Analyses of Proteinuria, Renal Infiltration of Leukocytes, and Renal Deposition of Proteins in Lupus-prone MRL/lpr Mice
Summary
The present protocol describes a method to track lupus progression in mice. Two additional procedures are presented to characterize lupus nephritis based on cell infiltration and protein deposition in the kidneys.
Abstract
Systemic lupus erythematosus (SLE) is an autoimmune disorder with no known cure and is characterized by persistent inflammation in many organs, including the kidneys. Under such circumstances, the kidney loses its ability to clean waste from the blood and regulate salt and fluid concentrations, eventually leading to renal failure. Women, particularly those of childbearing age, are diagnosed nine times more often than men. Kidney disease is the leading cause of mortality in SLE patients. The present protocol describes a quick and simple method to measure excreted protein levels in collected urine, tracking lupus progression over time. In addition, an approach to isolate kidney mononuclear cells is provided based on size and density selection to investigate renal infiltration of leukocytes. Furthermore, an immunohistochemical method has been developed to characterize protein deposition in the glomeruli and leukocyte infiltration in the tubulointerstitial space. Together, these methods can help investigate the progression of chronic inflammation associated with the kidneys of lupus-prone MRL/lpr mice.
Introduction
The kidney's primary function is the elimination of toxic substances through urine while maintaining the homeostasis of water and salts1. This function is threatened in patients with systemic lupus erythematosus (SLE), leading to so-called lupus nephritis (LN). LN is a consequence of the immune system attacking the kidney, leading to persistent kidney inflammation, therefore losing its ability to clean waste from the blood and regulate salt and fluid concentrations. This will eventually lead to renal failure, which can be fatal. During the nephritic process, circulating B cells, T cells, and monocytes are recruited to the kidney, secreting chemokines, cytokines, and immune complex-forming autoantibodies. This ultimately results in endothelial cell damage, membranous injuries, renal tubular atrophy, and fibrosis2.
MRL/Mp-Faslpr (MRL/lpr) lupus-prone mice is a classical mouse model exhibiting lupus-like clinical signs that resemble human SLE3. This model has been instrumental in understanding one of the leading causes of mortality in SLE patients, lupus nephritis (LN)4. In both human and mouse SLE, LN is characterized by gradual inflammation triggered by renal deposition of immune complexes, followed by complement activation, recruitment of inflammatory cells, and loss of renal function5. The immune complex deposition is the first step to induce chemokine and cytokine production by intrinsic renal cells, which expands the inflammatory response by recruiting immune cells6. The current protocol presents several techniques to follow renal disease progression that analyze cell infiltration and immune complex deposition.
Urine collected every week allows for detection and visualization of the time course of proteinuria before, during, and after lupus onset. Proteinuria as a biomarker can determine the biological progression of LN. Other advantages of this technique are that it is non-invasive, cost-efficient, and easy to implement7. When the kidney is working perfectly, the proteinuria level is consistently low; however, in MRL/lpr mice, after 8-9 weeks of age, a gradual increase of the proteinuria level, that is eventually high enough to cause renal failure8, is observed. Multiple reagent strips and colorimetric reagents are commercially available to monitor the issue. However, the Bradford assay is cheap and very accurate in determining the onset of proteinuria and the course of lupus nephritis. This assay is quick, and the reagent is not affected by the presence of solvents, buffers, reducing agents, and metal-chelating agents that may be in your sample9,10,11.
One important aspect to consider is cell infiltration in the kidney. These infiltrates promote pathogenesis by triggering the secretion of soluble factors such as cytokines to worsen inflammation12. To better understand what cell populations are present in the infiltrates, a useful method is to isolate leukocytes13. Here, the detection of renal infiltration of B cells is used as an example. The procedure begins with a digestion process with deoxyribonuclease (DNase) and collagenase, followed by density gradient separation that removes debris, red blood cells, and dense granulocytes. The reason for isolating B cells (CD19+) and plasma cells (CD138+) is that lupus kidneys can concentrate these cells14. It is suggested that the presence of B cells in small aggregates in the kidney can indicate clonal expansion and, consequently, immunoglobulin (Ig) production. Plasma cells are well-known to be present in these aggregates as well15. Once leukocytes have been isolated, fluorescence-activated cell sorting (FACS) can be used to analyze the cells of interest upon staining with different fluorescence-conjugated antibodies.
Immunofluorescence is one of the immunohistochemistry (IHC) detection methods that allows for fluorescent visualization of proteins in 4 μm thick kidney tissue samples. Other IHC detection methods depend on the nature of analytes, binding chemistry, and other factors16. Immunofluorescence is a rapid identification method that exposes the antigen to its counterpart antibody labeled with a specific fluorochrome (or a fluorescent dye). When excited, it produces light that a fluorescence microscope can detect. This technique can be used to observe the deposition of complement C3 and IgG2a17. Excessive complement cascade activation could be associated with an uncontrolled immune response and loss-of-function18. Immune deposition of anti-double-stranded DNA (anti-dsDNA) autoantibodies in the kidney is a major concern19, where those with IgG2a isotype have been associated with LN20. Specifically, anti-dsDNA antibodies exhibit more pathogenicity and affinity to nuclear materials, forming immune complexes21. When IgG2a is present, the complement cascade, including C3, is activated to clear the immune complexes22. The C3 and IgG2a markers can be quantified individually or overlaid to establish their correlation.
Notably, serum creatinine measurement is another reliable technique that can be used together with microscopic hematuria and kidney biopsies to diagnose LN23. However, the presence of proteinuria is a strong indicator of glomerular damage. In that sense, monitoring the proteinuria level during lupus can detect disease onset and complement other methods for diagnosing lupus. In addition, immune complexes deposited in glomeruli can induce an inflammatory response, activate the complement system, and recruit more inflammatory cells. Another noteworthy point of this protocol is B cell infiltration in the kidney. This, together with the infiltrated T cells, amplifies local immune responses that trigger organ damage. Importantly, the classification of LN is not only based on glomerular morphologic changes seen in microscopy but also immune deposits observed with immunofluorescence. Therefore, in this protocol, accurate and cost-effective methods for the analysis of renal function are offered in laboratory settings.
Protocol
The present protocol is approved by the Institutional Animal Care and Use Committee (IACUC) at Virginia Tech. Since lupus disease has a higher incidence in females, only female MRL/lpr mice were used. The sample collection was started at 4 weeks of age and finished at 15 weeks. The mice were obtained from commercial sources (see Table of Materials) and were bred and maintained in a specific pathogen-free environment following the institutional guidelines.
1. Proteinuria test
- Collect urine following the steps below.
- Sterilize the hood by spraying 70% ethanol. Place a sterile plastic sheet on the surface. Prepare tips, 1.5 mL tubes, and a pipette to collect about 20 μL of the liquid.
NOTE: Tips and tubes must be sterile and without any pre-treatment. - Take the cage and spray 70% ethanol before putting it inside the hood.
- Hold the mouse with one hand and use the other hand to massage the ventral area from top to bottom gently.
- Use a 1.5 mL tube or pipette to collect the urine directly, or if the sample drops, collect it from the plastic sheet. Use fresh gloves, sheets, and tips for each sample.
NOTE: If this does not work, use a sterile beaker to isolate the mouse, then collect the urine from the beaker after the mouse urinates. - Put the mouse back in the cage. Take out another mouse and repeat steps 1.1.3-1.1.4.
NOTE: Always clean the plastic sheet and the beaker with 70% ethanol after collecting the sample for each mouse. At least five mice per group are necessary for statistical significance. Urine samples can be stored at -80 °C until use for up to 12 months.
- Sterilize the hood by spraying 70% ethanol. Place a sterile plastic sheet on the surface. Prepare tips, 1.5 mL tubes, and a pipette to collect about 20 μL of the liquid.
- Quantify the proteins by the Bradford method24.
- Allow urine samples, Albumin standard, and the Bradford reagent to come to room temperature (RT) by keeping them outside the freezer for 10-20 min.
NOTE: Bradford reagent and Albumin standard are included in a commercially available kit (see Table of Materials). The starting concentration of the Albumin standard is 2 mg/mL. Serial dilutions (1:2 six times) must be done with water. - Add Bradford reagent to a 96-well plate (200 μL/well), undiluted.
- Pipette 5 µL of undiluted urine sample per well and use the tip to pipette up and down several times to mix properly.
- Add standards (400, 200, 100, 50, 25, 12.5 mg/dL) in duplicates. Leave a couple of wells as blanks.
NOTE: The addition of samples and standards needs to be done quickly. - Let it stand for 5-10 min at RT.
- Read the plate at 595 nm in a plate reader as per the manufacturer's protocol (see Table of Materials).
- Allow urine samples, Albumin standard, and the Bradford reagent to come to room temperature (RT) by keeping them outside the freezer for 10-20 min.
2. Isolation of the kidney cells
- Euthanize the mouse with CO2 (flow rate: 30%-70% volume/min, to achieve unconsciousness or death) in a chamber followed by cervical dislocation. Proceed with the dissection to collect both kidneys. Place one-and-a-half-kidneys in an ice-cold C10 medium. Put the other half kidney in OCT (step 3.1.1).
NOTE: C10 is made with RPMI 1640 supplemented with 10% fetal bovine serum, 1 mM of sodium pyruvate, 1% of 100x MEM nonessential amino acids, 10 mM of HEPES, 55 µM of 2-mercaptoethanol, and 100 U/mL of penicillin-streptomycin (see Table of Materials). - Cut the kidneys into grain size (1-2 mm3 pieces) and digest in 5 mL of digestion buffer containing 1 mg/mL of collagenase and 0.2 mg/mL of DNase I (see Table of Materials) in RPMI 1640 medium containing 10 mmol/L of HEPES for 1 h with continuous gentle shaking at 37 °C.
NOTE: Add 1 mL digestion buffer in a sterile 2 mL tube and use sterile scissors to chop the kidney. - Add 10 mL of 1x ice-cold PBS containing 10 mM of ethylenediaminetetraacetic acid (EDTA) and incubate for 10 min on ice. Then vortex the suspension twice.
- Filter the cell suspension through a 100 µM strainer and wash with 10 mL of HBSS-full.
NOTE: HBSS-full contains 5 mM of EDTA, 0.1% BSA, and 10 mM of HEPES. - Centrifuge at 350 x g for 10 min at RT.
- Prepare 30%, 37% and 70% Stock Isotonic Percoll (SIP) solution.
NOTE: Mix 1 part volume of 10x PBS and 9 parts volume of Percoll to prepare SIP (see Table of Materials); use HBSS-full to dilute SIP to 30%, 37%, and 70% SIP. - Resuspend cells in 5 mL of 30% SIP and load 37% of 5 mL (top) and 70% of 5 mL (bottom), making SIP gradient, slowly with a paster pipette.
- Centrifuge with Up9 down0 (or brake 0) at 1000 x g for 30 min at RT.
- Collect leukocytes from the 37%-70% interface and resuspend them in 5 mL of C10.
- Centrifuge at 350 x g for 10 min at 4 °C.
- Resuspend the cell pellet in 3 mL of FACS buffer. Cells are ready for FACS staining.
NOTE: FACS buffer contains 1x PBS and 0.1% Bovine Serum Albumin (BSA). - Centrifuge at 350 x g for 5 min at 4 °C and discard the supernatant (SN), leaving about 50 µL.
NOTE: To remove the supernatant, carefully pour or pipette the solution away from the solid or use a semiautomatic vacuum to aspirate the supernatant. - Add 50 µL of FcR block (see Table of Materials) per tube (prediluted 1/50 in 1x PBS); mix and incubate on ice in the dark for 10 min.
- Add 2 mL of FACS buffer to wash.
- Centrifuge at 350 x g for 5 min at 4 °C and discard SN, leaving about 50 µL.
- Add 20 µL of the appropriate fluorescent dye (dilute 1/100 with 1x PBS, see Table of Materials) and let it stand for 15-30 min on ice.
- Add 50 µL of antibody15 mix per tube: CD138-BV711 (1/200 in 1x PBS) and CD45-AF700 (1/200 in 1x PBS) (see Table of Materials).
- Incubate on ice in the dark for 15-30 min and add 3 mL of FACS buffer.
- Centrifuge at 350 x g for 5 min at 4 °C and vacuum SN, leaving about 50 µL. Resuspend in 200 µL of 1x PBS. This is now ready to be analyzed by flow cytometry.
3. Immunofluorescent staining
- Perform the sample collection.
- Embed the half kidney in the small plastic cryomold with the Optimal Cutting temperature (OCT) compound (see Table of Materials).
- Add dry ice into a polystyrene foam box (see Table of Materials), and carefully put the cryomold on top of the dry ice. Ensure that the cryomolds are placed flat.
NOTE: It takes 3-4 min for the OCT compound to solidify and turn white. - Take the cryomold out and wrap it in aluminum foil. Store it at -80 °C until further use.
NOTE: This can be stored at -80 °C for at least 1 year.
- Perform sectioning and fixing of the sample.
- Cut the samples into 4 µm sections, wait for at least 2-3 h to dry, and then fix with cold acetone (at 4 °C) for 10 min.
NOTE: Be careful when using acetone, which requires a specialized chemical hood. - After fixing the slides, air dry for 0.5-1 h and store them for a short time at -20 °C, or for a long time at -80 °C.
- Cut the samples into 4 µm sections, wait for at least 2-3 h to dry, and then fix with cold acetone (at 4 °C) for 10 min.
- Stain the samples.
- Refix the slides in cold acetone for 5-10 min.
- Allow the sections to warm up to RT. Use a PAP pen (see Table of Materials) around the section and allow it to dry.
NOTE: Nail enamel can be used as well. This step prevents antibody dilution from floating all over the slide. - Place slides in 1x Tris-buffered saline (TBS) in the Coplin jar (see Table of Materials) for 20 min, put the jar on the shaker, and shake gently.
- Pour off 1x TBS and fill the jar with 1x TBS, 5% BSA, and 0.1% Tween 20. Incubate for 20 min on the shaker, shake gently.
- Take the slides out and wipe the bottom of the slides dry. If necessary, shake the slides dry. Add 100 µL of conjugated antibodies25 C3-FITC (1/100) and IgG2a-PE (1/100) to the section (see Table of Materials). Incubate at RT in a humidified chamber for 1 h.
- Wash the section 3 times in 1x TBS, 5% BSA, and 0.1 % Tween 20 for 10 min on the shaker.
- Shake dry the slides and mount the section with an antifade reagent (see Table of Materials) then a coverslip.
- Store the slides at 4 °C until viewing under the microscope (e.g., confocal microscope or imaging system microscope). Quantify images using software and subtract background signals when comparing the fluorescence intensity between regions.
- For image analysis using the ImageJ software (see Table of Materials), outline the desired region.
- Set standard parameters by clicking on Analyze, then Set Measurements and Measure Area to get the results.
- Transfer the data obtained from the measure windows to a spreadsheet.
NOTE: A small area can be selected from the image as a background; it shouldn't have any fluorescence. - Once done, click on Analyze to measure the region and transfer data again to the previous spreadsheet.
- Repeat steps 3.3.11-3.3.12 for several regions.
- Calculate the mean fluorescence as Corrected Cell Fluorescence (CCF) = Integrated density – (Selected cell region x Mean background fluorescence).
- Calculate for each region and analyze data using the graphing and statistics software (see Table of Materials).
Representative Results
The protocol uses multiple methods to assess MRL/lpr mice for lupus nephritis. First, a procedure is described to study increased proteinuria levels due to kidney dysfunction over time. As shown in Figure 1, female mice were treated with oral gavage of 200 µL of phosphate-buffered saline (1x PBS) as the control group and probiotic Lactobacillus reuteri as the treatment group, at a concentration of 109 cfu/mL, twice a week. Treatment started at 3 weeks old and finished at 15 weeks old. The mice group treated with L. reuteri had a more aggressive proteinuria progression than the control group.
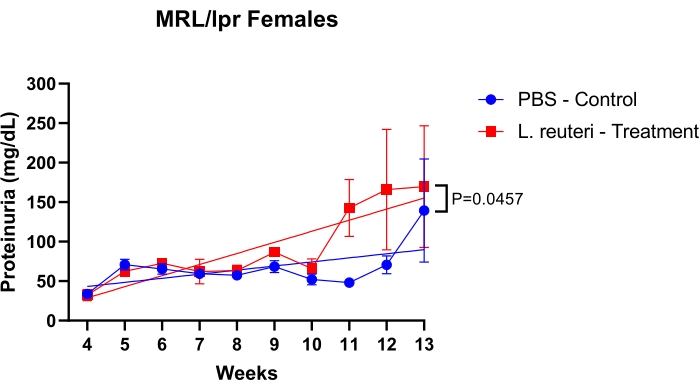
Figure 1: Detection of protein levels in the urine of MRL/lpr mice over time. n = 5 mice per group. The statistics were performed with the simple linear regression test. *P < 0.05, **P < 0.01. Please click here to view a larger version of this figure.
Figure 2 shows the FACS analysis of the isolated kidney leukocytes. The mice were treated with vancomycin (2 g/L) or vancomycin plus E. coli dsDNA (80 µg) in this experiment. Vancomycin was given along with the drinking water during the indicated period of time. Oral gavage of bacterial DNA was administered to the vancomycin-treated mice once a week for four consecutive weeks at 4, 5, 6, and 7 weeks of age. E. coli dsDNA was expected to trigger the renal infiltration of plasma cells leading to compromised kidney function, but no significant difference was found.
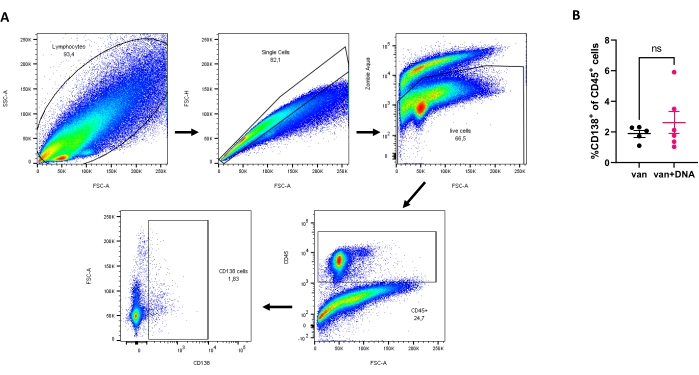
Figure 2: Analysis of plasma cells in isolated kidney leukocytes. (A) Sequential gating strategy: total cells, single cells, live cells, CD45+ cells, and finally CD138+ cells. (B) Percentage of plasma cells after treatment with van (Vancomycin) or van + DNA (Vancomycin + E. coli dsDNA) for 3 weeks (n ≥ 5 mice per group). No statistical significance ('ns') was observed. Please click here to view a larger version of this figure.
Figure 3 shows immunofluorescence staining of complement C3 and IgG2a on female MRL/lpr kidney sections. In this experiment, the effect of environmental factors was compared concerning the renal deposition of C3 and IgG2a. In-house mice were bred in the animal facility for several generations, and they were compared to recently purchased mice from a vendor. The immunohistochemical analysis did not show any difference between different facilities.
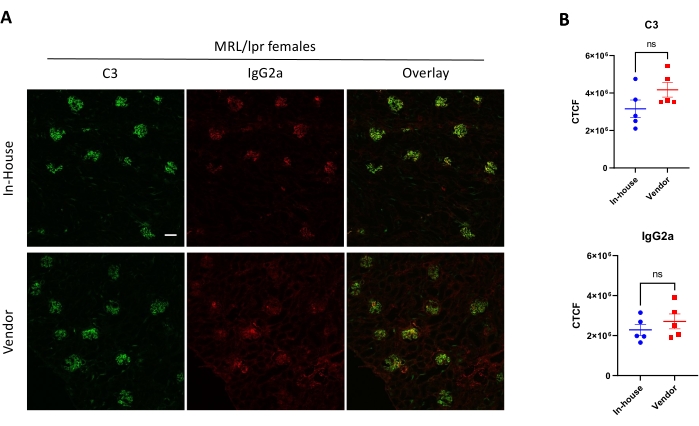
Figure 3: Detection of complement C3 and IgG2a in kidney sections via immunofluorescence staining. (A) Representative pictures of kidney sections stained with anti-C3-FITC (Green) and anti-IgG2a-PE (Red). (B) Comparison of corrected cell fluorescence (CCF) between the two groups (n ≥ 5 mice per group). Scale bar = 100 µM. No statistical significance ('ns') was observed. Please click here to view a larger version of this figure.
Discussion
LN is a leading cause of mortality in SLE patients, and factors aggravating the disease remain unclear. The application of this protocol is to characterize renal function using multiple methods, including measurement of proteinuria, FACS analysis of isolated kidney leukocytes, and immunofluorescence staining of frozen kidney sections.
One important point to consider while collecting urine is that one has to be consistent with the time of the day and the location of urine collection. Mice are nocturnal animals, so the most accurate samples are collected in the late afternoon before mice start being active. Another reason for choosing this time is that MRL/lpr mice tend to drink a lot of water at night, leading to diluted urine. Thus, collection at the wrong time may increase the urine's concentration and/or volume variabilities. It is also important to collect as much urine as possible to get accurate results.
The Bradford assay gives us an estimate of how much protein is present. It is not specific for any SLE marker proteins but good enough to give an idea of disease progression26. In addition, this method is cost-effective compared to others. Notably, the Bradford assay is time-sensitive; therefore, it must be completed within half an hour. Samples tend to have increased values or high false results if one waits too long. Another benefit compared to the commercial reagent strips for urinalysis is that it gives accurate numbers rather than a range within the urine sample. Scoring strips can give ambiguous results, particularly in the higher range9,10,24.
For the isolation of kidney leukocytes, the first key step is to perform the dissection as fast as possible. The viability of kidney cells greatly impacts the success of subsequent FACS analysis. When kidneys are being processed, each step is time- and temperature-sensitive. It needs to be noted that DNase I and collagenase concentrations and temperature are important for maximum efficiency, as they eliminate DNA and allow the disintegration of the organ, respectively27. The second key step is isolating cells, divided into two important processes. The first one is using the strainer that allows the removal of tissue debris; the second process involving Percoll is the most crucial step and requires focus and patience. When doing the density gradient with Percoll, it is important to maintain clear interfaces between different concentrations of SIP. It is recommended to use a Pasteur pipette since the flux and pressure can be controlled manually. Additional phases can be better built with the slow dispense of the SIP solution or even drop by drop. Once phases are established, the tube needs to be handled gently; otherwise, the phases will mix immediately, and samples will be lost, as it is impossible to separate the phases again. In addition, it is important to centrifuge the tube without a break because breaking could be too aggressive, leading to the loss of separate phases. The leukocytes are then recovered from interphase between 30% and 37% SIP.We analyzed one cell population in this example, plasma cells (CD45+ and CD138+). However, this technique is not limited to B cells; it can be used for other cell types and intracellular staining28.
The third method is a powerful tool for visualizing intracellular localizations of given protein depositions. Once tissues are fixed with acetone, it is important to perform adequate washing, so antibodies only attach to the target. Next, blocking with BSA reduces nonspecific binding and false positives. Direct immunofluorescence is usually recommended, as it is faster than indirect immunofluorescence with a secondary antibody, which is time-consuming, especially considering the extra washing and incubation steps. However, direct immunofluorescence restricts the flexibility of antibody selection compared to indirect immunofluorescence29. Therefore, both can be valuable methods depending on the purpose. Furthermore, it needs to be mentioned that dilution factors are key while staining for IHC. If the dilution is not well done, increased background (not enough antibody dilution) or weak staining (too much dilatation) will show on the microscopy images. It is important to start with serial dilutions of the antibody to optimize the antibody concentration for a better target-to-background ratio. Another step to consider is the amount of incubation time with the antibodies. The longer the antibody is in contact with the sample, the more nonspecific binding can be created.
The limitations of the present methods are as follows. Certain clinical parameters for LN, such as the albumin/creatinine ratio, cannot be revealed using these methods. In addition, some samples may have poor acid solubility leading to a high concentration reading above the standard curve. This requires further dilutions of samples or the addition of surfactants to precipitate the dye30. Moreover, this protocol may not be suitable if many cells are needed after kidney leukocyte isolation. The current kidney cell isolation protocol involves several washing steps, digestion, and isolation steps to maximize the removal of other cells, so even though the cell purity is high at the end, the cell number is usually low. Furthermore, it is a long procedure, so cell viability can be compromised. Other ready-to-use assays are more suitable for clinical use; however, the present methods may provide accurate results with a smaller budget for research laboratories.
In summary, three different efficient and accurate techniques are presented in this protocol to characterize the LN progression. Combining the Bradford method for the proteinuria level, FACS analysis for renal infiltration of the leukocytes, and IHC analysis for the renal deposition of IgG2a and C3, a clear picture of renal dysfunction in female MRL/lpr mice as a model of human LN is successfully established.
Declarações
The authors have nothing to disclose.
Acknowledgements
We thank the Flow Cytometry Core Facility, the Histopathology Laboratory, the Fralin Imaging Center at Virginia Polytechnic Institute, and State University for technical support. This work is supported by various NIH and internal grants.
Materials
| 10x Tris-Buffered Saline (TBS) | Thermo Fisher Scientific | J60764.K2 | |
| 2-mercaptoethanol | Thermo Fisher Scientific | 21985-023 | |
| Anti-Human/Mouse C3 | Cedarlane | CL7632F | |
| Anti-Mouse CD138 BV711 | Biolegend | 142519 | |
| Anti-Mouse CD45 AF700 | Biolegend | 103127 | |
| Bovine Serum Albumin | Sigma-Aldrich | A9418-100G | |
| Collagenase D | Sigma-Aldrich | 11088882001 | |
| Confocal Microscope LSM 880 | Zeiss | LSM 880 | |
| Coplin jar | Fisher Scientific | 50-212-281 | |
| Cryomold | Fisher Scientific | NC9511236 | |
| Density gradient medium | GE Healthcare | 17-1440-02 | Percoll |
| DEPC-Treated water | Thermo Fisher Scientific | AM9906 | |
| DNase I | Sigma-Aldrich | D4527 | |
| dsDNA-EC | InvivoGen | tlrl-ecdna | |
| Ethylenediaminetetraacetic Acid | Fisher Scientific | S311-500 | EDTA |
| EVOS M5000 Microscope imaging system | Thermo Fisher Scientific | AMF5000 | |
| FACS Fusion Cell sorter | BD Biosciences | FACS Fusion | |
| Fetal Bovine Serum – Premium, Heat Inactivated | R&D systems | S11150H | |
| Fisherbrand 96-Well Polystyrene Plates | Fisher Scientific | 12-565-501 | |
| Graphpad prism | GraphPad | N/A | |
| Hank’s Balanced Salt Solution | Thermo Fisher Scientific | 14175-079 | |
| HEPES | Thermo Fisher Scientific | 15630-080 | |
| ImageJ software | National Institutes of Health | N/A | |
| Lactobacillus reuteri Kandler et al. | ATCC | 23272 | |
| MEM non-essential amino acids | Thermo Fisher Scientific | 11140-050 | |
| MRL/MpJ-Fas lpr /J Mice (MRL/lpr) | Jackson Lab | 485 | |
| Nail enamel | N/A | N/A | Any conventional store |
| O.C.T compound | Tisse-Tek | 4583 | |
| PAP pen | Sigma-Aldrich | Z377821 | |
| Peel-A-Way Disposable Embedding Molds | Polysciences | R-30 | |
| Penicillin-Streptomycin | Thermo Fisher Scientific | 15140-122 | |
| Phosphate-buffered saline (PBS) | Thermo Fisher Scientific | 70011069 | |
| Pierce 20x TBS Buffer | Thermo Fisher Scientific | 28358 | |
| Pierce Coomassie Plus (Bradford) Assay kit | Thermo Fisher Scientific | 23236 | Albumin standard included |
| ProLon Gold Antifade Mountant | ThermoFisher | P36934 | |
| Purified Rat Anti-Mouse CD16/CD32 | BD Biosciences | 553141 | FcR block |
| RPMI 1640 | Thermo Fisher Scientific | 11875-093 | |
| Sodium pyruvate | Thermo Fisher Scientific | 11360-070 | |
| SpectraMax M5 | Molecular Devices | N/A | SoftMax Pro 6.1 software |
| Sterile cell Strainers 100 µM | Fisher Scientific | 22363549 | |
| Tween 20 | Fisher Scientific | BP337-500 | |
| Vancomycin Hydrochloride | Goldbio | V-200-1 | |
| Zombie Aqua | Biolegend | 423102 | fluorescent dye for flow cytometry analysis |
Referências
- Tsokos, G. C. Systemic Lupus Erythematosus. New England Journal of Medicine. 365 (22), 2110-2121 (2011).
- Davidson, A., Aranow, C. Lupus nephritis: lessons from murine models. Nature Reviews Rheumatology. 6 (1), 13-20 (2010).
- Maldonado, M. A., et al. The role of environmental antigens in the spontaneous development of autoimmunity in MRL-lpr mice. Journal of Immunology. 162 (11), 6322-6330 (1999).
- Lech, M., Anders, H. J. The pathogenesis of lupus nephritis. Journal of the American Society of Nephrology. 24 (9), 1357-1366 (2013).
- Bagavant, H., Fu, S. M. Pathogenesis of kidney disease in systemic lupus erythematosus. Current Opinion in Rheumatology. 21 (5), 489-494 (2009).
- Li, Q. Z., et al. Identification of autoantibody clusters that best predict lupus disease activity using glomerular proteome arrays. Journal of Clinical Investigation. 115 (12), 3428-3439 (2005).
- Aragón, C. C., et al. Urinary biomarkers in lupus nephritis. Journal of Translational Autoimmunity. 3, 100042 (2020).
- Mu, Q., et al. Control of lupus nephritis by changes of gut microbiota. Microbiome. 5 (1), 73 (2017).
- Lu, T. -. S., Yiao, S. -. Y., Lim, K., Jensen, R. V., Hsiao, L. -. L. Interpretation of biological and mechanical variations between the Lowry versus Bradford method for protein quantification. North American Journal of Medical Sciences. 2 (7), 325-328 (2010).
- Lamb, E. J., MacKenzie, F., Stevens, P. E. How should proteinuria be detected and measured. Annals of Clinical Biochemistry. 46, 205-217 (2009).
- Viswanathan, G., Upadhyay, A. Assessment of proteinuria. Advances in Chronic Kidney Disease. 18 (4), 243-248 (2011).
- Hsieh, C., et al. Predicting outcomes of lupus nephritis with tubulointerstitial inflammation and scarring. Arthritis Care & Research. 63 (6), 865-874 (2011).
- Hill, G. S., Delahousse, M., Nochy, D., Mandet, C., Bariéty, J. Proteinuria and tubulointerstitial lesions in lupus nephritis. Kidney International. 60 (5), 1893-1903 (2001).
- Chang, A., et al. In situ B cell-mediated immune responses and tubulointerstitial inflammation in human lupus nephritis. Journal of Immunology. 186 (3), 1849-1860 (2011).
- Schrezenmeier, E., Jayne, D., Dörner, T. Targeting B Cells and plasma cells in glomerular diseases: translational perspectives. Journal of the American Society of Nephrology. 29 (3), 741 (2018).
- Fitzgibbons, P. L., et al. Principles of analytic validation of immunohistochemical assays: Guideline from the college of American pathologists pathology and laboratory quality center. Archives of Pathology & Laboratory Medicine. 138 (11), 1432-1443 (2014).
- Lewis, M. J., Botto, M. Complement deficiencies in humans and animals: links to autoimmunity. Autoimmunity. 39 (5), 367-378 (2006).
- Watanabe, H., et al. Modulation of renal disease in MRL/lpr mice genetically deficient in the alternative complement pathway factor B. Journal of Immunology. 164 (2), 786-794 (2000).
- Yin, Z., et al. IL-10 regulates murine lupus. Journal of Immunology. 169 (4), 2148-2155 (2002).
- Singh, R. R., et al. Differential contribution of IL-4 and STAT6 vs STAT4 to the development of lupus nephritis. Journal of Immunology. 170 (9), 4818-4825 (2003).
- van Bavel, C. C., Fenton, K. A., Rekvig, O. P., vander Vlag, J., Berden, J. H. Glomerular targets of nephritogenic autoantibodies in systemic lupus erythematosus. Arthritis & Rheumatism. 58 (7), 1892-1899 (2008).
- Zuniga, R., et al. Identification of IgG subclasses and C-reactive protein in lupus nephritis: the relationship between the composition of immune deposits and FCgamma receptor type IIA alleles. Arthritis & Rheumatism. 48 (2), 460-470 (2003).
- Wang, S., Wang, F., Wang, X., Zhang, Y., Song, L. Elevated creatinine clearance in lupus nephritis patients with normal creatinine. International Journal of Medical Sciences. 18 (6), 1449-1455 (2021).
- Bradford, M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Analytical Biochemistry. 72 (1), 248-254 (1976).
- Cabana-Puig, X., et al. Phenotypic drift in lupus-prone MRL/lpr Mice: potential roles of micrornas and gut microbiota. Immunohorizons. 6 (1), 36-46 (2022).
- Wang, S., Wang, F., Wang, X., Zhang, Y., Song, L. Elevated creatinine clearance in lupus nephritis patients with normal creatinine. International Journal of Medical Sciences. 18 (6), 1449-1455 (2021).
- Kienzle, N., Young, D., Zehntner, S., Bushell, G., Sculley, T. B. DNaseI treatment is a prerequisite for the amplification of cDNA from episomal-based genes. Biotechniques. 20 (4), 612-616 (1996).
- Park, J. G. -., et al. Immune cell composition in normal human kidneys. Scientific Reports. 10 (1), 15678 (2020).
- Im, K., Mareninov, S., Diaz, M. F. P., Yong, W. H. An introduction to performing immunofluorescence staining. Methods in Molecular Biology. 1897, 299-311 (2019).
- Okutucu, B., Dınçer, A., Habib, &. #. 2. 1. 4. ;., Zıhnıoglu, F. Comparison of five methods for determination of total plasma protein concentration. Journal of Biochemical and Biophysical Methods. 70 (5), 709-711 (2007).

