Isolation and Culture of Oculomotor, Trochlear, and Spinal Motor Neurons from a Mouse Embryo
Abstract
Fujiki, R., et al. Isolation and Culture of Oculomotor, Trochlear, and Spinal Motor Neurons from Prenatal Islmn:GFP transgenic mice. J. Vis. Exp. (2019)
The video demonstrates the isolation of oculomotor, trochlear, and spinal motor neurons from a transgenic mouse embryo. The embryo expresses motor neurons labeled with a green fluorescent protein (GFP) for easy visualization and identification. The motor neurons are surgically isolated under a fluorescence microscope and maintained in a culture medium.
Protocol
1. Ventral midbrain and Spinal Cord Dissection
NOTE: Perform all of the following steps except for steps 1.1.1−1.1.3 and 1.1.5−1.1.6 under a fluorescence dissection stereomicroscope. Total dissection time per experiment is typically 3−5 h, depending on the proficiency at the dissection technique and the number of motor neurons required for each experiment.
- Ventral midbrain dissection
- Euthanize a pregnant mouse approximately 11.5 days postfertilization by carbon dioxide gas and cervical dislocation.
- Spray the abdomen thoroughly with ethanol and remove the uterus using sterile micro dissecting scissors and thumb dressing forceps. Wash the uterus briefly in sterile phosphate-buffered saline (PBS), then transfer to the dissection plate filled with prechilled sterile PBS.
- Remove the IslMN:GFP (Islet motor neuron: green fluorescent protein)-positive embryos carefully from the uterus using sterile micro dissecting scissors, thumb dressing forceps, and Dumont #5 tweezers in ice-cold sterile phosphate-buffered saline (PBS) under the bright light of the microscope. Using a sterile Moria mini-perforated spoon, transfer each embryo to a separate well of a 24 well plate filled with prechilled Hibernate-E low fluorescence medium supplemented with 1x B27. Keep the 24 well plate on ice.
- Transfer one embryo to a sterile dissection plate and cover it completely with ice-cold sterile Hank's balanced salt solution (HBSS).
- Ensure that the dissection steps are performed under fluorescein isothiocyanate (FITC) illumination of the microscope. Using tweezers, remove the tail and the face of the embryo without damaging the midbrain (Figure 1Ba). Place the embryo prone with limbs straddled underneath and tail pointing toward the front of the microscope, toward the dissector (indicated by an asterisk, Figure 1Bb).
- Using tweezers, slit open the roof of the fourth ventricle in order to generate a small opening. Use this opening to hook tweezers into the space created between the fourth ventricle and its roof. Dissect along the dorsal surface of the embryo rostral to the cortex and lateral to the floor plate and motor column (Figure 1Ca,b). Open the dissected tissue in an open-book manner to reveal the GFP-positive oculomotor neuron (cranial nerve 3 [CN3]) and trochlear neuron (cranial nerve 4 [CN4]) nuclei.
NOTE: A small piece of tissue from the ventral midbrain containing mesenchyme, CN3, and CN4 will now be exposed. - Carefully separate the ventral midbrain from the embryo and remove meningeal tissue using tweezers and a micro dissecting knife. Dissect the bilateral GFP-positive CN3 and CN4 nuclei away from the floor plate and other GFP-negative surrounding tissue using tweezers and a micro dissecting knife (Figure 1D). Maximize the number of GFP-positive motor neurons in the excised tissue but avoid touching or damaging them.
- If a collection of separate CN3 and CN4 nuclei is desired, cut along the midline of these two nuclei (yellow dotted line in Figure 1D). Using a P1000 pipette, collect the dissected ventral midbrain tissue with minimal HBSS and place it in a labeled 1.7 mL microcentrifuge tube filled with dissection medium. Store on ice until dissociation.
- Continue pooling ventral midbrains from additional embryos in the same tube until the total number meets the experimental requirement (refer to step 4 for ideal cell numbers).
NOTE: A pooled collection of at least 10 ventral midbrains yielding approximately 1 x 104 CN3/CN4 motor neurons is recommended because tissues are subject to stress during the dissociation and sorting.
- Ventral spinal cord dissection
- Keep the embryo prone with the head facing the front of the microscope, toward the dissector. Hold the embryo with one pair of tweezers and insert the tip of the other pair of tweezers into the unopened caudal part of the fourth ventricle.
- Open the rest of the hindbrain and spinal cord dorsally over the whole rostrocaudal extent of the embryo. Open by cutting dorsal tissue, starting from the fourth ventricle and working toward the central canal of the caudal spinal cord using the forceps as scissors (Figure 1Ca,b). Take care to avoid touching or damaging the ventral spinal cord during this procedure.
- Hold the embryo with one pair of tweezers and pinch off the flap of the dorsal tissue on each side with the other pair of tweezers (Figure 1Ea,b).
NOTE: Excised dorsal tissues contain dorsal skin, mesenchyme, dorsal root ganglia (DRGs), dorsal hindbrain, and spinal cord. Remove as much of these tissues as possible without damaging the spinal motor neuron (SMN) nuclei, because they are adhesive and can trap SMNs during filtering or cause clogging during fluorescence-activated Cell Sorting (FACS). - Remove the ventral spinal cord using the microdissection knife to pierce directly below the GFP-positive SMN. Lift the ventral spinal cord with saw-like movements on both sides (Figure 1Fa,b). Cut the floating ventral spinal cord transversely directly above C1, where the first GFP-positive anterior horn projects (Figure 1G). Also, cut transversely at the upper boundary of the lower limb (Figure 1G). Remove the cervical (C1)-lumbar (L2-L3) portion of the ventral spinal cord after this procedure.
- Place the ventral spinal cord dorsal side up and hold by pressing the GFP-negative tissue between the GFP-positive SMN columns with one pair of tweezers. Remove the remaining attached mesenchyme, DRGs, and dorsal spinal cord by trimming both sides of the GFP-positive SMN column with the microdissection knife (Figure 1H). Take care to maximize GFP-positive motor neurons without damaging them.
- Using a P1000 pipette, collect the dissected ventral spinal cord tissue with minimal HBSS and place in the SMN-labeled 1.7 mL microcentrifuge tube filled with dissection medium. Store on ice until dissociation. Continue pooling ventral spinal cords from additional embryos in the same tube until the total number meets the experimental requirements.
NOTE: Collecting at least three ventral spinal cords yielding approximately 2.1 x 104 SMN is recommended because tissues are subject to stress during dissociation and sorting. - Collect facial motor neurons and extremities of the IslMN:GFP mouse embryos as GFP-positive and GFP-negative controls for fluorescence-activated cell sorting (FACS), respectively. Extremities are GFP-negative because the GFP-positive axons of the SMNs have not yet extended into the extremities at this embryonic age.
2. Tissue Dissociation
NOTE: Total dissociation time is typically 1.5 h per experiment.
- Warm papain and albumin-ovomucoid inhibitor solution aliquots to 37 °C 30 min prior to dissociation.
- Briefly spin down micro dissected tissues at a low speed.
- Using a P100 pipette, carefully remove as much Hibernate E as possible without aspirating tissues.
NOTE: Be sure to remove all residual Hibernate E after this step to avoid reducing the efficacy of papain dissociation in the next step. - Add the appropriate volume of papain solution (Table 1) to each of the 1.7 mL microcentrifuge tubes containing the microdissected tissue samples.
NOTE: The appropriate volume of papain for the dissociation was determined in order to maximize the effective dissociation while minimizing stress on the cells. - Gently triturate 8x with a P200 pipette. Perform all trituration steps gently to preserve motor neuron viability.
- Incubate the tubes containing the tissues for 30 min at 37 °C, agitating by finger flicking 10x every 10 min. Gently triturate each suspension 8x with a P200 pipette after incubation. Spin down the cells at 300 x g for 5 min.
- To ensure the efficacy of ovomucoid inhibition in the next step, use a P1000 pipette to remove and discard as much supernatant as possible without aspirating the tissues.
- Resuspend pellets in the appropriate volume of albumin-ovomucoid inhibitor solution (Table 1) by gently triturating 8x with a P200 pipette.
- Wait for 2 min to allow any remaining pieces of undissociated tissue to settle to the bottom of the tube.
- Collect as much supernatant as possible without aspirating undissociated tissues using a P200 pipette. Transfer the supernatant to fresh 1.7 mL microcentrifuge tubes.
- If some chunks of tissue remain undissociated after step 2.10, repeat steps 2.8−2.10 for the undissociated tissues that remain in the original 1.7 mL microcentrifuge tubes to maximize the final yield of dissociated cells while minimizing stress on the cells dissociated previously, contained in the supernatant of step 2.10.
- Spin down the cells at 300 x g for 5 min. Carefully remove and discard the supernatant using a P1000 pipette.
- Resuspend the pellet in the appropriate volume of dissection medium (Table 1) by pipetting 8x using a P1000 pipette. The appropriate volume of final suspension was determined so that cell density does not exceed 107 cells/mL, which can block the stream of the flow cytometry machine, but also so that cells are not excessively diluted, which results in a slowed sorting speed.
- Filter the suspensions through 70 µm cell strainers to eliminate any large clumps or undigested tissue. Transfer the suspensions into 5 mL round bottom polystyrene test tubes and store on ice until required.
3. Fluorescence-activated Cell Sorting (FACS)
NOTE: This protocol was optimized using a FACS sorter equipped with a 15 mw 405 nm violet laser, a 100 mw 488 nm blue laser, a 75 mw 594 nm orange laser, and a 40 mw 640 nm red laser. Cells were sorted as sheath fluid in sterile PBS under aseptic conditions through a 100 µm nozzle. In order to minimize cell stress, the flow rate was set to a sample pressure of 1−3, such that a maximum of 1,000−4,000 events per second were acquired. Total FACS time is typically 1−2 h per experiment.
- Set up voltages for forward and side scatter so that the cell population can be visualized properly. Setting up appropriate voltages for cell sorting is complex and requires an experienced FACS operator.
- To distinguish different cell populations, plot cells based on size as determined by the Forward Scatter Area (FSC-A) versus internal complexity as determined by the Side Scatter Area (SSC-A). Draw a gate around the live cells as indicated in Figure 2Aa and Figure Ba to exclude debris and dead cells. Group the cells within the gated region as population 1 (P1).
- To exclude cell clumps and doublets, plot P1 cells next based on the Side Scatter Width (SSC-W) versus the SSC-A. Gate the population of single cells as population 2 (P2) (Figure 2Ab and Figure Bb).
- Plot P2 cells based on the Forward Scatter Width (FSC-W) versus the FSC-A and gate the population of single cells as population 3 (P3) (Figure 2Ac and Figure Bc).
NOTE: Use of two consecutive gates in 3.3 and 3.4 excludes cell clumps and doublets (high FSC-W and high FSC-A). - Gate P3 cells based on GFP versus allophycocyanin (APC). The APC channel detects autofluorescence. Gating on this channel avoids capturing autofluorescent cells. Use GFP-negative cells to adjust the voltage for FITC/GFP fluorescent channels. Ideally, position gates for these cell populations around 102. Select gate thresholds for GFP-positive population 4 (P4) individually for each type of motor neuron (Figure 2Ad and Figure Bd).
NOTE: Set the GFP gate much higher for SMNs than for CN3s/CN4s in order to obtain a pure culture (Figure 2Ad and Figure Bd). A lower GFP gate for SMN cultures leads to contamination of the cultures by glia and non-motor neurons. This is likely because there is low-level GFP expression in some glia and non-motor neurons due to a leaky promoter. The percentage of GFP-positive cells as compared to total cells is typically 0.5−1.5% for CN3s/CN4s and 1.5−2.5% for SMNs. If the dissection was successful, these numbers can be used as a benchmark to determine the appropriate position for the GFP-positive gate (Figure 2Ae and Figure Be). - Perform FACS according to the manufacturer's protocol. Collect P4 cells into fresh 1.7 mL microcentrifuge tubes filled with 500 µL of motor neuron culture medium. Store on ice until plating.
NOTE: Although the cells can be sorted directly into the wells, this results in an uneven number of cells per well. Sort the cells into 1.75 mL microcentrifuge tubes and then plate manually in order to achieve a more even plating distribution.
4. Culture of Purified Primary Motor Neurons
- Dilute FACS-isolated CN3/CN4 and SMN suspensions with motor neuron culture medium prewarmed to 37 °C to densities of 5 x 103 and 1 x 104 cells/mL, respectively.
NOTE: One E11.5 embryo yields approximately 1 x 103 CN3/CN4 and 7 x 103 SMN. However, these yields rely heavily on the purity of dissected tissues, the thoroughness of cell dissociation, and the appropriate thresholding of GFP gates during FACS. - Transfer 96 well plates precoated with poly D-lysine (PDL) and laminin from the 37 °C tissue culture incubator to the laminar flow hood and aspirate laminin from each well. Use plates and coverslips immediately without washing.
- Add 200 µL of diluted CN3/CN4 and SMN suspensions into the each well of PDL/laminin-coated 96 well plates. Final cell densities should be 1 x 103 and 2 x 103 cells/well for CN3/CN4 and SMN, respectively.
NOTE: Initial plating density of SMNs in 96 well plates (2 x 103 cells/well) is double that of CN3s/CN4s (1 x 103 cells/well) in order to obtain similar final motor neuron numbers and densities at 2 and 9 days in vitro (DIV) (4−6 x 102 and 2−4 x 102 cells per well, respectively). - Culture neurons in a 37 °C, 5% CO2 incubator.
- Feed neurons every 5 days by removing half of the old media (100 µL) and replacing with the same volume of fresh motor neuron culture medium. Ensure that neuronal processes become visible on 1 DIV and become thicker and longer by 14 DIV (Figure 3). Neuronal cell bodies become enlarged and tend to aggregate in long-term cultures, particularly for SMNs (Figure 3).
NOTE: Perform all medium and solution changes by leaving half of the original medium volume in order to avoid detaching cultured cells. This includes fixation and immunocytochemistry (ICC) steps. If all media is removed, regardless of how gently, most of the cells will detach and be washed away.
Representative Results
Table 1: Appropriate volumes of papain, albumin-ovomucoid, and final suspension used in dissociation steps. The appropriate volumes of papain and albumin-ovomucoid to be used with various numbers of ventral midbrain and ventral spinal cord tissues were modified from the manufacturer's instructions after several rounds of optimization. Because tissues are subject to stress during dissociation and sorting, a pooled collection of more than 10 ventral midbrains and more than three ventral spinal cords is recommended. The volume of papain was determined by considering the balance between effective dissociation and the stress of this procedure. The volume of albumin-ovomucoid inhibitor solution is half of that of papain. The appropriate volume of Hibernate E final suspension was determined such that cell density does not exceed 107 cells/mL, but the cells do not become excessively diluted.
| Number of midbrains (X) | Papain | Albumin-ovomucoid | Hibernate E |
| 10 ≤ X ≤20 | 200 μL | 100 μL | 600 μL |
| 20 < X ≤30 | 300 μL | 150 μL | 700 μL |
| 30 < X ≤40 | 400 μL | 200 μL | 800 μL |
| Number of spinal cords (Y) | Papain | Albumin-ovomucoid | Hibernate E |
| 3 ≤ Y ≤5 | 200 μL | 100 μL | 500 μL |
| 5 < Y ≤10 | 400 μL | 200 μL | 800 μL |
| 10 < Y ≤15 | 600 μL | 300 μL | 1200 μL |
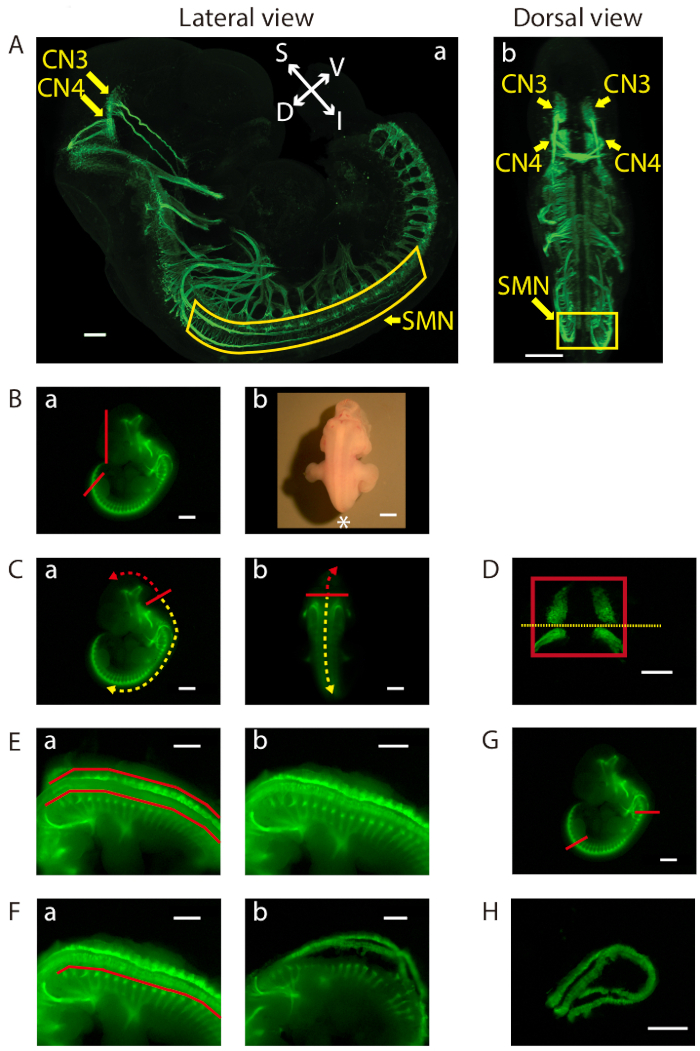
Figure 1: Dissection of the ventral midbrain and the cervical (C1)-lumbar (L2-L3) portion of the ventral spinal cord. (A) Lateral (a) and dorsal (b) views of GFP-positive motor neurons in an E11.5 IslMN:GFP transgenic mouse embryo under fluorescein isothiocyanate (FITC) illumination. A whole mount E11.5 embryo was prepared in order to make the embryo transparent. Subsequently, the embryo was analyzed by immunofluorescence labeling with anti-GFP staining (green). Images were captured under a confocal microscope. Scale bars = 200 µm (lateral view) and 400 µm (dorsal view). Abbreviations: S = superior; I = inferior; V = ventral; D = dorsal. (B-H) Dissection steps highlighted on images of E11.5 ventral midbrain and ventral spinal cord tissues taken with an equipped camera under bright light (Bb) or FITC illumination using a fluorescence dissection stereomicroscope. Scale bars = 200 µm (D), and 1 mm (A−C, E−H). (B) (a) Removal of the face and tail of the embryo by cutting along the red lines. (b) Embryo positioned for dissection. Positioning of the front of the microscope is indicated by an asterisk. (C) Cutting along the solid red line in order to slit open the roof of the fourth ventricle (a) lateral view and (b) dorsal view. Use of this opening to cut along the surface of the embryo dorsal to the brain (trajectory indicated by dashed red arrow). This exposes the tissue containing mesenchyme, CN3, and CN4, which can be lifted out of the cranium. For SMN dissection, insertion of forceps into the same opening between the fourth ventricle and its roof, then cutting toward the caudal side of the embryo (trajectory indicated by dashed yellow arrow). (D) Final view of the ventral midbrain containing bilateral GFP-positive CN3 and CN4 nuclei. The edges of the tissue are highlighted by a red rectangle. Cutting along yellow dotted line to collect CN3 and CN4 nuclei separately, if desired. (E) After opening the rest of the hindbrain and spinal cord, flapping dorsal tissues pinched off above the red lines on both sides with tweezers (a) before, and (b) after. (F) Bilaterally removal of excess tissue ventral to the spinal cord along the red line (a) before, and (b) after. (G) Cutting of the ventral spinal cord at the two locations indicated by the red lines. On the rostral side, cutting of the floating ventral spinal cord transversely above C1 where the first GFP-positive anterior horn projects. Cutting of the caudal end of the spinal cord transversely at the upper boundary of the lower limb. Once these cuts are made, the cervical (C1) through lumbar (L2-L3) portion of the ventral spinal cord can be dissected away. (H) Final view of the ventral spinal cord containing GFP-positive SMN columns.
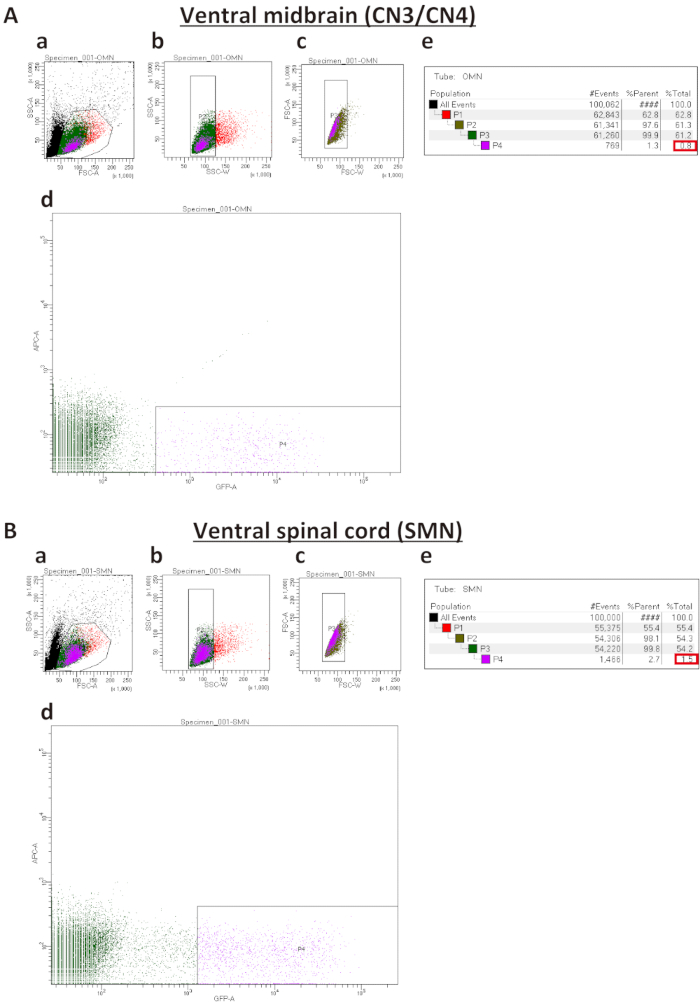
FIGURE 2. Representative sort plots of ventral midbrains (A) and ventral spinal cords (B). (Aa and Ba) Forward Scatter Area (FSC-A) versus Side Scatter Area (SSC-A) sorted plot before exclusion of debris and dead cells. (Ab, Bb, Ac, Bc) Sorted plots for exclusion of cell clumps (b) and doublets (c) based on Width (SSC-W) versus SSC-A and Forward Scatter Width (FSC-W) versus FSC-A, respectively. (Ad and Bd) Sorted plots to isolate IslMN:GFP -positive motor neurons. In order to obtain a pure culture, the GFP gate must be set higher for SMNs (Bd) than for CN3s/CN4s (Ad). (Ae and Be) Percentages of cells gated for collection by FACS sorting. %Parent represents the percentage of cells in the current gated population relative to the number of cells in the previous gated cell population, whereas %Total represents the percentage of gated cells relative to total cells. Expected percentages of GFP-positive cells as compared to total cells (boxed in red) are 0.5−1.5% for CN3/CN4 and 1.5−2.5% for SMN. If the dissection was performed successfully, these percentages can be used as a benchmark to set up the GFP-positive gate in (Ad and Bd).
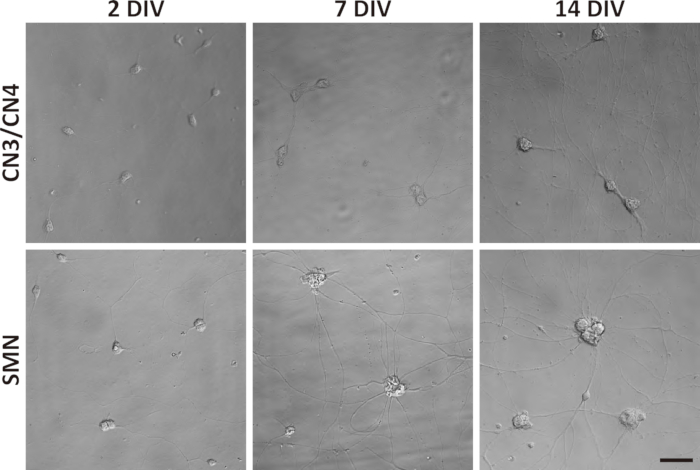
Figure 3: Phase-contrast images of primary CN3/CN4 and SMN monocultures at 2, 7, and 14 DIV. Representative differential interference contrast images of primary CN3/CN4 and SMN cultures were captured at 2, 7, and 14 DIV with inverted fluorescence microscope using corresponding image acquisition and processing software and 40x objectives. Neuronal processes became thicker and longer by 14 DIV. Neuronal cell body sizes became enlarged and tended to aggregate in long-term cultures, especially for SMNs. Both cultures can be maintained at least 14 DIV. Scale bar = 50 µm.
Declarações
The authors have nothing to disclose.
Materials
| B27 Supplement (50X), serum free | Thermo Fisher Scientific | 17504-044 | |
| BD FACSAria llu SORP Flow Cytometer | BD Bioscience | – | This has 4 laser system equipped with 405, 488, 594, and 640 nm lasers. |
| BD Falcon 70μm Nylon Cell Strainers | CORNING | 352350 | For filtering the dissociating cells before FACS. |
| BD Falcon Round Bottom Test Tubes With Snap Cap | CORNING | 352054 | |
| BDNF Human | ProSpec-Tany TechnoGene, Ltd. | CYT-207 | |
| Cell Culture microplate, 96 well, PS, F-bottom (Chimney Well) | Greiner Bio-One International | 655090 | We tried multiple 96-well dishes and this was the best one for culture and analyses after ICC |
| CNTF Human | ProSpec-Tany TechnoGene, Ltd. | CYT-272 | |
| 4′,6-diamidino-2-phenylinodole (DAPI) | Thermo Fisher Scientific | D1306 | |
| Dimethyl sulfoxide | Sigma-Aldrich | D2650 | DMSO |
| Dumont #5 Forceps Inox Tip Size .05 x .01 mm Biologie Tips | Roboz Surgical Instrument | RS-5015 | |
| Forskolin | Thermo Fisher Scientific | BP25205 | |
| GDNF Human | ProSpec-Tany TechnoGene, Ltd. | CYT-305 | |
| GlutaMAX supplement | Thermo Fisher Scientific | 35050-061 | |
| Hanks' Balanced Salt Solution (HBSS) | Thermo Fisher Scientific | 14175-095 | |
| Hibernate E | BrainBits | HE | |
| Hibernate E low fluorescence | BrainBits | HELF | Fluorescence which hinders observation of embryo's GFP expressions should be low. |
| Horse serum, heat inactivated, New Zealand origin | Thermo Fisher Scientific | 26050-070 | |
| IBMX | Tocris Cookson | 2845 | Isobutylmethylxanthine |
| Laminin | Thermo Fisher Scientific | 23017-015 | |
| Leibovitz's L15 medium | Thermo Fisher Scientific | 11415064 | |
| 2-Mercaptoethanol | Sigma-Aldrich | M6250 | |
| Micro Dissecting Scissors | Roboz Surgical Instrument | RS-5913 | |
| Micro Knife 4.75" 1.7 x 27 mm blade | Roboz Surgical Instrument | RS-6272 | |
| Moria Mini Perforated Spoon | Fine Science Tools | 10370-19 | |
| Olympus 1.7ml Microtubes, Clear | Genesee Scientific | 22-281 | These are the tubes that we described "1.7 mL microcentrifuge tubes" in the context. |
| Papain Dissociation System | Worthington Biochemical Corp | LK003150 | Papain solution and alubumin- ovomucoid inhibitor solution are prepared from this kit. |
| Penicillin-streptomycin (10,000 U/ ml) | Thermo Fisher Scientific | 15140-122 | |
| Phosphate buffered saline (PBS) | Thermo Fisher Scientific | 10010-023 | |
| Poly D-lysin (PDL) | MilliporeSigma | A-003-E | |
| SMZ18 and SMZ1500 zoom stereomicroscopes with DS-Ri1 camera | Nikon | – | Dissection was performed and images of dissected embryos and tissues are captured under these fluorescence microscopes. |
| Sylgard 170 Black Silicone Encapsulant – A+B 0.9 Kg kit | Dow Corning | 1696157 | We make dissection dishes using this kit. |
| TC treated Dishes, 100 x 20 mm | Genesee Scientific | 25-202 | We make dissection dishes using this dish. |
| Thum Dressing Forceps 4.5" Serrated 2.2 mm Tip Width | Roboz Surgical Instrument | RS-8100 | |
| Zeiss LSM 700 series laser scanning confocal microscope and Zen Software | Carl Zeiss | – | Confocal image of the embryo was captured with these equipments |

