resin | Bio Rad | 142-2822 | |
| MBEC-plates | Innovotech | 19111 | |
| Resazurin Sodium Salt | Sigma | R7017 | 800µg/ml in DI water Filter sterile |
| Micro Plate Shaking Platform | Heidolph Titramax 1000 | ||
| Cytation 3 Plate Reader | Biotek | ||
| Gen5 software | Biotek | Recording and analysis of resazurin conversion | |
| Neocuproine | Sigma | N1501 | prepare 10 mM stock in 100% Ethanol, store at -80ºC |
| Copper sulfate | Acros Organics | 7758-99-8 | prepare a 100 mM stock solution in water, store at 4ºC |
| Cu-Neocuproine | Self-Made | Generated by mixing equal molarities of neocuproine and copper sulfate. Mix was diluted in CRPMI medium to desired concentration. | |
| Gentamicin | Sigma | G3632-1G |
Referanslar
- Bjarnsholt, T., Ciofu, O., Molin, S., Givskov, M., Hoiby, N. Applying insights from biofilm biology to drug development – can a new approach be developed. Nat Rev Drug Discov. 12, 791-808 (2013).
- Song, Z., et al. Prosthesis infections after orthopedic joint replacement: the possible role of bacterial biofilms. Orthop Rev (Pavia). 5, 65-71 (2013).
- Hall-Stoodley, L., Costerton, J. W., Stoodley, P. Bacterial biofilms: from the natural environment to infectious diseases). Nat Rev Microbiol. 2, 95-108 (2004).
- Bordi, C., de Bentzmann, S. Hacking into bacterial biofilms: a new therapeutic challenge. Ann Intensive Care. 1, 19 (2011).
- Fux, C. A., Shirtliff, M., Stoodley, P., Costerton, J. W. Can laboratory reference strains mirror ‘real-world’ pathogenesis?. Trends Microbiol. 13, 58-63 (2005).
- Kwasny, S. M., Opperman, T. J. Static biofilm cultures of Gram-positive pathogens grown in a microtiter format used for anti-biofilm drug discovery. Curr Protoc Pharmacol. Chapter 13, Unit 13A 18 (2010).
- Ceri, H., et al. The MBEC Assay System: multiple equivalent biofilms for antibiotic and biocide susceptibility testing. Methods Enzymol. 337, 377-385 (2001).
- Ceri, H., et al. The Calgary Biofilm Device: new technology for rapid determination of antibiotic susceptibilities of bacterial biofilms. J Clinical Microbiol. 37, 1771-1776 (1999).
- Baker, J., et al. Copper stress induces a global stress response in Staphylococcus aureus and represses sae and agr expression and biofilm formation. Appl Environ Microbiol. 76, 150-160 (2010).
- Haeili, M., et al. Copper complexation screen reveals compounds with potent antibiotic properties against methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 58, 3727-3736 (2014).
- O’Brien, J., Wilson, I., Orton, T., Pognan, F. Investigation of the Alamar Blue (resazurin) fluorescent dye for the assessment of mammalian cell cytotoxicity. Eur J Biochem. 267, 5421-5426 (2000).
- Mah, T. F. Establishing the minimal bactericidal concentration of an antimicrobial agent for planktonic cells (MBC-P) and biofilm cells (MBC-B). J Vis Exp. (83), e50854 (2014).
- Junker, L. M., Clardy, J. High-throughput screens for small-molecule inhibitors of Pseudomonas aeruginosa biofilm development. Antimicrob Agents Chemother. 51, 3582-3590 (2007).
- Shah, D., et al. Persisters: a distinct physiological state of E. coli. BMC Microbiol. 6, 53 (2006).
- Lewis, K. Riddle of biofilm resistance. Antimicrob Agents Chemother. 45, 999-1007 (2001).

