In vitro Investigation of the MexAB Efflux Pump From Pseudomonas aeruginosa
Summary
We present a protocol for the in vitro investigation of efflux pumps from Pseudomonas aeruginosa. This protocol allows for the generation of a robust, reversible, and tunable proton gradient within the liposome membrane and hence should be adaptable to any membrane protein energized by the protomotive force.
Abstract
There is an emerging scientific need for reliable tools for monitoring membrane protein transport. We present a methodology leading to the reconstitution of efflux pumps from the Gram-negative bacteria Pseudomonas aeruginosa in a biomimetic environment that allows for an accurate investigation of their activity of transport. Three prerequisites are fulfilled: compartmentation in a lipidic environment, use of a relevant index for transport, and generation of a proton gradient. The membrane protein transporter is reconstituted into liposomes together with bacteriorhodopsin, a light-activated proton pump that generates a proton gradient that is robust as well as reversible and tunable. The activity of the protein is deduced from the pH variations occurring within the liposome, using pyranin, a pH-dependent fluorescent probe. We describe a step-by-step procedure where membrane protein purification, liposome formation, protein reconstitution, and transport analysis are addressed. Although they were specifically designed for an RND transporter, the described methods could potentially be adapted for use with any other membrane protein transporter energized by a proton gradient.
Introduction
Integral membrane proteins represent up to 30% of the genes encoded in eukaryotic genomes. They share pivotal roles such as gate keeping (receptors), transporting nutrients, ions or noxious compounds (transporters) or maintaining of the permeability across membrane bilayers (channels) amongst all living cells1. Efflux pumps have a central role in resistance against antibiotic therapy2. In the Gram-negative bacteria Pseudomonas aeruginosa, which is protected by an outer membrane, efflux transporters are organized as tripartite systems where MexB, the efflux pump located in the inner membrane, works in conjunction with MexA, a periplasmic protein, and OprM, an outer membrane channel. The cytoplasmic inner membrane protein acts as an energy-dependent pump with broad substrate specificity. The outer membrane protein acts as a porin whereas the third one is located in the periplasmic space and is thought to stabilize the whole complex3. In the following, we focus on the design of a functional assay for the MexA MexB assembly.
Structural information has accumulated over the last 5 years thanks to the X-ray determination of the homologous protein AcrB from Escherichia coli4-7. Although it is clearly important to couple such information with dynamic and kinetic data, designing a functional assay for this transporter is a real challenge8. Indeed, the membrane proteins must be maintained in a membranous environment and a closed compartment must be maintained for a vectorial transport of substrate to be achieved.
Reconstitution of membrane proteins into proteoliposomes has been extensively presented and reviewed (see Rigaud and Levy9). Such protocols allow for the monitoring of membrane proteins activity, for instance by following the substrates across the liposome membrane. In this case limitations arise from the availability of titratable substrates (fluorescent, radioactive, etc.) and from the fact that very often these substrates are hydrophobic and have a tendency to cross membranes irrespective of the presence of the transporter. As an alternative, one can follow the spectroscopic variations of a reporting dye sensitive to chemical changes that occur as a consequence of transport. As stated above, protons energize transport via MexB. Hence, a relevant option is to monitor the spectroscopic variations of a pH-sensitive reporting dye to follow pH variations due to proton-driven substrate transport. The choice of a fluorescent dye avoids any artifactual effects due to the hydrophobic nature of the substrate.
An additional difficulty is the generation of a proton gradient. Several protocols can be found in the literature. Among them, the use of valinomycin technique is widespread but we have shown that it is tedious to perform10-11. Moreover it is not reproducible and it is not reversible. We have resorted to the use of bacteriorhodopsin (BR), a light-activated proton pump from Halobacter salinarium, a halophilic marine Gram-negative obligate aerobic archaeon. This protein is coreconstituted into liposomes with MexB and, upon illumination, BR pumps protons inside of the liposomes and hence creates the ∆pH (acidic inside) needed by the protein to transport the substrate12-13.
Protocol
1. Protein Production and Purification
- MexA, MexB from Pseudomonas aeruginosa
- Transform the plasmids harboring the MexA and MexB genes in an E. coli production strain (e.g. C43 DE3). Plate on LB Agar medium supplemented with the appropriate antibiotic. Pick a single colony to inoculate an O/N preculture. The next morning, inoculate the preculture in 1 L of LB and grow at 37 °C under agitation (240 rpm).
- Once the cells have reached the exponential phase (OD600 = 0.6-0.8) induce with 1 mM IPTG. Perform induction for 2-3 hr at 30 °C under agitation.
- Centrifuge cells (9,000 x g for 20 min) and resuspend in 30 ml Tris-HCl 20 mM, pH 8, NaCl 150 mM.
- Disrupt cells using a French press (10,000 psi, 4 pass). Discard unbroken cells and inclusion bodies by centrifuging at 9,000 x g for 20 min.
- Pellet membranes 1 hr at 100,000 x g and resuspend each pellet in 30 ml Bis-Tris 10 mM pH 7.4, glycerol 20% (w/v), imidazole 10 mM, NaCl 500 mM.
- Determine the total membrane protein concentration using the BCA colorimetric assay and solubilize the membranes O/N with 2% dodecyl maltoside (DDM) in a final volume determined so that detergent: protein is 1:1.5 (w/w).
- Ultracentrifuge at 100,000 x g for 1 hr to discard non solubilized and aggregated material.
- Incubate the supernatant for 2 hr at 4 °C with a nickel resin (MexA purification) or a cobalt resin (MexB purification) equilibrated with Bis-Tris 10 mM pH 7.4, glycerol 5% (w/v), imidazole 10 mM, NaCl 500 mM, DDM 0.2%.
- Pour the resin in an empty column. Collect the Flow Through.
- Wash the resin with 50 ml of Bis-Tris 10 mM pH 7.4, glycerol 20% (w/v), imidazole 10 mM, NaCl 500 mM, DDM 0.2%, then with 25 ml of Bis-Tris 10 mM pH 6, glycerol 5% (w/v), imidazole 10 mM, NaCl 500 mM, DDM 0.2%.
- Elute the protein with 20 ml Bis-Tris 10 mM pH 7.4, glycerol 5% (w/v), imidazole 300 mM, NaCl 500 mM, DDM 0.2%.
- Concentrate the protein with an ultrafiltration device down to 3 ml and load on a desalting column equilibrated with an imidazole-free buffer. Just before reconstitution (see below), ultracentrifuge the solubilized proteins (100,000 x g for 20 min) to get rid of lipids and unsolubilized proteins. Then concentrate the protein again to 0.3 ml.
- Bacteriorhodopsin from Halobacter salinarium
- Grow Halobacter salinarium cells under illumination at 37 °C for 10 days in a liquid growth medium containing NaCl 4 M, MgSO4 150 mM, citrate 10 mM, KCl 30 mM, yeast extract 5 g/L, and peptone 5 g/L.
- Disrupt cells by dialyzing against deionized water.
- Purify the purple membrane on a 30-40% (w/v) sucrose gradient (100,000 x g for 17 hr).
- Solubilize the membrane suspension with 2% octylglucoside for 24 hr at 37 °C (volume for the solubilization in order to resuspend the membrane at a 2:1 (w/w) detergent : protein ratio). Estimate the concentration of solubilized BR using ε (570 nm) = 54,000 M-1 cm-1. Just before reconstitution (see below), ultracentrifuge the solubilized BR (100,000 x g for 20 min) to get rid of lipids and unsolubilized proteins.
Note: The native membranes are naturally enriched in BR so no further purification is needed.
2. Preparation of Proteoliposomes
- Liposome formation: Put 400 μl of DOPC dissolved in chloroform (25 mg/ml) in a glass beaker and add 1.5 mg of cholesterol stored as a powder. Dry them just before manipulation under a steam of nitrogen or vacuum for at least 1 hr in a glass beaker. The DOPC:cholesterol molar ratio is 3.3:1.
- After they have dried, hydrate the lipids with 1 ml of HEPES 25 mM pH 7, K2SO4 100 mM, MgCl2 2 mM, pyranine 2 mM. The suspension should appear turbid at this stage.
- Heat the solution for 10 min at 37 °C. Sonicate for 10 min at 40 W with 30 sec pulse, 30 sec pause cycles at RT. The suspension should appear clear at this stage.
- To obtain a monodisperse population of liposomes, perform two cycles of extrusion and for each cycle, pass the liposome suspension through the filter at least 11x. For the first cycle, use a membrane pore size of 200 nm and for the second cycle, use a membrane pore size of 100 nm. Dynamic Light scattering measurements can be performed at this step to check the homogeneity of the suspension.
- Protein incorporation
Principle: membrane proteins can be reconstituted in liposomes thanks to the solubilizing effect of detergents. Detergent-solubilized liposomes are incubated with the detergent-solubilized membrane protein and formation of the proteoliposomes is triggered by rapid elimination of the detergent with polystyrene beads9.- Add 28 mg of Triton X-100 (0.56% final) and incubate O/N at 4 °C (the solubilization temperature can be adapted to the protein being studied).
- Add the detergent-solubilized protein (BR, MexB and MexA) to the solubilized liposomes and incubate 15 min at 4 °C. Add the proteins to the solubilized liposome suspension at the following ratios (w/w): lipids/MexB = 20, MexB/MexA = 2.5, and lipids/BR = 30.
- Add polystyrene beads at a 30:1 beads:detergent ratio (w/w, considering the total weight of detergent, including detergent added with the purified proteins) and incubate 5 hr, in the dark (because of the BR) at RT under gentle stirring. Prior to this procedure, activate the biobeads with methanol and ethanol incubation and extensively wash with water.
- Purify the proteoliposomes protein and free substrates using a PD-10 desalting column equilibrated with HEPES 25 mM pH 7, K2SO4 100 mM and MgCl2 2 mM.
- Store the proteoliposomes at 18 °C in the dark for up to 2-3 weeks.
- Assessment of the reconstitution efficacy on sucrose gradient.
To check that both the MexB and the BR have been reconstituted in liposomes, purify the proteoliposome suspension on a discontinuous sucrose gradient.- Gently settle the proteoliposomes on a five-layer gradient of sucrose (60%, 20%, 10%, 5%, and 2.5%, w/v).
- Ultracentrifuge for 17 hr at 100,000 x g (with minimal acceleration and deceleration rates).
- Carefully collect the various gradient fractions (especially the interfaces between each layer of sucrose) and analyze them on SDS-PAGE (10%) using Coomassie staining or Western blotting. Aggregated proteins are found at the bottom of the tube, while nonincorporated, detergent-solubilized proteins are found at the top of the tube. Proteoliposomes and liposomes are found at sucrose interfaces that correspond to their intrinsic density. Empty liposomes are recovered farther up in the gradient than the proteoliposomes.
3. Fluorescence Measurement
- Perform fluorescence measurements at 25.0 °C using a spectrofluorometer allowing for dual-wavelength measurements (Xe lamp, 150 W). Alternate illumination periods (to activate BR with excitation / emission wavelengths of 550 nm / 550 nm) and fluorescence measurements with excitation / emission wavelengths of 455 nm / 509 nm for 2 sec to measure the pyranine fluorescence. Set the excitation and emission bandwidths to 5 nm. Perform the measurements in the presence of 50 nM valinomycin to prevent the formation of a reverse membrane potential ΔΨ.
Note: Indeed, because the BR pumps protons, there are more positive charges inside of the liposome than outside. This charge gradient can be discarded using valinomycin, which is a K+ selective ionophore. Once added to the liposomes suspension valinomycin, as a hydrophobic molecule, will insert into the liposome membrane, passively transport K+, and therefore dissipate the membrane potential ΔΨ.
Representative Results
The assay consists of monitoring pyranine fluorescence as a function of time while a proton gradient is generated. To that purpose, samples are subjected alternatively to illumination (hence proton pumping by the BR is triggered) and then to fluorescence measurement using the excitation and emission wavelengths of pyranine (λex = 455 nm and λem = 509 nm).
Figure 1 shows a representative control obtained with the assay, in the absence of MexA/MexB, verifying that the proton gradient generated by the BR is reversible. After one cycle of acidification, proteoliposomes are kept in the dark for 45 min in order for the BR to stop pumping protons (protons diffuse slowly and passively across lipid bilayers following their concentration gradient). After this recovery time, activation of BR is possible, simply by illuminating the same suspension again.
Figure 2 corresponds to an actual transport measurement. A negative control with protein free liposomes shows that, as expected, the pH is constant upon illumination (Figures 2A and 2B, orange circles). However, proteoliposomes containing BR in their membranes do pump proton, thus the pH inside the liposome decreases and a stable gradient is built (Figures 2A and 2B, purple squares). Grey triangles and red diamonds (Figure 2A) represent pH inside of proteoliposomes containing BR and MexB, in the presence or in the absence of Hoechst 33342, respectively. We observe a substrate-independent activity where the proton gradient is only partially dissipated by MexB counter-transport. We attribute this observation to a basal activity of MexB.
In order to test the effect of MexA on the activity of MexB, MexA was added to the reconstitution, first in the absence of any substrate (Figure 2B, blue diamonds). Again, the proton gradient generated by the BR is only partially dissipated. The actual transport measurement is realized on the very same sample. Beforehand, the proton gradient must be reinitialized and the substrate must be added in the suspension in order to reach its binding site in the protein. To that purpose the suspension is incubated in the dark in the presence of the substrate for 45 min. Upon subsequent illumination, one can see that the proton gradient generated by the BR is now totally dissipated by MexAB (Figure 2B, green triangles), as a consequence of substrate transport by the pump.
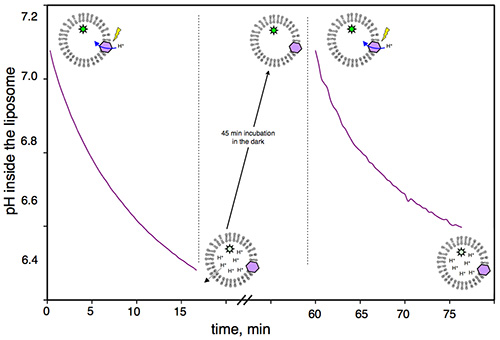
Figure 1. Reversibility of the proton gradient built by BR illumination: Pyranine fluorescence of BR proteoliposomes measured as a function of time. From 0-15 min, BR pumps protons as a result of light activation. From 15-60 min, proteoliposomes are incubated in the dark; during this time, protons passively move out of the liposomes until the intravesicular pH and extravesicular pH are equal. From 60-75 min, BR is still functional and pumps protons upon illumination.
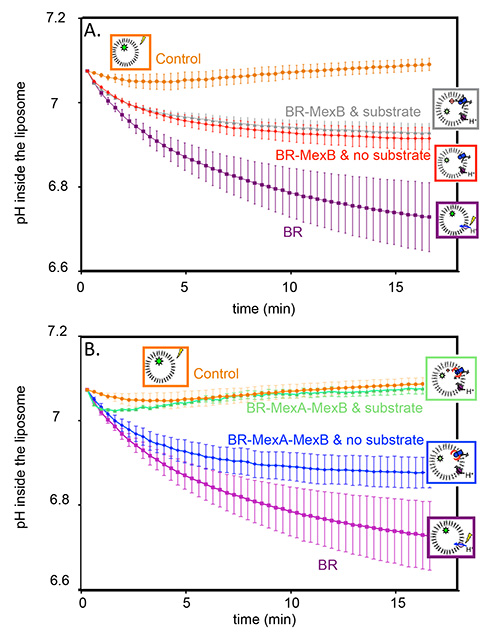
Figure 2. Transport assay: pyranine fluorescence, converted to the corresponding pH variations, as a function of time. A) pH inside of control liposomes (orange circles); pH inside of proteoliposomes containing BR in the membrane (purple squares); pH inside of proteoliposomes containing BR, and MexB in the membrane without Hoechst 33342 (red diamonds); pH inside of proteoliposomes containing BR and MexB in the membrane with Hoechst 33342 (grey triangles). B) pH inside of control liposomes (orange circles); pH inside of proteoliposomes containing BR in the membrane (purple squares); pH inside of proteoliposomes containing BR, MexB and MexA in the membrane without Hoechst 33342 (blue diamonds); pH inside of proteoliposomes containing BR, MexB and MexA in the membrane with Hoechst 33342 (green triangles).
Discussion
We describe a reconstitution procedure designed for assaying membrane protein transport. Once established, the protein reconstitution procedure leads to measurements that are very reproducible. However, the exact conditions for reconstituting the protein will vary from one protein to the other. Much care must be taken in optimizing the following parameters: i) quality of the purification (purity and absence of aggregated material); ii) efficiency of the detergent solubilizing step (when possible, low cmc detergent should be preferred because they are easier to get rid of); iii) desorption of the detergent (it is for instance possible to combine the use of polystyrene beads with a dialysis step but this may affect the rate of desorption, a critical parameter for the subsequent activity of the protein, see ref [9]); iv) lipid reconstitution (the chemical nature of the lipid, the lipid to protein ratio, the presence of additional amphiphiles are critical parameters that must be varied and optimized). In addition temperature and incubation time affect all these issues.
Quality control is thus primordial at each step of the reconstitution procedure. From this point of view, light scattering is a very convenient technique because it is rapid, nondestructive, and it only requires 20 μl of sample at a concentration in the micromolar range. Once the proteoliposomes are formed, sucrose gradient is also of great help because it opens the way towards systematic verification of the reconstitution: qualitative (is the protein indeed embedded into the liposome membrane) as well as quantitative (how many protein in one liposome). The latter would be addressed by precisely measuring the protein and lipid concentrations.
Our assay does not rely on the titration of the substrate itself and this avoids uneasy considerations regarding possible artifactual passive diffusion of the molecule due to hydrophobic partitioning in the membranes. The assay exploits properties of pyranine as a reliable and quantitative probe for monitoring pH changes. Our results are in agreement with results obtained with another procedure where proton gradient formation was performed using valinomycin10, but it allows a more robust and reproducible gradient11. In addition, the proton gradient can be precisely tuned, simply by varying the concentration of the buffer (for further detail see Verchere et al.12).
In the procedure a control measurement is first performed in the absence of substrate (this gives access to the passive activity of the protein). The actual membrane protein transporter activity is then measured after the substrate has been added in the same cuvette. This is truly an asset because the experiment can be considered as a genuine, non ambiguous, substrate-induced outcome.
The protocol could be adapted to high-medium throughput (for instance 96-wells measurements) automatization because illuminating the sample triggers the reaction. Automation and parallelization would consist in preparing a large batch of proteoliposome, and in dispensing aliquots in a 96-wells plate. Substrates (and also possible inhibitors) would then be added and after incubation in the dark for 45 min the plate would be subjected to illumination / pyranine fluorescence measurement cycles on a microplate reader.
For additional information on the protocol, readers are invited to read Verchere et al.12,13
Declarações
The authors have nothing to disclose.
Acknowledgements
This study was supported by the Agence Nationale de la Recherche (project ANR-2010-BLAN-1535) and by a grant from Région Ile-de-France (DIM-Malinf 110058).
Materials
| DOPC (1,2-Dioleoyl-sn-glycero-3-phosphocholine) | Avanti Polar Lipids | 850375C |
| Cholesterol | Sigma Aldrich | C8667 |
| Hoechst 33342 | Sigma Aldrich | B2261 |
| Triton X-100 | Sigma Aldrich | 93427 |
| Valinomycin | Invitrogen | V-1644 |
| Pyranine (8-hydroxypyrene-1,3,6-trisulphonic acid) | Invitrogen | H-348 |
| DDM (n-Dodecyl-β-D-maltopyranoside) | Affymetrix | D310LA |
| Extruder kit (extruder, Hamilton gas-tight syringes) | Avanti Polar Lipids | 610000 |
| Biobeads SM-2 Adsorbents | Bio-Rad | 152-3920 |
| Econo-Pac Chromatography empty column | Biorad | 732-1010 |
| Desalting columns | Bio-Rad | 54805 |
| Dynamic light scattering instrument | Wyatt Technology | DynaPro NanoStar |
| Fluorometer JASCO FP-6200 | JASCO | 6816-J002A |
Referências
- Granseth, E., et al. Membrane protein structural biology – How far can the bugs take us? (Review). Molecular Membrane Biology. 24 (5-6), 329 (2007).
- Nikaido, H. Multidrug resistance in bacteria. Annu Rev Biochem. 78, 119 (2009).
- Pos, K. M. Drug transport mechanism of the AcrB efflux pump. Biochim Biophys Acta. 1794 (5), 782 (2009).
- Murakami, S., et al. Crystal structures of a multidrug transporter reveal a functionally rotating mechanism. Nature. 443 (7108), 173 (2006).
- Seeger, M. A. Structural Asymmetry of AcrB Trimer Suggests a Peristaltic Pump Mechanism. Science. 313 (5791), 1295 (2006).
- Sennhauser, G., Bukowska, M. A., Briand, C., Grutter, M. G. Crystal structure of the multidrug exporter MexB from Pseudomonas aeruginosa. J Mol Biol. 389 (1), 134 (2009).
- Su, C. C., et al. Crystal structure of the CusBA heavy-metal efflux complex of Escherichia coli. Nature. 470 (7335), (2011).
- Nikaido, H., Takatsuka, Y. Mechanisms of RND multidrug efflux pumps. Biochim Biophys Acta. 1794 (5), 769 (2009).
- Rigaud, J. L., Levy, D. Reconstitution of membrane proteins into liposomes. Methods in enzymology. 372, 65 (2003).
- Aires, J. R., Nikaido, H. Aminoglycosides are captured from both periplasm and cytoplasm by the AcrD multidrug efflux transporter of Escherichia coli. J Bacteriol. 187 (6), 1923 (2005).
- Picard, M., Verchere, A., Broutin, I. Monitoring the active transport of efflux pumps after their reconstitution into proteoliposomes: caveats and keys. Anal Biochem. 420 (2), 194 (2012).
- Verchere, A., Dezi, M., Broutin, I., Picard, M. Basic Methods in Protein Purification and Analysis, edited by iConcept Press. , (2012).
- Verchere, A., Broutin, I., Picard, M. Photo-induced proton gradients for the in vitro investigation of bacterial efflux pumps. Sci Rep. 2, 306 (2012).

